Abstract
OBJECTIVE
We hypothesized that interaction between PPARG2 Pro12Ala and variants in the promoter region of HNF4A are associated with type 2 diabetes–related quantitative traits in Mexican-American families of a proband with previous gestational diabetes.
RESEARCH DESIGN AND METHODS
The BetaGene project genotyped PPARG2 Pro12Ala and nine HNF4A single nucleotide polymorphisms (SNPs) in 473 individuals in 89 families. Members of the proband generation had fasting glucose <126 mg/dl and were phenotyped by oral and intravenous glucose tolerance tests.
RESULTS
Neither PPARG2 Pro12Ala nor any of the nine HNF4A SNPs were independently associated with type 2 diabetes–related quantitative traits. However, the interaction between PPARG2 Pro12Ala and HNF4A rs2144908 was significantly associated with both insulin sensitivity (SI) (Bonferroni P = 0.0006) and 2-h insulin (Bonferroni P = 0.039). Subjects with at least one PPARG2 Ala allele and homozygous for the HNF4A rs2144908 A allele had 40% higher SI compared with individuals with at least one G allele. SI did not vary by rs2144908 genotype among PPARG2 Pro/Pro. The interaction result for SI was replicated by the Insulin Resistance Atherosclerosis Family Study (P = 0.018) in their San Antonio sample (n = 484) where subjects with at least one PPARG2 Ala allele and homozygous for the HNF4A rs2144908 A allele had a 29% higher SI compared with individuals with at least one G allele. However, the interaction was not replicated in their San Luis Valley sample (n = 496; P = 0.401).
CONCLUSIONS
Together, these results suggest that variation in PPARG2 and HNF4A may interact to regulate insulin sensitivity in Mexican Americans at risk for type 2 diabetes.
Mexican-American women with previous gestational diabetes mellitus (GDM) exhibit significant β-cell dysfunction and are at high risk for developing type 2 diabetes (1–3). Moreover, their risk for future type 2 diabetes can be significantly reduced by improving insulin sensitivity and reducing insulin secretory demands (4). This propensity for β-cell failure in the face of chronic insulin resistance led us to design a family-based study, the BetaGene Study, to identify possible genetic determinants underlying this β-cell defect. In BetaGene, we performed detailed phenotyping of Mexican-American probands with recent GDM and their family members to obtain quantitative estimates of insulin sensitivity (SI), acute insulin response (AIR), and β-cell compensation (disposition index), which are traits that we have shown to be heritable in Mexican-American families (5,6).
PPARG2 is a lipid-activated transcription factor that has a key role in the expression of genes involved in adipocyte differentiation and function, as well as regulation of genes in several other tissues (7,8). The common Pro12Ala polymorphism in PPARG2 has been reported to be associated with type 2 diabetes (9–12) and with changes in plasma insulin levels (9,12,13) and insulin sensitivity (9) and thus is an accepted diabetes susceptibility variant. (HNF4A) is a transcription factor that regulates a vast network of genes involved in insulin secretion and glucose regulation (14). Mutations in the coding region of HNF4A have been shown to confer susceptibility to maturity-onset diabetes of the young (15). Common variants in the promoter region of HNF4A were initially shown to be associated with type 2 diabetes in Finns (16) and Ashkenazi Jews (17); some subsequent studies have replicated the association (18,19), although others have not (20,21). In Finns, variation in the P2-promoter region was also associated with measures of insulin secretion and β-cell function in nondiabetic offspring of patients with type 2 diabetes (16).
Given the evidence that variants of both PPARG2 and HNF4A play a role in diabetes risk and variation in diabetes-related traits, we tested whether these variants are associated with type 2 diabetes–related quantitative traits in BetaGene. We then tested our positive results in two similarly phenotyped Mexican-American samples from the Insulin Resistance Atherosclerosis Family Study (IRASFS), one from San Antonio, Texas, and the other from the San Luis Valley in Colorado (22).
RESEARCH DESIGN AND METHODS
Subject recruitment
Subject recruitment for BetaGene is ongoing and is briefly described in the online appendix (available at http://dx.doi.org/10.2337/db07-0848). For the purposes of this report, we describe only those clinical protocols and assays relevant to the results presented herein. Participation in BetaGene is restricted to Mexican Americans with fasting glucose <126 mg/dl (7 mmol/l) from families of a proband with GDM diagnosed within the previous 5 years who have available for study either two nondiabetic siblings and three nondiabetic first cousins from a single nuclear family or at least five siblings. The probands, siblings, and cousins have extensive phenotyping for diabetes-related traits (below) and form the basis for the primary analysis of this report.
IRASFS subject characteristics and ascertainment have been previously described (22). Briefly, probands were identified from the IRAS cohort study (23). IRASFS probands and their family members were recruited without regard to diabetes or glucose tolerance status. Mexican-American participants in IRASFS were from San Antonio, Texas, or the San Luis Valley in Colorado.
All protocols for BetaGene and the IRASFS were approved by the Institutional Review Boards of participating institutions, and all participants provided written informed consent before participation.
Clinical protocols
Phenotyping for BetaGene is performed on two separate visits to the General Clinical Research Center. Visit 1 consists of a physical examination, DNA collection, a 75-g oral glucose tolerance test (OGTT), and fasting blood for lipid measurements. Visit 2 consists of a duel-energy X-ray absorptiometry scan for determination of body fat and an insulin-modified intravenous glucose tolerance test (IVGTT) performed as previously described (4). Probands, their siblings, and their cousins undergo the full phenotyping protocol, whereas parents, uncles, aunts, spouses, and offspring had only a physical exam, fasting glucose measurement, and DNA collection.
Details of the IRASFS clinical exams and phenotype measurements have been published (22). Of particular relevance for this report, the IVGTTs for IRASFS were performed using the same protocol as was used in the BeteGene study, but no OGTT was performed in IRASFS.
Assays
In both BetaGene and IRASFS, plasma glucose was measured on an autoanalyzer using the glucose oxidase method (YSI Model 2300; Yellow Springs Instruments, Yellow Springs, OH). In BetaGene, insulin was measured by two-site immunoenzymometric assay that has <0.1% cross-reactivity with proinsulin and intermediate split products; whereas in IRASFS, insulin was measured by radioimmunoassay with dextran-charcoal separation (24).
Molecular analysis
In BetaGene, we attempted to genotype the PPARG2 Pro12Ala variant (rs1801282) and 10 HNF4A single nucleotide polymorphisms (SNPs) showing evidence for association with type 2 diabetes in Finns (16): rs4810424, rs1884613, rs1884614, rs2144908, rs6031551, rs6031552, rs2425637, rs2425640, rs3212183, and rs1885088. The assay for rs3212183 failed, but genotype data for the remaining nine SNPs were obtained. SNP genotyping was performed using the Applied Biosystems TaqMan system (25). Genotyping assays were either selected through the Assays on Demand database (Applied Biosystems; http://myscience.appliedbiosystems.com/navigation/mysciapplications.jsp) or custom designed using the Assays by Design service (Applied Biosystems). Based on 22 blinded duplicate samples, the discrepancy rate for genotyping was 0%, and the overall genotype success rate was >97.6%.
IRASFS genotyped the PPARG2 Pro12Ala and 23 SNPs that mapped to unique locations in and around HNF4A, 16 of which were chosen from the dbSNP database (rs2868093, rs6073418, rs717248, rs717247, rs736820, rs736822, rs736824, rs745975, rs736823, rs1885088, rs1885089, rs3212198, rs1028583, rs1028584, rs2273618, and rs911358) and 7 of which were selected on the basis of previous reports of association with type 2 diabetes in Finns (16) and/or Ashkenazim (17). SNPs were genotyped using a MassARRAY system (Sequenom, San Diego, CA) (26). Seven of 23 HNF4A SNPs were common to both IRASFS and BetaGene (rs4810424, rs1884613, rs1884614, rs2144908, rs2425637, rs2425640, and rs1885088). Based on 90 blinded duplicate samples, the genotyping discrepancy rate was 0%, and overall genotype success rate was >90.0%.
Data analysis
In BetaGene, we calculated two measures of insulin response to glucose: 1) the difference between the 30′ and fasting OGTT insulin concentrations (30′ Δinsulin) and 2) the incremental area under the insulin curve during the first 10 min of the IVGTT (AIR). For both BetaGene and IRASFS, IVGTT glucose and insulin data were analyzed using the minimal model (MINMOD Millennium V5.18) to derive measures of glucose effectiveness (SG) and SI. The disposition index is computed as the product of SI and AIR and measures β-cell compensation for insulin resistance (27).
For both studies, the observed genotype frequencies were assessed for deviation from Hardy-Weinberg equilibrium, and allele frequencies for each SNP were estimated using family data and MENDEL (V5.7). Linkage disequilibrium and haplotype block structure were assessed using Haploview V3.2 (28) and the method of Gabriel et al. (29). Quantitative trait data were statistically transformed to approximate univariate normality before analyses. The measured genotypes approach under a variance components framework was used to test SNP associations with continuous phenotypes and implemented using SOLAR (V2.1.4).
In BetaGene, because of underlying linkage disequilibrium among the nine HNF4A SNPs, we tested five of the HNF4A SNPs for association with type 2 diabetes–related quantitative traits under additive, dominant, and recessive genetic models. Because of the low frequency of the PPARG2 Ala allele, association with the Pro12Ala variant was only tested under a dominant genetic model for Ala. We tested the interaction between rs2144908 and Pro12Ala for association with type 2 diabetes–related traits due to observed univariate effects of HNF4A rs2144908 and the previously reported associations with type 2 diabetes (16,17). The multiplicative interaction effect between PPARG2 Pro12Ala and HNF4A rs2144908 was tested using a likelihood ratio test.
We had 80% power to detect an association between an SNP with 50% allele frequency (as observed for HNF4A rs2144908) that explains 1.7% of the variation in SI, assuming an additive model and α = 0.05. Similarly, we had 80% power to detect an association between an SNP with 10% allele frequency (as observed for PPARG2 P12A) that explains 1.7% of the variation in SI, assuming a dominant genetic model and α = 0.05. Finally, assuming a multiplicative interaction between two SNPs as described above, we had 80% power to detect an interaction that explains at least 1.6% of the variation in SI.
Because BetaGene families were ascertained through a proband with previous GDM, we corrected for ascertainment bias in each model by conditioning on the proband’s phenotype value. All models were adjusted for age, sex, and, where appropriate, BMI. Results were similar when BMI was not included as a model covariate. Linear modeling results are reported as means and SD, adjusted for age, sex, and BMI. All other results are reported as unadjusted medians and interquartile ranges. All BetaGene P values for univariate tests of association between SNPs and quantitative traits are Bonferroni-adjusted for the number of SNPs (n = 6), models (n = 1 for PPARG2 and n = 3 for HNF4A), and traits (n = 10; Table 1). Because our a priori hypothesis concerning variant interaction only included PPARG2 Pro12Ala and HNF4A rs2144908, P values for tests of interaction effects are Bonferroni adjusted for the number of models and traits only. Statistical significance was defined as a corrected P < 0.05. Tests in the IRASFS are a priori hypotheses based on BetaGene results, thus IRASFS P values were not corrected for multiple testing.
TABLE 1.
Subject characteristics
| BetaGene
|
IRASFS
|
|||||
|---|---|---|---|---|---|---|
| Probands | Siblings | Cousins | All | San Antonio | San Luis Valley | |
| n | 71 | 231 | 171 | 473 | 490 | 496 |
| Men/women | 0/71 | 84/147 | 79/92 | 163/310 | 191/299 | 213/283 |
| Age (years) | 34.7 (7.8) | 33.8 (10.5) | 32.7 (11.8) | 33.8 (10.6) | 38.8 (20.1) | 40.0 (16.8) |
| BMI (kg/m2) | 31.3 (8.7) | 29.0 (6.4)* | 27.4 (6.4)*† | 28.8 (7.0) | 28.7 (8.2) | 26.4 (6.8)‡ |
| Fasting glucose (mmol/l) | 5.4 (0.9) | 5.1 (0.6)* | 5.1 (0.6)* | 5.2 (0.6) | 5.1 (0.7) | 5.1 (0.6)‡ |
| 2-h glucose (mmol/l) | 8.4 (3.5) | 7.3 (2.5)* | 6.4 (2.2)*† | 7.2 (2.6) | — | — |
| Fasting insulin (pmol/l) | 76 (62) | 49 (49)* | 42 (42)* | 49 (56) | 78 (72) | 66 (60)‡ |
| 2-h insulin (pmol/l) | 660 (556) | 431 (437)* | 333 (403)* | 424 (465) | — | — |
| 30′ Δinsulin (pmol/l) | 431 (285) | 379 (295) | 410 (410) | 399 (319) | — | — |
| SG (×10−2 min−1) | 1.29 (0.42) | 1.55 (0.71)* | 1.74 (0.77)* | 1.53 (0.74) | 1.86 (1.00) | 2.24 (1.02)‡ |
| SI (×10−3 min−1 per pmol/l) | 2.17 (1.24) | 2.62 (1.94) | 2.94 (1.95) | 2.65 (1.77) | 2.47 (2.92) | 3.17 (3.83) |
| AIR (pmol/l × 10 min) | 3,467 (4,313) | 4,280 (4,677)* | 4,783 (6,050)* | 4,340 (5,070) | 5,506 (5,945) | 6,262 (6,475)‡ |
| Disposition index | 7,890 (7,799) | 11,401 (11,458)* | 13,249 (10,654)* | 11,409 (11,225) | 14,315 (18,101) | 19,898 (26,954)‡ |
Data are median (interquartile range).
Uncorrected P < 0.05 vs. probands after adjusting for age, sex, and BMI (where appropriate).
Uncorrected P < 0.05 vs. siblings after adjusting for age, sex, and BMI (where appropriate).
Uncorrected P < 0.05 vs. San Antonio after adjusting for age, sex, and BMI (where appropriate).
RESULTS
We report results from 473 individuals in 89 BetaGene families with complete OGTTs and IVGTTs. The size of the proband generation (probands, siblings, and cousins) for each family ranged from 1 to 12 with a mean of 5.3. Descriptive characteristics of BetaGene subjects are shown in Table 1. In general, probands, siblings, and cousins were matched on age, although there was a tendency for cousins to be younger than probands and their siblings. The median BMI exceeded the threshold for overweight among all subjects, although siblings and cousins had a significantly lower BMI compared with probands (P = 0.001 and P = 2.1 × 10−5, respectively). The correlations among the quantitative traits are presented in the online appendix (Supplemental Table S1).
From the IRASFS, this report includes 490 individuals in 60 families from San Antonio and 496 individuals in 30 families from the San Luis Valley. The San Antonio sample had a significantly higher mean BMI (P = 0.0006) and lower mean SG, AIR, and disposition index (all P < 0.005) compared with San Luis Valley (Table 1).
Table 2 shows allele frequencies estimated from Beta-Gene subjects compared with frequencies for the IRASFS San Antonio and San Luis Valley subjects. The minor alleles reported for HNF4A SNPs in Finns (16) were used as reference alleles. In BetaGene, PPARG2 Pro12Ala and each of the nine HNF4A SNPs had allele frequencies that differed considerably from those observed in Finns and Ashkenazim (16,17). The PPARG2 Ala allele frequency was ~10% in Mexican Americans compared with ~17% in Finns (16). The observed frequencies of HNF4A rs4810424, rs1884613, rs1884614, and rs2144908 reference alleles were all estimated at roughly 50% compared with ~20% in the Finns and Ashkenazim (16,17). Allele frequency differences for other HNF4A variants were smaller, ranging from 7 to 18%. Allele frequency estimates from the IRASFS were consistent with those estimated in BetaGene for the seven SNPs common to both studies.
TABLE 2.
SNP characteristics among BetaGene and IRASFS participants
| Chromosome | kb Position* | Reference allele† | Reference allele frequency‡
|
|||
|---|---|---|---|---|---|---|
| BetaGene | IRASFS
|
|||||
| San Antonio | San Luis Valley | |||||
| PPARG2 | ||||||
| Pro12Ala (rs1801282) | 3 | 12368125 | Ala | 0.103 | 0.118 | 0.100 |
| HNF4A | ||||||
| rs4810424 | 20 | 42408437 | C | 0.500 | 0.459 | 0.481 |
| rs1884613 | 20 | 42413829 | G | 0.501 | 0.451 | 0.481 |
| rs1884614 | 20 | 42413933 | T | 0.499 | 0.456 | 0.481 |
| rs2144908 | 20 | 42419131 | A | 0.503 | 0.453 | 0.472 |
| rs6031551 | 20 | 42423128 | C | 0.116 | — | — |
| rs6031552 | 20 | 42423208 | A | 0.107 | — | — |
| rs2425637 | 20 | 42457463 | T | 0.608 | 0.640 | 0.622 |
| rs2425640 | 20 | 42461451 | A | 0.202 | 0.139 | 0.149 |
| rs1885088 | 20 | 42472454 | A | 0.199 | 0.169 | 0.238 |
Based on National Center for Biotechnology Information build 36.1.
Minor allele for Finns in FUSION (16).
Allele frequency of BetaGene reference allele.
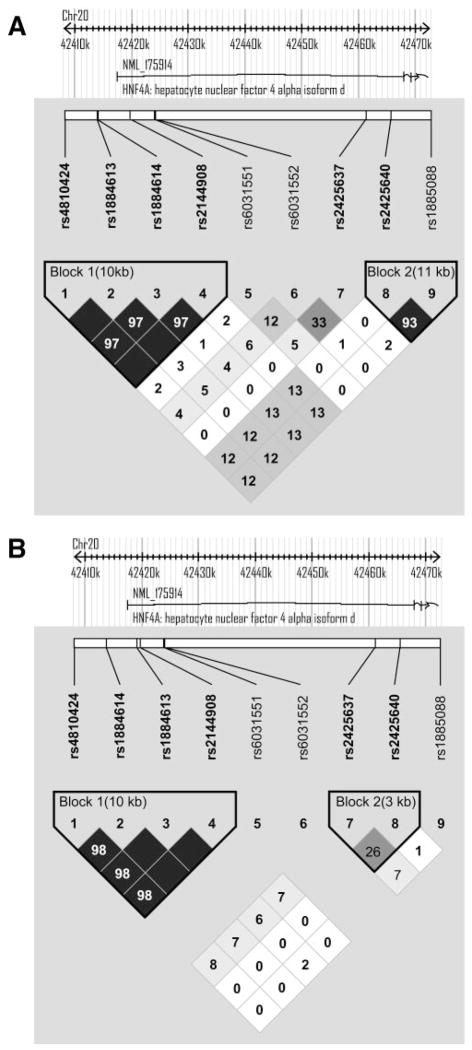
Figure 1A shows the pairwise linkage disequilibrium and haplotype block structure for the nine HNF4A SNPs in BetaGene. The four SNPs in the P2-promoter region, rs4810424, rs1884613, rs1884614, and rs2144908, were in near perfect linkage disequilibrium (D′ = 1.0, r2 ≥ 0.98), forming a single 10.7-kb haplotype block; SNPs rs6031551 and rs6031552 were also in strong linkage disequilibrium (D′ = 1.0, r2 ≥ 0.94), forming an independent 80-bp haplotype block. We chose to test rs2144908 from the first haplotype block, rs6031551 from the second block, and the remaining SNPs (rs2425637, rs2425640, and rs1885088) for association with phenotypes of interest in BetaGene. A similar block structure was observed in the IRASFS using a combination of San Antonio and San Luis Valley subjects (Fig. 1B). The linkage disequilibrium structure using all 23 HNF4A SNPs genotyped in the IRASFS is presented as Supplemental Fig. 1.
FIG. 1.
HNF4A pairwise linkage disequilibrium and haplotype block structure. A: Linkage disequilibrium and haplotype block structure based on the nine SNPs genotyped in all BetaGene subjects. Haplotype blocks were determined using the method of Gabriel et al. (29) as implemented in Haploview V3.2. B: Linkage disequilibrium and haplotype block structure based on the seven SNPs genotyped in the IRASFS that were common to BetaGene. The linkage disequilibrium and haplotype block structure based on the combined San Antonio and San Luis Valley data are shown, because the results were nearly identical between the two samples. The linkage disequilibrium and block structure based on all 23 SNPs genotyped in the IRASFS is presented as Supplemental Fig. 1. Linkage disequilibrium is displayed as pairwise r2 values, where white indicates r2 = 0, varying shades of gray indicate 0 < r2 < 1, and black indicates r2 = 1.
Neither PPARG2 Pro12Ala nor any of the five HNF4A SNPs alone showed significant evidence for association with type 2 diabetes–related quantitative traits in Beta-Gene after correction for multiple comparisons. However, without multiple comparisons correction, HNF4A rs2144908 showed marginal association with the disposition index under a dominant model, where subjects with at least one copy of the HNF4A A allele had a 14% higher adjusted mean disposition index compared with those homozygous for the G allele (13,493 ± 2,911 vs. 11,538 ± 2,767; uncorrected P = 0.034). Pro12Ala also showed marginal association with triglycerides under a dominant model, where subjects with at least one PPARG2 Ala allele had a 12% lower adjusted mean triglyceride level compared with those homozygous for PPARG2 Pro (0.90 ± 0.27 vs. 1.04 ± 0.25 mmol/l; uncorrected P = 0.05). Univariate association results for rs1801282 (PPARG2 Pro12Ala) and rs2144908 (HNF4A) are presented in the online appendix (Supplemental Table S2).
Consistent with BetaGene, none of the SNPs tested in the IRASFS showed significant evidence for association with type 2 diabetes–related quantitative traits after multiple comparisons correction. HNF4A rs2144908 showed nominal association with SI (uncorrected P = 0.024) and disposition index (uncorrected P = 0.047) under an additive model in the San Antonio sample. Pro12Ala showed nominal association with SI in the San Luis Valley sample under a dominant genetic model (uncorrected P = 0.01) but no association with triglycerides.
Although none of the PPARG2 or HNF4A variants individually showed significant association with diabetes-related quantitative traits, the multiplicative interaction between PPARG2 Pro12Ala and HNF4A rs2144908 was significantly associated with SI (P = 0.001) and 2-h insulin (P = 0.054), assuming an additive genetic model for rs1244908 (Table 3). The interaction was also nominally associated with fasting insulin (uncorrected P value = 0.01) and AIR (uncorrected P value = 0.07), but these associations did not remain significant after correction for multiple comparisons.
TABLE 3.
Trait association with interaction between PPARG2 and HNF4A among BetaGene subjects
|
PPARG2 Pro12Ala genotype
|
Interaction P value
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Ala/Pro or Ala/Ala
|
Pro/Pro
|
|||||||
|
HNF4A rs2144908 genotype
| ||||||||
| A/A | A/G | G/G | A/A | A/G | G/G | Additive* | Recessive* | |
| n | 30 | 44 | 23 | 102 | 190 | 84 | ||
| BMI (kg/m2) | 26.3 (0.7) | 27.4 (0.8) | 27.8 (0.9) | 27.8 (1.0) | 27.8 (1.0) | 27.9 (1.1) | 1 | 1 |
| Fasting glucose (mmol/l) | 5.1 (0.2) | 5.1 (0.3) | 5.0 (0.2) | 5.1 (0.2) | 5.1 (0.2) | 5.1 (0.2) | 1 | 1 |
| 2-h glucose (mmol/l) | 6.3 (0.6) | 7.0 (0.9) | 7.0 (0.8) | 7.0 (0.8) | 7.1 (0.8) | 7.0 (0.8) | 1 | 1 |
| Fasting insulin (pmol/l) | 36 (15) | 57 (46) | 79 (77) | 53 (28) | 59 (42) | 54 (32) | 0.636 | 0.294 |
| 2-h insulin (pmol/l) | 279 (93) | 467 (212) | 625 (292) | 405 (142) | 413 (179) | 383 (155) | 0.054 | 0.039 |
| 30′ Δinsulin (pmol/l) | 334 (59) | 432 (98) | 588 (173) | 406 (93) | 412 (111) | 396 (118) | 0.222 | 0.021 |
| SG (×10−2 min−1) | 1.79 (0.18) | 1.56 (0.22) | 1.47 (0.21) | 1.69 (0.19) | 1.62 (0.21) | 1.59 (0.20) | 1 | 1 |
| SI (×10−3 min−1 per pmol/l) | 3.92 (0.88) | 2.73 (0.92) | 1.95 (0.72) | 2.74 (0.73) | 2.71 (0.85) | 2.89 (0.85) | 0.0011 | 0.0006 |
| AIR (pmol/l × 10 min) | 4,480 (921) | 5,198 (1,353) | 6,832 (2,707) | 5,230 (1,410) | 5,060 (1,538) | 4,567 (1,534) | 0.969 | 1 |
| Disposition index | 16,113 (2,686) | 13,026 (3,152) | 12,426 (2,829) | 13,561 (2,882) | 12,742 (2,805) | 12,257 (2,899) | 1 | 1 |
Genotype-specific trait values are shown as mean (SD) adjusted for age, sex, and BMI (except in the case of BMI). The interaction P value is Bonferroni-corrected for 30 tests, as described in RESEARCH DESIGN AND METHODS.
Genetic model assumed for HNF4A rs2144908 in the interaction with PPARG2. PPARG2 Pro12Ala was assumed to follow a dominant genetic model for Ala.
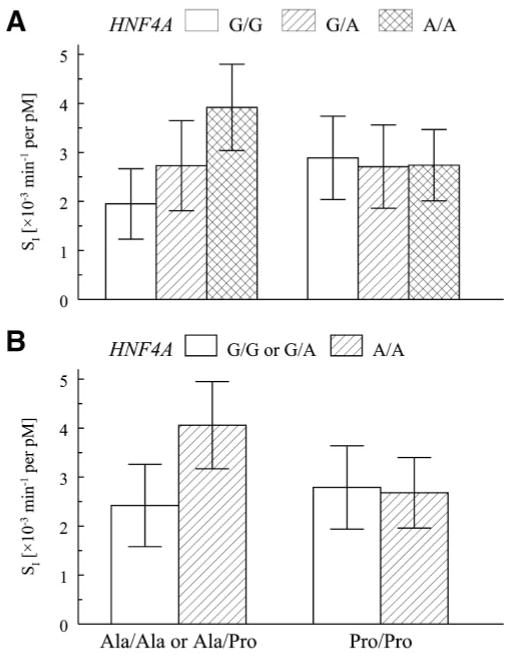
Figure 2A shows the age, sex, and BMI adjusted mean SI in BetaGene stratified by HNF4A rs2144908 and PPARG2 Pro12Ala genotypes. Mean SI increased progressively with each copy of the A allele for HNF4A rs2144908 among subjects with at least one copy of PPARG2 Ala. In contrast, mean SI did not differ across HNF4A rs2144908 genotypes among PPARG2 Pro homozygotes. Figure 2B shows the same interaction result under a recessive genetic model for the HNF4A rs2144908 A allele. Among subjects with at least one PPARG2 Ala allele, subjects homozygous for HNF4A rs2144908 A allele had a 68% higher adjusted mean SI than those with at least one G allele (P = 0.0001). In contrast, among subjects homozygous for the PPARG2 Pro allele, mean SI was not significantly different between individuals homozygous for the HNF4A rs2144908 A allele and those with a G allele (P = 0.30).
FIG. 2.
Association with SI and the interaction between PPARG2 and HNF4A among BetaGene subjects. A: Genotype-specific means ± SD, adjusted for age, sex, and BMI stratified by PPARG2 Pro12Ala and HNF4A rs2144908. Mean values were generated by computing the predicted value under the additive model and removing the transformation to maintain physiological interpretation. A: Values when PPARG2 was assumed to follow a dominant genetic model for Ala and HNF4A rs2144908 was assumed to follow an additive genetic model for the A allele. The test of association for the interaction was significant (P = 0.0011). B: The same data when PPARG2 remains modeled under a dominant model, but HNF4A rs2144908 was assumed to follow a recessive genetic model for the A allele. The test for association was significant (P = 0.0006).
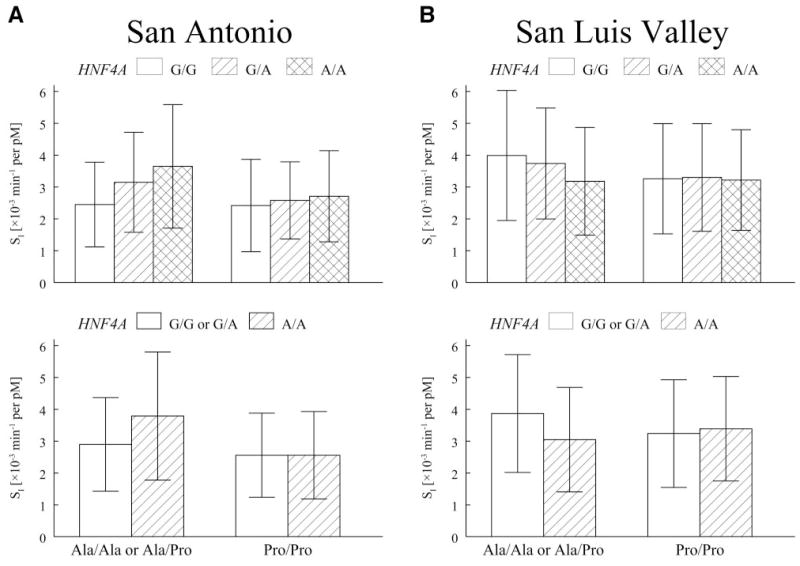
The interaction result for SI was replicated in the IRASFS San Antonio sample (additive P = 0.056, recessive P = 0.018; Table 4) but not in the San Luis Valley sample (Table 4; Fig. 3). The patterns observed in the San Antonio sample (Fig. 3A) were similar to those observed in Beta-Gene (compare with Fig. 2). Under an additive genetic model for HNF4A rs2144908 in BetaGene, mean SI increased by 40 and 44% with each copy of the A allele for HNF4A rs2144908 among subjects with at least one copy of the PPARG2 Ala allele. No increase was seen among subjects homozygous for the PPARG2 Pro allele. In the San Antonio sample from the IRASFS, the increases in mean SI with each copy of the HNF4A rs2144908 A allele were 29 and 16% among subjects with at least one copy of the PPARG2 Ala allele, with no increase among subjects homozygous for PPARG2 Pro (compare with Fig. 3A). Under a recessive genetic model for the HNF4A rs2144908 A allele, subjects with at least one PPARG2 Ala allele and homozygous for rs2144908 A allele had adjusted mean SI values that were 31% higher than those with at least one G allele in the San Antonio sample (compare with Fig. 3A). The association between SI and the interaction between PPARG2 and HNF4A was not observed in the San Luis Valley sample (Fig. 3B)
TABLE 4.
SI association with interaction between PPARG2 and HNF4A among IRASFS subjects
|
PPARG2 Pro12Ala genotype
|
Interaction P value
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Ala/Pro or Ala/Ala
|
Pro/Pro
|
|||||||
|
HNF4A rs2144908 genotype
| ||||||||
| A/A | A/G | G/G | A/A | A/G | G/G | Additive* | Recessive* | |
| San Antonio | ||||||||
| n | 32 3.65 (1.94) |
63 3.15 (1.57) |
23 2.45 (1.33) |
75 2.71 (1.43) |
187 2.58 (1.21) |
104 2.42 (1.45) |
0.056 | 0.018 |
| San Luis Valley | ||||||||
| n | 19 3.18 (1.69) |
44 3.74 (1.74) |
25 3.99 (2.04) |
85 3.22 (1.58) |
214 3.30 (1.69) |
109 3.26 (1.73) |
0.401 | 0.176 |
Genotype-specific trait values are shown as mean (SD) adjusted for age, sex, and BMI.
Genetic model assumed for HNF4A rs2144908 in the interaction with PPARG2. PPARG2 Pro12Ala was assumed to follow a dominant genetic model for Ala.
FIG. 3.
Association with SI and the interaction between PPARG2 and HNF4A among IRASFS subjects. A: Genotype-specific means ± SD, adjusted for age, sex, and BMI stratified by PPARG2 Pro12Ala and HNF4A rs214408 for the San Antonio samples. B: The same data for the San Luis Valley samples. Mean values were generated by computing the predicted value under the additive model and removing the transformation to maintain physiological interpretation. Figures at the top of each panel show values when PPARG2 was assumed to follow a dominant genetic model for Ala and HNF4A rs2144908 was assumed to follow an additive genetic model for the A allele. Figures at the bottom show the same data assuming a recessive genetic model for the HNF4A rs2144908 A allele. The test of association for the interaction was significant in the San Antonio samples but not in the San Luis Valley samples (see text for details).
DISCUSSION
We found no significant association between PPARG2 Pro12Ala or individual P2-promoter variants in HNF4A and type 2 diabetes–related quantitative traits in the BetaGene sample of Mexican-American families of a proband with previous GDM. By contrast, the interaction between PPARG2 Pro12Ala and HNF4A rs2144908 was strongly associated with SI. More importantly, this interaction was independently replicated in the San Antonio sample from the IRASFS but not in the San Luis Valley sample. In carriers of the PPARG2 Ala allele, which has been reported to be protective from type 2 diabetes, SI increased with each copy of the A allele for HNF4A rs2144908, whereas SI remained low among all HNF4A genotypes in individuals with the “risk” PPARG2 Pro allele. This pattern suggests that among individuals with the PPARG2 Ala, the A allele for HNF4A may afford some protection from insulin resistance but not in the presence of the G allele for HNF4A rs2144908 or PPARG2 Pro. The reduced SI observed in PPARG2 Pro homozygotes or in individuals with at least one PPARG2 Ala and one HNF4A rs2144908 G allele may place them at increased risk for type 2 diabetes.
The interaction first observed in BetaGene was replicated in the San Antonio sample from the IRASFS but not in the San Luis Valley sample. There are several possible explanations for the observed difference in results, including low statistical power or type 1 statistical error, ethnic admixture, and cryptic stratification. Although our power calculations indicated that we had sufficient power to detect the association in both studies, it is noteworthy that both BetaGene and the San Antonio sample from IRASFS had ~30 subjects who were homozygous for the HNF4A rs2144908 A allele and had at least one PPARG2 Ala, whereas there were only 19 subjects with the same genotype combination in the San Luis Valley sample. Replication in a larger sample would provide additional support for our observation, but we are not aware of additional genetic studies in Mexican Americans with frequently sampled IVGTT–derived measures of SI of equal or greater size than BetaGene or IRASFS. Within the individual analyses of the three samples, we do not believe cryptic stratification or ethnic admixture is contributing to our results. First, both BetaGene and IRASFS use family-based designs, which afford some protection against cryptic stratification, even in the absence of applying a family-based statistic like the quantitative transmission disequilibrium test. Second, BetaGene participants were required to have Mexican ancestry by self-reported birthplace going back two generations (compare with the online appendix), which should minimize admixture due to other Latino groups such as Central or South Americans.
However, the observed difference in outcome between BetaGene and San Antonio versus San Luis Valley could be due to differences in admixture. The two IRASFS Mexican-American samples differ substantially in terms of environment and relevant metabolic characteristics. First, San Antonio is an urban environment, whereas the San Luis Valley is a rural, high-elevation (~7,500 feet above sea level) environment. Second, the San Luis Valley families tend to be leaner and have better glucose homeostasis profiles, with significantly lower BMI and higher SG, AIR, and disposition index, compared with the San Antonio sample (compare with Table 1). Although it is not possible to directly compare metabolic parameters between San Antonio and BetaGene participants because of differences in assays, these two groups are more similar compared with the San Luis Valley participants based on demographic parameters, e.g., age and BMI. This led us to analyze the two IRASFS samples separately and may explain the very similar results in BetaGene and San Antonio samples compared with the San Luis Valley sample. In addition, the ancestry of San Luis Valley Mexican Americans may differ from the urban Mexican Americans in San Antonio or BetaGene. Mexican Americans in the San Luis Valley tend to self-identify as “Spanish” (30), and admixture analysis suggests a large proportion of “Spanish” (~60%) and less Native American (~30%) admixture in this population (31). This is in contrast to recent estimates of 48% European and 40% Native American for Mexican Americans from Los Angeles (32) and even higher Native American admixture estimates for Southwestern Hispanics (33).
It is important to note that our analysis simply denotes a statistical interaction between variants and cannot be used to accurately characterize the underlying biology. Although additional studies will be required to characterize the underlying biology of this interaction, one can envision two general scenarios to explain how these two genes may interact to alter SI. The first presumes that both genes are expressed in the same tissues where they could interact directly (although there currently is no evidence that they do) or co-regulate transcription of genes in the same or interacting biochemical pathways. PPARG2 is expressed in adipose tissue (7,8) where it regulates a number of genes (34), and the Pro variant could result in increased activation of genes in pathways, such as adipogenesis, that contribute to lower SI (9). HNF4A regulates a large network of genes in both the pancreas and liver (14). rs2144908 lies near the P2-promoter of HNF4A, which is believed to be primarily active in pancreatic β-cells and thought to regulate insulin secretion (35). However, there is evidence of PPARG2 expression in liver with development of hepatic steatosis, a condition typically accompanied by obesity, impaired glucose tolerance, and type 2 diabetes (36–38). Also, studies by Thomas et al. show there may be low-level activity of the HNF4A P2-promoter in hepatocytes (35). Thus, the presence of PPARG2 Pro and HNF4A rs2144908 G allele could result in dysregulation of hepatic gene transcription, leading to hepatic insulin resistance and thereby reducing SI.
The other possibility is that these two genes may act independently in different tissues, but the net integrated physiological effect of these genes alters SI. PPARG2 Pro12Ala showed marginal association with triglyceride levels in BetaGene, with individuals homozygous for Pro having modestly elevated triglycerides. Stumvoll et al. (39) reported an association between the Ala allele and increased SI compared with the Pro/Pro genotype, due to enhanced insulin action on the suppression of lipolysis resulting in decreased release of free fatty acids. This suggests an effect of PPARG2 Pro allele on adipose tissue that could result in elevated fatty acid flux, which could contribute to impaired ability to suppress hepatic glucose output (40). As noted above, variation in HNF4A rs2144908 could disrupt gene transcription within the liver, which could further contribute to hepatic insulin resistance.
To better understand the potential mechanisms underlying this interaction, we applied the prioritizing disease genes by analysis of common elements (PDG-ACE) algorithm (online appendix). When applied to our PPARG2-HNF4A interaction, “triglyceride” was identified as a common, over-represented term (Bonferroni-corrected P = 0.016) in the annotation of these genes. Additional examination showed literature citing both PPARG2 and HNF4A as potential regulators of human microsomal triglyceride transfer protein (MTTP) (41,42), which is involved in lipoprotein assembly in the intestine and VLDL in the liver (43). Expression of MTTP is elevated in type 2 diabetes (43), which can lead to dyslipidemia. Variation in MTTP has also been shown to be associated with type 2 diabetes and postprandial insulin levels (44). Thus, the interaction between PPARG2 and HNF4A, as described under the above scenarios, may work through MTTP to change triglyceride levels, which could alter SI.
In conclusion, we did not observe association between diabetes-related quantitative traits and either PPARG2 Pro12Ala or variation in the HNF4A promoter region in Mexican-American families of a proband with previous GDM. We did observe a strong association between SI and the interaction between PPARG2 Pro12Ala and HNF4A rs2144908 in BetaGene. This interaction was independently replicated in the San Antonio subjects from the IRASFS but not in the San Luis Valley sample. The characteristics of the interaction suggest that having PPARG2 Pro leads to relatively low SI regardless of HNF4A genotype, while the impact of the type 2 diabetes “protective” PPARG2 Ala depends at least in part on the HNF4A genotype. The biological nature of this interaction requires further investigation.
Supplementary Material
Acknowledgments
T.E.F. is supported by an American Diabetes Association (ADA) Junior Faculty Award. T.A.B. is partly supported by an ADA Distinguished Clinical Scientist Award. R.M.W. is partly supported by ADA Research Grant 1-05-RA-140. The BetaGene study is supported by National Institutes of Health (NIH) Grant DK-61628 awarded to T.A.B. A portion of the work for BetaGene was conducted in a facility constructed with support from Research Facilities Improvement Program Grant Number C06 (RR-10600-01, CA-62528-01, and RR-14514-01) from the National Center for Research Resources. The IRAS Family Study (IRASFS) is supported by NIH Grants HL-60944-02, HL-61210-02, HL-61019-02, HL-60894, and HL-60931-02. The Prioritizing Disease Genes by Analysis of Common Elements (PDG-ACE) project is supported by NIH Grant U54-DA-021519.
We thank the families who volunteered to participate in the BetaGene study and IRASFS. We also gratefully acknowledge the tremendous dedication and work performed by the staff for both studies. The BetaGene study also acknowledges the support of the University of Southern California General Clinical Research Center (M01-RR-00043).
Glossary
- AIR
acute insulin response
- GDM
gestational diabetes mellitus
- IRASFS
Insulin Resistance Atherosclerosis Family Study
- IVGTT
intravenous glucose tolerance test
- MTTP
microsomal triglyceride transfer protein
- OGTT
oral glucose tolerance test
- SNP
single nucleotide polymorphism
Footnotes
Additional information for this article can be found in an online appendix at http://dx.doi.org/10.2337/db07-0848.
References
- 1.Buchanan TA, Metzger BE, Freinkel N, Bergman RN. Insulin sensitivity and B-cell responsiveness to glucose during late pregnancy in lean and moderately obese women with normal glucose tolerance or mild gestational diabetes. Am J Obstet Gynecol. 1990;162:1008–1014. doi: 10.1016/0002-9378(90)91306-w. [DOI] [PubMed] [Google Scholar]
- 2.Kjos SL, Peters RK, Xiang A, Henry OA, Montoro M, Buchanan TA. Predicting future diabetes in Latino women with gestational diabetes. Diabetes. 1995;44:586–591. doi: 10.2337/diab.44.5.586. [DOI] [PubMed] [Google Scholar]
- 3.Kautzky-Willer A, Prager R, Waldhäusl W, Pacini G, Thomaseth K, Wagner OF, Ulm M, Streli C, Ludvik B. Pronounced insulin resistance and inadequate β-cell secretion characterize lean gestational diabetes during and after pregnancy. Diabetes Care. 1997;20:1717–1723. doi: 10.2337/diacare.20.11.1717. [DOI] [PubMed] [Google Scholar]
- 4.Buchanan TA, Xiang AH, Peters RK, Kjos SL, Marroquin A, Goico J, Ochoa C, Tan S, Berkowitz K, Hodis HN, Azen SP. Preservation of pancreatic β-cell function and prevention of type 2 diabetes by pharmacological treatment of insulin resistance in high-risk Hispanic women. Diabetes. 2002;51:2796–2803. doi: 10.2337/diabetes.51.9.2796. [DOI] [PubMed] [Google Scholar]
- 5.Watanabe RM, Xiang A, Trigo E, Hernandez S, Berrios F, Langefeld C, Kjos S, Ouzunian J, Sacks D, Lawrence J, Buchanan T. Evidence of genetic predisposition for B-cell dysfunction in Mexican-American families of probands with gestational diabetes (Abstract) Diabetes. 2003;52:A257. [Google Scholar]
- 6.Bergman RN, Zaccaro DJ, Watanabe RM, Haffner SM, Saad MF, Norris JM, Wagenknecht LE, Hokason JE, Rotter JI, Rich SS. Minimal model-based insulin sensitivity has greater heritability and a different genetic basis than homeostasis model assessment or fasting insulin. Diabetes. 2003;52:2168–2174. doi: 10.2337/diabetes.52.8.2168. [DOI] [PubMed] [Google Scholar]
- 7.Fajas L, Auboeuf D, Raspe E, Schoonjans K, Lefebvre AM, Saladin R, Najib J, Laville M, Fruchart JC, Deeb S, Vidal-Puig A, Flier J, Briggs MR, Staels B, Vidal H, Auwerx J. The organization, promoter analysis, and expression of the human PPARγ gene. J Biol Chem. 1997;272:18779–18789. doi: 10.1074/jbc.272.30.18779. [DOI] [PubMed] [Google Scholar]
- 8.Braissant O, Foufelle F, Scotto C, Dauca M, Wahli W. Differential expression of peroxisome proliferator-activated receptors (PPARs): tissue distribution of PPAR-α, -β, and -γ in the adult rat. Endocrinology. 1996;137:354–366. doi: 10.1210/endo.137.1.8536636. [DOI] [PubMed] [Google Scholar]
- 9.Deeb SS, Fajas L, Nemoto M, Pihlajamäki J, Laakso M, Fujimoto W, Auwerx J. A Pro12Ala substitution in PPARγ2 associated with decreased receptor activity, lower body mass index and improved insulin sensitivity. Nat Genet. 1998;20:284–287. doi: 10.1038/3099. [DOI] [PubMed] [Google Scholar]
- 10.Altshuler D, Hirschhorn JN, Klannemark M, Lindgren CM, Vohl M-C, Nemesh J, Lane CR, Schaffner SF, Bolk S, Brewer C, Tuomi T, Gaudet D, Hudson TJ, Daly M, Groop L, Lander ES. The common PPARγ Pro12Ala polymorphism is associated with decreased risk of type 2 diabetes. Nat Genet. 2000;26:76–80. doi: 10.1038/79216. [DOI] [PubMed] [Google Scholar]
- 11.Cole SA, Mitchell BD, Hsueh W-C, Pineda P, Beamer BA, Shuldiner AR, Comuzzie AG, Blangero J, Hixson JE. The Pro12Ala variant of peroxisome proliferator-activated receptor-gamma2 (PPAR-gamma2) is associated with measures of obesity in Mexican Americans. Int J Obes. 2000;24:522–524. doi: 10.1038/sj.ijo.0801210. [DOI] [PubMed] [Google Scholar]
- 12.Douglas JA, Erdos MR, Watanabe RM, Braun A, Johnston CL, Oeth P, Mohlke KL, Valle TT, Ehnholm C, Buchanan TA, Bergman RN, Collins FS, Boehnke M, Tuomilehto J. The peroxisome proliferator-activated receptor-γ2 Pro12Ala variant: association with type 2 diabetes and trait differences. Diabetes. 2001;50:886–890. doi: 10.2337/diabetes.50.4.886. [DOI] [PubMed] [Google Scholar]
- 13.Hsueh W-C, Cole SA, Shuldiner AR, Beamer BA, Blangero J, Hixson JE, MacCluer JW, Mitchell BD. Interactions between variants in the β3-adrenergic receptor and peroxisome proliferator-activated receptor-γ2 genes and obesity. Diabetes Care. 2001;24:672–677. doi: 10.2337/diacare.24.4.672. [DOI] [PubMed] [Google Scholar]
- 14.Odom DT, Zizlsperger N, Gordon DB, Bell GW, Rinaldi NJ, Murray HL, Volkert TL, Schreiber J, Rolfe PA, Gifford DK, Fraenkel E, Bell GI, Young RA. Control of pancreas and liver gene expression by HNF transcription factors. Science. 2004;303:1378–1381. doi: 10.1126/science.1089769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamagata K, Furuta H, Oda N, Kaisaki PJ, Menzel S, Cox NJ, Fajans SS, Signorini S, Steffel M, Bell GI. Mutations in the hepatocyte nuclear factor-4α gene in maturity-onset diabetes of the young (MODY1) Nature. 1996;384:458–460. doi: 10.1038/384458a0. [DOI] [PubMed] [Google Scholar]
- 16.Silander K, Mohlke KL, Scott LJ, Peck EC, Hollstein P, Skol AD, Jackson AU, Deloukas P, Hunt S, Stavrides G, Chines PS, Erdos MR, Narisu N, Conneely KN, Li C, Fingerlin TE, Dhanjal SK, Valle TT, Bergman RN, Tuomilehto J, Watanabe RM, Boehnke M, Collins FS. Genetic variation near the hepatocyte nuclear factor-4α gene predicts susceptibility to type 2 diabetes. Diabetes. 2004;53:1141–1149. doi: 10.2337/diabetes.53.4.1141. [DOI] [PubMed] [Google Scholar]
- 17.Love-Gregory LD, Wasson J, Ma J, Jin CH, Glaser B, Suarez BK, Permutt MA. A common polymorphism in the upstream promoter region of the hepatocyte nuclear factor-4α gene on chromosome 20q is associated with type 2 diabetes and appears to contribute to the evidence for linkage in an Ashkenazi Jewish population. Diabetes. 2004;53:1134–1140. doi: 10.2337/diabetes.53.4.1134. [DOI] [PubMed] [Google Scholar]
- 18.Damcott CM, Hoppman N, Ott SH, Reinhart LJ, Wang J, Pollin TI, O’Connell JR, Mitchell BD, Shuldiner AR. Polymorphisms in both promoters of hepatocyte nuclear factor 4-α are associated with type 2 diabetes in the Amish. Diabetes. 2004;53:3337–3341. doi: 10.2337/diabetes.53.12.3337. [DOI] [PubMed] [Google Scholar]
- 19.Hansen SK, Rose CS, Glümer C, Drivsholm T, Borch-Johnsen K, Jørgensen T, Pedersen O, Hansen T. Variation near the hepatocyte nuclear factor (HNF)-4α gene associates with type 2 diabetes in the Danish population. Diabetologia. 2005;48:452–458. doi: 10.1007/s00125-005-1671-0. [DOI] [PubMed] [Google Scholar]
- 20.Bagwell AM, Bento JL, Mychaleckyj JC, Freedman BI, Langefeld CD, Bowden DW. Genetic analysis of HNF4A polymorphisms in Caucasian-American type 2 diabetes. Diabetes. 2005;54:1185–1190. doi: 10.2337/diabetes.54.4.1185. [DOI] [PubMed] [Google Scholar]
- 21.Winckler W, Graham RR, de Bakker PIW, Sun M, Almgren P, Tuomi T, Gaudet D, Hudson TJ, Ardlie KG, Daly MJ, Hirschhorn JN, Groop L, Altshuler D. Association testing of variants in the hepatocyte nuclear factor 4α gene with risk of type 2 diabetes in 7,883 people. Diabetes. 2005;54:886–892. doi: 10.2337/diabetes.54.3.886. [DOI] [PubMed] [Google Scholar]
- 22.Henkin L, Bergman RN, Bowden DW, Ellsworth DL, Haffner SM, Langefeld CD, Mitchell BD, Norris JM, Rewers M, Saad MF, Stamm E, Wagenknecht LE, Rich SS. Genetic epidemiology of insulin resistance and visceral adiposity: the IRAS family study design and methods. Am J Epidemiol. 2003;13:211–217. doi: 10.1016/s1047-2797(02)00412-x. [DOI] [PubMed] [Google Scholar]
- 23.Wagenknecht LE, Mayer EJ, Rewers M, Haffner S, Selby J, Borok GM, Henkin L, Howard G, Savage PJ, Saad MF, Bergman RN, Hamman R. The Insulin Resistance Atherosclerosis Study (IRAS) objectives, design, and recruitment results. Ann Epidemiol. 1995;5:464–472. doi: 10.1016/1047-2797(95)00062-3. [DOI] [PubMed] [Google Scholar]
- 24.Herbert V, Law K, Gottlieb C, Bleicher S. Coated charcoal immunoassay of insulin. J Clin Endocrinol Metab. 1965;25:1375–1384. doi: 10.1210/jcem-25-10-1375. [DOI] [PubMed] [Google Scholar]
- 25.Livak KJ. SNP genotyping by the 5′-nuclease reaction. Methods Mol Biol. 2003;212:129–147. doi: 10.1385/1-59259-327-5:129. [DOI] [PubMed] [Google Scholar]
- 26.Beutow KH, Edmonson M, MacDonald R, Clifford R, Yip P, Kelley J, Little DP, Strausberg R, Koester H, Cantor CR, Braun A. High-throughput development and characterization of a genomewide collection of gene-based single nucleotide polymorphism markers by chip-based matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Proc Natl Acad Sci U S A. 2001;98:581–584. doi: 10.1073/pnas.021506298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bergman RN, Phillips LS, Cobelli C. Physiologic evaluation of factors controlling glucose tolerance in man: measurement of insulin sensitivity and β-cell glucose sensitivity from the response to intravenous glucose. J Clin Invest. 1981;68:1456–1467. doi: 10.1172/JCI110398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 29.Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, Liu-Cordero SN, Rotimi C, Adeyemo A, Cooper R, Ward R, Lander ES, Daly MJ, Altshuler D. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 30.Shetterly SM, Bater J, Morgenstern NE, Grigsby J, Hamman RF. Higher instrumental activities of daily living disability in Hispanics compared with non-Hispanic Whites in rual Colorado. Am J Epidemiol. 1998;147:1019–1027. doi: 10.1093/oxfordjournals.aje.a009395. [DOI] [PubMed] [Google Scholar]
- 31.Bonilla C, Parra EJ, Pfaff CL, Dios S, Marshall JA, Hamman RF, Ferrell RE, Hoggart CL, McKeigue PM, Shriver MD. Admixture in the Hispanics of the San Luis Valley, Colorado, and its implications for complex trait gene mapping. Ann Intern Med. 2004;68:139–153. doi: 10.1046/j.1529-8817.2003.00084.x. [DOI] [PubMed] [Google Scholar]
- 32.Price AL, Patterson N, Yu F, Cox DR, Waliszewska A, McDonald GJ, Tandon A, Schirmer C, Neubauer J, Bedoya G, Duque C, Villegas A, Bortolini MC, Salzano FM, Gallo C, Mazzotti G, Tello-Ruiz M, Riba L, Aguilar-Salinas CA, Canizales-Quinteros S, Menjivar M, Klitz W, Henderson B, Haiman CA, Winkler C, Tusie-Luna T, Ruiz-Linares A, Reich D. A genomewide admixture map for Latino populations. Am J Hum Genet. 2007;80:1024–1036. doi: 10.1086/518313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Allard MW, Polanskey D, Wilson MR, Monson KL, Budowle B. Evaluation of variation in control region sequences for Hispanic individuals in the SWGDAM mtDNA data set. J Forensic Sci. 2006;51:566–573. doi: 10.1111/j.1556-4029.2006.00136.x. [DOI] [PubMed] [Google Scholar]
- 34.Auwerx J. PPARγ, the ultimate thrifty gene. Diabetologia. 1999;42:1033–1049. doi: 10.1007/s001250051268. [DOI] [PubMed] [Google Scholar]
- 35.Thomas H, Jaschkowitz K, Bulman M, Frayling TM, Mitchell SMS, Roosen S, Lingott-Frieg A, Tack CJ, Ellard S, Ryffel GU, Hattersley AT. A distant upstream promoter of the HNF-4α gene connects the transcription factors involved in maturity-onset diabetes of the young. Hum Mol Genet. 2001;10:2089–2097. doi: 10.1093/hmg/10.19.2089. [DOI] [PubMed] [Google Scholar]
- 36.Gavrilova O, Haluzik M, Matsusue K, Cutson JJ, Johnson L, Dietz KR, Nicol CJ, Vinson C, Gonzalez FJ, Reitman MI. Liver peroxisome proliferator-activated receptor γ contributes to hepatic steatosis, triglyceride clearance, and regulation of body fat mass. J Biol Chem. 2003;278:34268–34276. doi: 10.1074/jbc.M300043200. [DOI] [PubMed] [Google Scholar]
- 37.Schadinger SE, Rucher NLR, Schreiber BM, Farmer SR. PPARγ2 regulates lipogenesis and lipid accumulation in steatotic hepatocytes. Am J Physiol. 2005;288:E1195–E1205. doi: 10.1152/ajpendo.00513.2004. [DOI] [PubMed] [Google Scholar]
- 38.Luyckx FH, Lefebvre PJ, Scheen AJ. Non-alcoholic steatohepatitis: association with obesity and insulin resistance, and influence of weight loss. Diabete Metab. 2000;26:98–106. [PubMed] [Google Scholar]
- 39.Stumvoll M, Wahl HG, Löblein K, Becker R, Machicao F, Jacob S, Häring H. Pro12Ala polymorphism in the peroxisome proliferator-activated receptor-γ2 gene is associated with increased antilipolytic insulin sensitivity. Diabetes. 2001;50:876–881. doi: 10.2337/diabetes.50.4.876. [DOI] [PubMed] [Google Scholar]
- 40.Rebrin K, Steil GM, Mittelman SD, Bergman RN. Causal linkage between insulin suppression of lipolysis and suppression of liver glucose output in dogs. J Clin Invest. 1996;98:741–749. doi: 10.1172/JCI118846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sheena V, Hertz R, Berman I, Nousbeck J, Bar-Tana J. Transcriptional suppression of human microsomal triglyceride transfer protein by hypo-lipidemic insulin sensitizers. Biochem Pharmacol. 2005;70:1548–1559. doi: 10.1016/j.bcp.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 42.Sheena V, Hertz R, Nousbeck J, Berman I, Magenheim J, Bar-Tana J. Transcriptional regulation of human microsomal triglyceride transfer protein by hepatocyte nuclear factor-4α. J Lipid Res. 2005;46:328–341. doi: 10.1194/jlr.M400371-JLR200. [DOI] [PubMed] [Google Scholar]
- 43.Phillips C, Mullan K, Owens D, Tomkin GH. Intestinal microsomal triglyceride transfer protein in type 2 diabetic and non-diabetic subjects: the relationship to triglyceride-rich postprandial lipoprotein composition. Atherosclerosis. 2006;187:57–64. doi: 10.1016/j.atherosclerosis.2005.08.020. [DOI] [PubMed] [Google Scholar]
- 44.Rubin D, Helwig U, Pfeuffer M, Schreiber S, Boeing H, Fisher E, Pfeiffer A, Freitag-Wolf S, Foelsch UR, Doering F, Schrezenmeir J. A common functional exon polymorphism in the microsomal triglyceride transfer protein gene is associated with type 2 diabetes, impaired glucose metabolism, and insulin levels. J Hum Genet. 2006;51:567–574. doi: 10.1007/s10038-006-0400-y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.