Abstract
The phytopathogenic ascomycete Botrytis cinerea is known to produce abscisic acid (ABA), which is thought to be involved in host-pathogen interaction. Biochemical analyses had previously shown that, in contrast to higher plants, the fungal ABA biosynthesis probably does not proceed via carotenoids but involves direct cyclization of farnesyl diphosphate and subsequent oxidation steps. We present here evidence that this “direct” pathway is indeed the only one used by an ABA-overproducing strain of B. cinerea. Targeted inactivation of the gene bccpr1 encoding a cytochrome P450 oxidoreductase reduced the ABA production significantly, proving the involvement of P450 monooxygenases in the pathway. Expression analysis of 28 different putative P450 monooxygenase genes revealed two that were induced under ABA biosynthesis conditions. Targeted inactivation showed that one of these, bcaba1, is essential for ABA biosynthesis: ΔBcaba1 mutants contained no residual ABA. Thus, bcaba1 represents the first identified fungal ABA biosynthetic gene.
Fungi, especially phytopathogenic species, have been shown to produce all major classes of phytohormones (for a review, see reference 35). The best investigated fungal phytohormone system (and the only one in which the biosynthetic genes involved have been identified) is that of Gibberella fujikuroi. It has been shown that the genes involved in gibberellic acid (GA3) biosynthesis are arranged in a cluster (34) and that the biosynthesis in major respects is different from the higher plant pathway (12). In this case, therefore, the original hypothesis that fungal biosynthesis of phytohormones represents a good example for a horizontal gene transfer (6) is negated. This view was confirmed recently for ethylene production in the gray mold fungus Botrytis cinerea. Chagué et al. (5) presented evidence that ethylene is synthesized via α-keto-γ-methylthiobutyric acid, as in prokaryotic systems, and not via the 1-aminocyclopropane-1-carboxylic acid pathway used in higher plants.
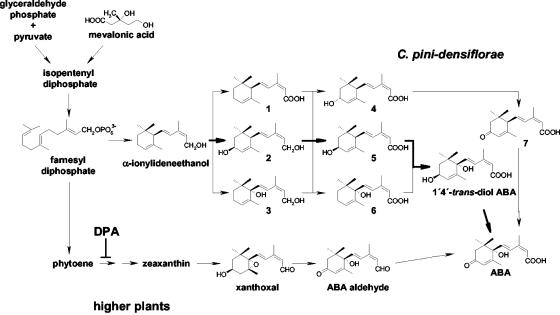
Strains of B. cinerea (like several other fungi) have also been shown to synthesize abscisic acid (ABA) (13, 18); overproducing strains are now used for biotechnological production of ABA (see the review by Tudzynski and Sharon [35]). Biosynthesis of ABA in fungi also seems to be distinct from the pathway used by higher plants (for a review, see references 19 and 23) (see Fig. 1). Biochemical analyses in various Cercospora species provide evidence that these fungi synthesize ABA directly from farnesyl diphosphate, via different oxidative steps, with either 1′-4′-dihydroxy-γ-ionylidene acetate, 1′-deoxy-ABA, or 1′-4′-trans-diol ABA as intermediates, and not via the carotenoid pathway used by higher plants (1, 20-22). However, the presence of the carotenoid pathway in fungi cannot be excluded (40). In B. cinerea, biosynthesis of ABA seems to follow a pathway which is similar to the major route in Cercospora pini-densiflorae, i.e., via 1′-4′-trans-diol-ABA (13). Thus far, no fungal ABA biosynthesis gene has been identified. The fact that ABA is produced mainly by pathogenic fungi and given the effects that ABA has on higher plants (induction of senescence, bud inhibition, etc.), it has been speculated that ABA, like gibberellins, might be involved in pathogenesis. It has been shown that ABA biosynthesis in B. cinerea is stimulated by the host plant (15) and that external application of ABA can enhance disease development caused by B. cinerea (29). Furthermore, Audenaert et al. (2) showed that ABA appears to negatively modulate the salicylic acid-dependent defense pathway in tomato plants during infection with B. cinerea. Thus, ABA could represent a virulence factor for the fungus. The unequivocal proof for a role of fungal ABA in the host-pathogen interaction would require defined mutants, which are absolutely unable to produce ABA in planta. Strains not producing detectable amounts of ABA in axenic culture have been described (21), but it is not clear whether they still have the capability to produce ABA in planta. Therefore, for several reasons the identification of genes involved in the ABA pathway in B. cinerea would be interesting: it would advance evolutionary research by allowing comparison of the pathways in higher plants and fungi, lead to advanced biotechnological methods for the generation of overproducing strains, and provide new insight into phytopathological investigations as indicated above. The objective of the present study was to find conclusive evidence for the postulated alternative ABA biosynthetic pathway in B. cinerea. We show here that P450 monooxygenases are involved in ABA biosynthesis, by targeted inactivation of a cytochrome P450 oxidoreductase gene, and we describe the identification of the first fungal gene involved in ABA biosynthesis, encoding a P450 monooxygenase.
FIG. 1.
Postulated biosynthetic pathways of ABA in higher plants and in C. pini-densiflorae (19, 22), respectively. Intermediates identified in C. pini-densiflorae are α-ionylideneacetic acid (structure 1), 4′S-OH-α-ionylideneethanol (structure 2), 1′-OH-α-ionylideneethanol (structure 3), (1′R)-4′R-OH-α-ionylideneacetic acid (structure 4), (1′R)-4′S-OH-α-ionylideneacetic acid (structure 5), 1′-OH-α-ionylideneacetic acid (structure 6), and 1′-deoxy-ABA (structure 7). The site of action of DPA is indicated.
MATERIALS AND METHODS
Fungal strains.
Strain ATCC 58025 of B. cinerea Pers.:Fr. (Botryotinia fuckeliana [de Bary] Whetz), a nonsporulating overproducer of ABA (18), and B05.10, a haploid strain were obtained after benomyl treatment of strain SAS56 (25).
Bacterial strains.
Escherichia coli strain TOP10F′ (Invitrogen, Groningen, The Netherlands) was used for propagation of plasmids. Propagation of lambda clones was performed in strains LE392 (Stratagene, La Jolla, Calif.) and XL1-Blue MRF′ (Stratagene), respectively.
Media and culture conditions.
B. cinerea strains were grown on 2% malt extract (Oxoid, Ltd., Basingstoke, Hampshire, England) amended with 0.5% glucose, 0.1% casein peptone (Difco Laboratories, Sparks, Md.), 0.1% Casamino Acids (Difco), 0.1% yeast extract (Duchefa Biochemie BV, Haarlem, The Netherlands), and 0.02% RNA sodium salt. For ABA production, fungi were cultivated in 300-ml Erlenmeyer flasks with 100 ml of a defined liquid medium (31; modified as described by Kettner [14]) containing 20 g of lactose/liter instead of glucose (Sprecher medium). Fungi and culture filtrates were harvested after 3 to 7 days of cultivation on a rotary shaker at 150 rpm and 20°C. For DNA isolation, mycelium was grown for 3 to 4 days at 20°C on complex medium agar (24) with a cellophane overlay.
DNA isolation.
Fungal genomic DNA was isolated as described by Cenis (4). Lambda DNA was isolated according to the standard method (28). Plasmid DNA was isolated by using a plasmid DNA preparation kit (Genomed, Bad Oeynhausen, Germany).
Southern blot analysis.
Genomic DNA was digested with restriction enzymes, size separated on a 1% agarose gel, and blotted onto Hybond-N+ membranes (Amersham Pharmacia, Freiburg, Germany) according to the method of Sambrook et al. (28). Hybridization was carried out in 6× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7]), 5× Denhardt solution, 0.1% sodium dodecyl sulfate, and 50 mM phosphate buffer (pH 6.6) at 65°C for 16 to 20 h in the presence of a random-primed [α-32P]dCTP-labeled probe. Membranes were washed in 2× SSPE-0.1% sodium dodecyl sulfate at 65°C before being exposed to an autoradiographic film.
RNA blot analysis.
RNA was isolated from mycelial samples by using the RNAgents total RNA isolation system (Promega, Mannheim, Germany). Samples of 10 to 15 μg of RNA were transferred on to Hybond-N+ filters after electrophoresis on a 1% agarose gel containing formaldehyde according to the method of Sambrook et al. (28). Blot hybridizations were carried out in 0.6 M NaCl, 0.16 M Na2HPO4, 0.06 M EDTA, 1% N-lauroylsarcosine (Sigma), and 10% dextran sulfate (Eppendorf AG, Hamburg, Germany) (pH 6.5) as described for the Southern blots.
Sequencing.
DNA sequencing of recombinant plasmid clones was performed with an automatic sequencer LI-COR 4200 (MWG Biotech, Munich, Germany) by using the ThermoSequenase fluorescence-labeled primer cycle sequencing kit (Amersham Pharmacia). For sequence analysis and construction of phylogenetic trees, the program DNAStar (Madison, Wis.) was used.
Cloning of bccpr1 gene.
bccpr1 was cloned in a PCR approach by amplifying B05.10 genomic DNA with degenerate primers CPR1 and CPR2 (kindly provided by C. Wasmann, University of Arizona). PCRs contained 25 ng of DNA, 10 pmol of each primer, 200 nM concentrations of deoxynucleoside triphosphates, and 1 U of Taq polymerase (Red Taq; Sigma-Aldrich, Deisenhofen, Germany) and were carried out at 94°C for 4 min, followed by 35 cycles of 94°C for 1 min, 48°C for 1 min, and 72°C for 1 min. The primers were CPR1 (5′-AAG YTG CAG CCY CGC TAC TAY TCS ATC TC-3′) and CPR2 (5′-CTT CCA YTC RTC CTT GTA SAR GAA RTC CTC-3′). The resulting 0.45-kb PCR fragment was used as a probe for screening a genomic EMBL3 library of B. cinerea strain SAS56 (25). Positive and purified phages were subcloned in pUC19 (41) and pBluescriptII SK(−) (Stratagene), respectively.
Construction of a replacement and a complementation vector for bccpr1.
For construction of the gene replacement vector pCPR1Rep, the plasmid pOliHP (27) carrying the E. coli hygromycin phosphotransferase gene hph under control of the Aspergillus nidulans oliC promoter and trpC terminator was used as a basis vector. A 0.4-kb PCR fragment was amplified from the 5′ region of bccpr1 by using the primers CPR5′F (primer 1; 5′-CAC TGA GAA GAT GCT CGA GTC C-3′) and CPR5′R (primer 2; 5′-TTC TTG TAG CGT CGA CCT TCT CG-3′) (artificial XhoI and SalI sites, respectively, are indicated in boldface). From the 3′ end of bccpr1, a 0.4-kb fragment was amplified by using the primers CPR3′F (primer 3; 5′-AAC CGG TGT TGA AGC TTT CAG G-3′) and CPR3′R (primer 4; 5′-AAT ATC CTC AGC CTT GAA TTC TGG-3′) (artificial HindIII and EcoRI sites, respectively, are indicated in boldface). Both fragments were cloned into pCR2.1-TOPO (Invitrogen), cut with XhoI/SalI and HindIII/EcoRI, respectively, and cloned into the corresponding sites of pOliHP. By cutting with XhoI and EcoRI, the 3.5-kb replacement cassette was isolated from the vector prior to transformation.
For construction of the complementation vector pComCPR1, the 2.7-kb HindIII fragment containing the 5′ region of bccpr1, including 1.5-kb promoter sequence, was cloned into pNR1 (17), a vector that carries the Streptomyces noursei nourseothricin acetyltransferase gene nat-1 under the control of the A. nidulans oliC promoter and a B. cinerea β-tubulin transcription terminator fragment. Using the internal ClaI site of the HindIII fragment (see Fig. 4), the 2.2-kb ClaI fragment comprising the 3′ region of bccpr1 was fused to this construct.
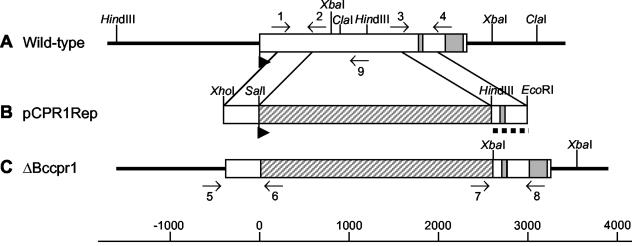
FIG. 4.
Scheme of gene replacement approach for bccpr1. Physical maps of bccpr1 from wild-type strain ATCC 58025 (A), the gene replacement fragment pCPR1Rep (B), and bccpr1 from a ΔBccpr1 replacement mutant (C) showing the organization of exons (□), introns (░⃞), the hygromycin resistance cassette (▨), and flanking regions of bccpr1 (heavy lines). Orientations of the genes are indicated by arrowheads. Binding sites of primers 1 to 9 used for PCR analysis of replacement mutants (see Materials and Methods) and 3′ flank of pCPR1Rep (dotted line) used as a probe for Southern and Northern analyses are indicated.
Cloning of the bcaba1 gene.
A cDNA fragment derived from an expressed sequence tag (EST) library of strain ATCC 58025 under ABA biosynthesis conditions (V. Siewers, D. Tapadar, P. Schreier, and P. Tudzynski, unpublished data) was used to screen the EMBL3 library of SAS56.
Construction of a replacement vector and a complementation vector for bcaba1.
The following primers were used to amplify the 5′ and the 3′ regions of bcaba1(see Fig. 7): ABA1-5′F (primer 10; 5′-ATC ACC ACT CGC GGA TCT GCG-3′), ABA1-5′R (primer 11; 5′-CAT AAT ATG GAT TGA GTC GAC CGG-3′), ABA1-3′F (primer 12; 5′-CAT AAT CTT TCA CAA AGC TTG AGC-3′), and ABA1-3′R (primer 13; 5′-ATA GCA GAT AGC AAA TAA ATG AGG-3′). Primer ABA1-5′R contains an artificial SalI site, and primer ABA1-3′F contains an artificial HindIII site (indicated in boldface). Both fragments were cloned into pCR2.1-TOPO, cut with XhoI/SalI and HindIII/EcoRI, respectively, and cloned into the corresponding sites of pOliHP to construct pABA1Rep. The replacement cassette was isolated from the vector prior to transformation by cutting both XhoI and EcoRI.
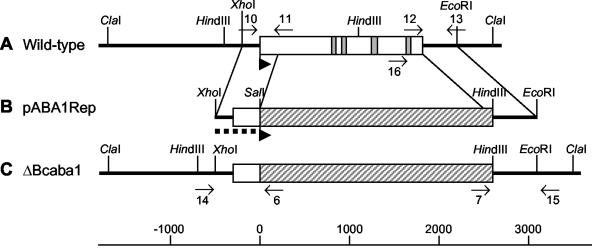
FIG. 7.
Scheme of gene replacement approach for bcaba1. Physical maps of bcaba1 from wild-type strain ATCC 58025 (A), the gene replacement fragment pABA1Rep (B), and bcaba1 from a ΔBccpr1 replacement mutant (C) showing organization of exons (□), introns (░⃞), the hygromycin resistance cassette (▨), and flanking regions of bcaba1 (heavy lines). Orientations of the genes are indicated by arrowheads. The binding sites of primers 6, 7, and 10 to 16 used for PCR analysis of replacement mutants (see Materials and Methods) and the 5′ flank of pABA1Rep (dotted line) used as a probe for Southern and Northern analysis are indicated.
In order to construct the complementation vector pComABA1, a 4.1-kb ClaI fragment containing the complete bcaba1 gene (see Fig. 7) was ligated into pNR1.
Transformation of B. cinerea.
Protocols for protoplast formation were adapted from an established procedure (32). Mycelium derived from cultures grown for 3 days on complex medium agar with a cellophane overlay was ground in a mortar and cultivated for 36 h in 1.5% malt extract medium (Difco) (amended as described for solid medium) at 150 rpm and 20°C. After a wash with KCl buffer (0.6 M KCl, 50 mM CaCl2), 1.5 g of mycelium was suspended in 10 ml of KCl buffer with 0.4% Novozyme 234 (Calbiochem-Novabiochem Corp., La Jolla, Calif.) and then agitated at 28°C and 90 rpm for 1 h 15 min. Protoplasts were collected by using sterile cheese cloth, pelleted by centrifugation for 10 min at 1,500 × g, and resuspended in KCl buffer. DNA (10 to 15 μg) was added to 107 protoplasts in 100 μl of KCl buffer.
Transformed protoplasts were added to 100 ml of liquid agar (1.2%) containing 0.6 M sucrose, 5 mM Tris (pH 6.5), and 1 mM (NH4)2HPO4 (SH agar) prior to pouring on plates. After 24 h, the plates were overlaid with SH agar supplemented with 10 μg of hygromycin B (Calbiochem)/ml or with 70 μg of hygromycin B and 140 μg of nourseothricin (Werner-Bioagents, Jena, Germany)/ml. Resistant colonies were transferred to agar plates containing Gamborg's B5 medium (Duchefa Biochemie BV, Haarlem, The Netherlands) and 2% glucose complemented with 70 μg of hygromycin B/ml or with 70 μg of hygromycin B and 140 μg of nourseothricin/ml.
Homologous integration events and complementation were identified by PCR with the following primers (see Fig. 4 and 7, respectively): pLOF-oliP (primer 6; 5′-GGT ACT GCC CCA CTT AGT GGC AGC TCG CG-3′), pAN-T (primer 7; 5′-CCC AGA ATG CAC AGG TAC AC-3′), CPR-Bc1 (primer 5; 5′-CGC CAC CAA TGG AAA CAA ACC CGC-3′), CPR-Bc2 (primer 8; 5′-GGT ATT GAT TGG CGG ATC GC-3′), CPR-Bc3 (primer 9; 5′-GCA TCG TAT GTA GTT GGG GTA GGG-3′), P16a (primer 14; 5′-GGA GCC AGA CTC TCA TTT GAC ATG TGG-3′), P16b (primer 15; 5′-CGG CCT GTT GTA GTC TCT GC-3′), and P16c (primer 16; 5′-CTT CTC TGT TGA GAT CCA CTG C-3′).
EIA for cis-(+)-abscisic acid.
Enzyme immunoassay (EIA) for detection of ABA in culture filtrates was performed on 96-well microplates (flat bottom; Sarstedt, Inc., Newton, N.C.) in triplicate measurements as described by Weiler (39). A rabbit polyclonal antibody raised against mouse immunoglobulin G (RAMIG), an anti-ABA mouse monoclonal antibody, and an ABA-labeled alkaline phosphatase (tracer) were kindly provided by E. W. Weiler, Ruhr University, Bochum, Germany. p-Nitrophenyl phosphate (Biomol Feinchemikalien GmbH, Hamburg, Germany) was used as phosphatase substrate. ABA standards ranged from 0.05 to 50 pmol/ml. For each sample, ABA was assayed in at least two dilutions. The optical density at 405 nm was measured by using a microplate reader (model 550; Bio-Rad Laboratories GmbH, Munich, Germany).
Liquid chromatography-mass spectrometry (LC-MS) analyses.
Fungi were grown on alkaloid-forming agar (26) for 7 days in the dark. Culture extracts were prepared by using a plug extraction method modified from Smedsgaard (30). Three 6-mm agar plugs were cut from each culture and transferred to a 1.5-ml disposable autosampler screw-cap vial. The plugs were extracted twice by sonication for 60 min with 500 μl of ethyl acetate with 0.5% formic acid for the first extraction. The ethyl acetate extract was transferred to a clean vial and evaporated to dryness in a rotary vacuum concentrator (Christ RVC). The plugs were reextracted with 500 μl of 2-propanol. The 2-propanol extract was transferred to the vial with the dry residues from the first extraction and again evaporated to dryness. The combined residues were redissolved by sonication for 10 min in 300 μl of methanol and filtered before analysis.
LC-MS analyses were performed on an Agilent 1100 system connected to a Micromass LCT (Waters, United Kingdom) time-of-flight mass spectrometer with an electrospray source and LockSpray interface. The separation was done on a Luna C18 (50 by 2 mm [inner diameter]) (2) column (Phenomenex) at a flow rate of 0.3 ml/min. A linear gradient of H2O (containing 10 mM ammonium acetate and 20 mM formic acid) and CH3CN (containing 20 mM formic acid) was used; the gradient was changed from 15% CH3CN to 100% over 20 min and then maintained at 100% CH3CN for 5 min before the return to starting conditions. Leucine-enkephaline dissolved in 50% acetonitrile with 0.1% formic acid was used as a lockmass at a flow rate of 5 μl/min. The instrument was tuned to maximum sensitivity at a resolution better than 5,000 full width half maximum and then calibrated on a mixture of polyethylene glycol 400, 600, and 1000. A sample cone voltage of 25 V was used. The mass accuracy was verified to be better than 5 ppm by injection of a known test sample. Mass spectra were collected from m/z 100 to 900 at 0.5 scan/s; every third scan was taken from the reference spray. UV spectra were collected in parallel from 200 to 700 nm.
Nucleotide sequence accession numbers.
The nucleotide sequence accession numbers were AJ609392 for bcaba1 and AJ609393 for bccpr1.
RESULTS
B. cinerea probably does not use the carotenoid pathway for ABA biosynthesis.
Okamoto et al. (21) used inhibitors of enzymes of the carotenoid pathway to show that this higher plant-specific pathway is obviously not the (major) pathway used by Cercospora (Fig. 1). We used the same approach to test whether B. cinerea utilizes (at least partially) this pathway. We performed these analyses with an ABA-overproducing isolate (ATCC 58025), which facilitated the biochemical analyses (field isolates of B. cinerea tend to produce variable but low amounts of ABA in axenic culture). The strain was cultivated in liquid medium in the presence of diphenylamine (DPA), an inhibitor of phytoene dehydrogenase (7) at a concentration of 5 × 10−5 M, i.e., below growth-limiting concentrations. After 7 days, the ABA concentration in the growth medium was analyzed by EIA (see Materials and Methods). Since the amount of ABA produced by the control and the inhibitor-treated culture did not differ significantly (data not shown), the inhibitor obviously had no effect on ABA synthesis by strain ATCC 58025. This supports the view that also in B. cinerea the carotenoid pathway is not involved in ABA biosynthesis.
Cytochrome P450-monooxygenases are involved in ABA biosynthesis in B. cinerea.
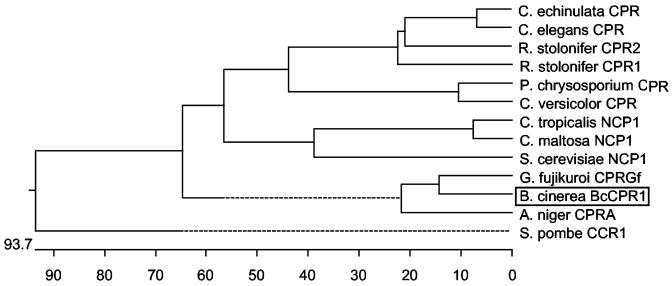
The postulated pathway for ABA biosynthesis in B. cinerea (Fig. 1) involves several hydroxylation and oxidation steps, which in fungi regularly are catalyzed by P450 monooxygenases (reviewed by van den Brink et al. [38]). Since B. cinerea probably possesses more than 50 enzymes of this class (42), we first used a general approach to test whether indeed P450 monooxygenases (and not dioxygenases) are likely to be involved in ABA biosynthesis. In spite of the high number of P450 monooxygenases, most fungi seem to have only one cytochrome P450 oxidoreductase which acts as the unspecific partner for all of the specific monooxygenases facilitating the electron transfer from NADPH via FAD and FMN to the heme group of the P450 enzyme (37). Cytochrome P450 oxidoreductase (CPR) genes have been isolated and functionally characterized thus far only from a few fungi such as Aspergillus niger (37) and G. fujikuroi (17). Based on these fungal cpr sequences, degenerate primers were used to amplify a PCR fragment from genomic DNA of B. cinerea showing significant homology to cpr genes of fungi (data not shown). The PCR fragment was used as probe to screen a genomic EMBL3 library of B. cinerea. Two positive plaques were purified and then subcloned. Sequence analysis revealed an open reading frame of 2,348 bp interrupted by two introns of 58 and 211 bp, respectively; it encodes a polypeptide of 692 amino acids corresponding to a molecular mass of 77 kDa. The derived gene product has significant overall homology to fungal CPRs, its closest homologues being the cpr genes of G. fujikuroi and A. niger with 75 and 67% identity, respectively (Fig. 2), and thus represents a bona fide cytochrome P450 oxidoreductase. The gene was therefore named bccpr1.
FIG. 2.
Cladogram of fungal cytochrome P450 oxidoreductases based on amino acid sequences. The accession numbers are as follows: Aspergillus niger CprA, CAA81550; Candida maltosa NCP1, P50126; Candida tropicalis NCP1, P37201; Coriolus versicolor CPR, BAB83588; Cunninghamella echinulata CPR, AAF89959; Cunninghamella elegans CPR, AAF89958; Gibberella fujikuroi CPRGf, AJ576025; Phanerochaete chrysosporium CPR, AAG31350; Rhizopus stolonifer CPR isoenzyme 1, AAG23833; Rhizopus stolonifer CPR isoenzyme 2, AAG23834; Saccharomyces cerevisiae NCP1, P16603; and Schizosaccharomyces pombe CCR1, P36587.
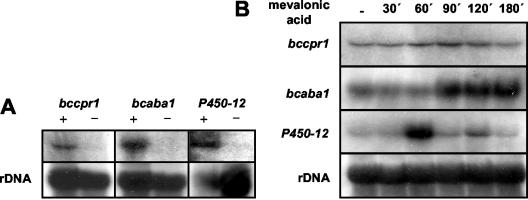
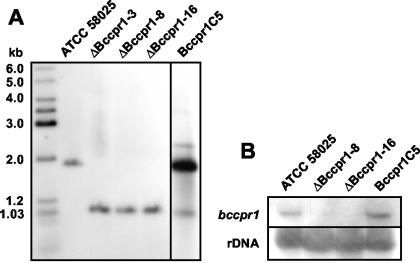
Since Malonek et al. (17) found that the single cpr gene of G. fujikuroi is induced under gibberellin biosynthesis conditions, we performed a Northern analysis to test the expression of bccpr1. As shown in Fig. 3A, the gene is induced under conditions of ABA biosynthesis. The addition of mevalonic acid, the ABA precursor, led to an increased expression of bccpr1 for up to 120 min (Fig. 3B). This expression pattern supports the possible role of P450 monooxygenases in ABA biosynthesis. To prove this role unequivocally, we performed a targeted gene inactivation by using a gene replacement approach as outlined in Fig. 4 (for details, see Materials and Methods). The gene replacement fragment was used for transformation of B. cinerea protoplasts. Hygromycin-resistant transformants were screened by PCR for the presence of a knockout allele, with primers 5 and 6 and primers 7 and 8, respectively (Fig. 4). Transformants yielding the corresponding diagnostic PCR fragments were genetically purified by protoplast subculturing (the standard method of single-spore isolation was not applicable because the recipient strain ATCC 58025 does not sporulate) and analyzed again by PCR. Transformants showing only the diagnostic PCR fragments, but not the wild-type-specific fragment of 1 kb generated by primers 5 and 9 (data not shown), were analyzed by Southern hybridization: genomic DNA of the wild type and the three different transformants was digested with XbaI, separated in an agarose gel, and transferred to a nylon membrane. As shown in Fig. 5A, transformants ΔBccpr1-3, ΔBccpr1-8, and ΔBccpr1-16 lack the wild-type fragment of 1.9 kb hybridizing to the labeled bccpr1 3′ flank and show the 1.03-kb fragment expected for the knockout situation (Fig. 4), proving that they are homokaryotic deletion mutants. A Northern analysis confirmed this conclusion: in the mycelia of the mutants grown under ABA biosynthesis conditions, no bccpr1 transcript could be detected (Fig. 5B). The mutants showed a slightly reduced growth rate in axenic culture but retained the wild-type morphology (data not shown). An EIA analysis showed that mutants ΔBccpr1-8 and ΔBccpr1-16 produced significantly less ABA than the wild type (Table 1).
FIG. 3.
Expresssion analysis of genes bccpr1, bcaba1, and P450-12. (A) Total RNA was extracted from ABA-producing B. cinerea strain ATCC 58025 (lanes +) and nonproducing strain B05.10 grown for 4 days (bccpr1 and bcaba1) or 11 days (P450-12) in Sprecher medium. (B) Total RNA was extracted from strain ATCC 58025 grown for 3 days in Sprecher medium. Mycelia were harvested 30, 60, 90, 120, and 180 min after the addition of 3.8 mM mevalonic acid lactone. Fragments of the 3′ region of bccpr1 (see Fig. 4), the 5′ region of bcaba1 (see Fig. 7), and an internal 720-bp PCR fragment of P450-12 were used for probing. Loading of lanes with RNA was checked by probing with ribosomal DNA.
FIG. 5.
(A) Southern blot analysis of DNA from wild-type strain ATCC 58025; replacement mutants ΔBccpr1-3, ΔBccpr1-8, and ΔBccpr1-16; and complemented mutant Bccpr1C5. DNA was digested with XbaI, blotted, and hybridized with the 3′ flank of the replacement vector pCPR1Rep. (B) Northern analysis. Total RNA from ATCC 58025, replacement mutants ΔBccpr1-8 and ΔBccpr1-16, and complemented mutant Bccpr1C5 was blotted and also probed with the 3′ flank of pCPR1Rep. Equal loading of the lanes was checked by hybridization with rDNA.
TABLE 1.
ABA production of B. cinerea wild-type and mutant strainsa
| Strain | ABA (nmol liter−1) | SD |
|---|---|---|
| ATCC 58025 | 2,927 | 621 |
| ΔBccpr1-8 | 3.77 | 3.75 |
| ΔBccpr1-16 | 3.14 | 1.84 |
| Bccpr1C5 | 3,374 | 582 |
| ΔBcaba1-2 | 0.14 | 0.14 |
| ΔBcaba1-27 | 0.38 | 0.45 |
| Bcaba1C5 | 3,174 | 349 |
| Control | 0.39 | 0.03 |
ABA was determined in 7-day-old liquid cultures by EIA with noninoculated Sprecher medium as a control.
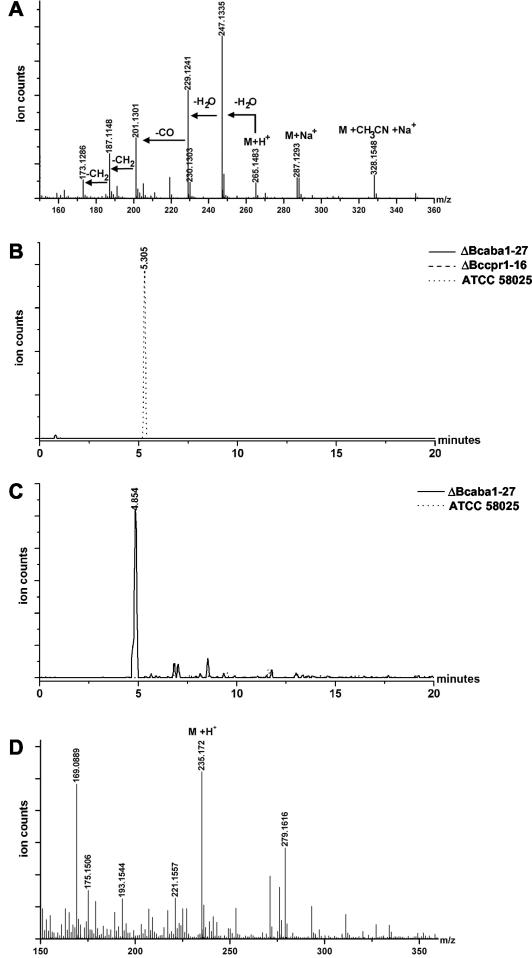
This finding was confirmed by LC-MS analysis (Fig. 6) by using accurate mass and analysis of a standard. ABA elutes at 5.305 min, and the mass spectrum shows a significant ion at m/z 247.1335 due to the loss of water from the protonated molecular ion at m/z 265.1483 (Fig. 6A). Using narrow ion traces with a width of 30 ppm around the significant ions, shown for m/z 247.1335 in Fig. 6B, the presence of ABA was confirmed in the wild type, but it is not found in the mutants. Both mutants show a drastic reduction of other (nonidentified) metabolites produced, as expected since P450 monooxygenases are involved in many pathways (data not shown).
FIG. 6.
LC-MS analyses. (A) Mass spectrum of ABA standard showing a predominant peak at m/z 247.1335 (M+H+-H2O). (B) Ion traces for m/z 247.1335 with a width of 30 ppm of metabolites extracted from wild-type ATCC 58025 (dotted line) and replacement mutants ΔBccpr1-16 (dashed line) and ΔBcaba1-27 (full line). (C) Ion traces for m/z 235.172 with a width of 30 ppm of metabolites extracted from wild-type ATCC 58025 (dotted line) and replacement mutant ΔBcaba1-27 (full line). (D) Mass spectrum of the unknown metabolite from replacement mutant ΔBcaba1-27 eluting at 4.854 min.
To be sure that the loss of ABA production is the result of deletion of bccpr1, we performed a complementation test: mutant ΔBccpr1-8 was transformed with vector pComCPR1 containing a full-length genomic copy of bccpr1 (see Materials and Methods). One of the complemented mutants obtained, Bccpr1C5, was shown to contain obviously more than one correctly sized wild-type copies of the gene (Fig. 5A). Northern analysis confirmed the transcription of the bccpr1 gene copies (Fig. 5B). EIA analyses proved a complementation of the mutant phenotype, i.e., ABA production in the complemented mutant Bccpr1C5 reached the level of the wild type again (Table 1), finally proving that the loss of ABA production in the mutants was caused by the deletion of bccpr1.
Taken together, these data strongly suggest that P450 monooxygenases are involved in ABA biosynthesis. Therefore, a screening for candidate ABA biosynthesis genes within the P450 monooxygenase gene family of B. cinerea was performed. Altogether, 28 different putative P450 monooxygenase genes identified in different EST libraries of B. cinerea were probed in a dot blot analysis with labeled cDNAs from ABA-producing and -nonproducing mycelia, respectively. Two of them, bcP450-12 and bcaba1, showed increased expression under ABA biosynthesis conditions (data not shown). This was confirmed by Northern analysis (Fig. 3A): both genes are strongly induced under ABA biosynthesis conditions and show almost no constitutive expression. The addition of the precursor mevalonic acid (Fig. 3B) resulted in a significant and persistent induction of bcaba1 after 90 min, whereas P450-12 showed a transient induction at 60 min. Thus, both genes are candidates for ABA biosynthetic genes and, because of the persistent expression under mevalonic acid, we focused on bcaba1. The genomic copy of bcaba1 was isolated from the genomic EMBL3 library of strain SAS56 by using the cDNA clone as probe. Sequencing of subcloned lambda fragments hybridizing to the probe revealed an open reading frame of 1,769 bp, interrupted by four introns of 48, 49, 83, and 59 bp, respectively, with a coding capacity of 509 amino acids, yielding a calculated molecular mass of 57.2 kDa. As expected from the EST data, the derived amino acid sequence of bcaba1 showed significant homology to fungal P450 monooxygenases.
bcaba1 is part of the ABA biosynthetic pathway.
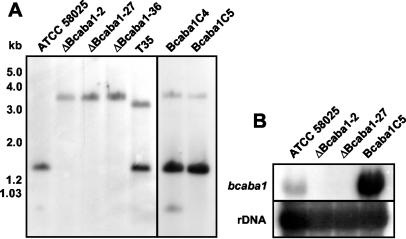
To clarify the role of this candidate gene unequivocally, a gene disruption approach was performed, comparable to the one described for bccpr1. The gene replacement fragment pABA1Rep (see Fig. 7 and Materials and Methods) was used to generate transformants of strain ATCC 58025. As described above, these transformants were checked by PCR and Southern analysis. Transformants ΔBcaba1-2, ΔBcaba1-27, and ΔBcaba1-36 showed the diagnostic PCR fragments of 0.8 and 0.7 kb generated by primer pairs composed of primers 14 and 6 and of primers 7 and 15, respectively, and lacked the wild-type fragment of 0.8 kb generated by primers 16 and 15 (data not shown). Hybridization of HindIII-digested genomic DNA with the labeled bcaba1 5′ flank showed lack of the 1.5-kb wild-type fragment and the presence of the expected 3.5-kb replacement fragment (Fig. 8A). Transformant T35, on the other hand, is an ectopic transformant: it contains the wild-type fragment and an additional hybridizing fragment of 3.0 kb. Northern analyses confirmed this interpretation. In the mycelia of mutants ΔBcaba1-2 and ΔBcaba1-27 grown under ABA biosynthesis conditions, no bcaba1 transcript could be detected (Fig. 8B). All mutants showed a normal growth rate and morphology in axenic culture. Mutants ΔBcaba1-2 and ΔBcaba1-27 were subjected to a detailed biochemical analysis. EIA tests showed that both contain no detectable ABA, even less than the bccpr1 mutants (Table 1). LC-MS analyses confirmed these findings: both mutants lack the specific ABA peak, proving unequivocally that BcABA1 is involved in ABA biosynthesis (as shown in Fig. 6C for mutant ΔBcaba1-27).
FIG. 8.
(A) Southern blot analysis of wild-type strain ATCC 58025; replacement mutants ΔBcaba1-2, ΔBcaba1-27, and ΔBcaba1-36; ectopic transformant T35; and complemented mutants Bcaba1C4 and Bcaba1C5. DNA was digested with HindIII and hybridized with the 5′ flank of the replacement vector pABA1Rep. (B) Northern analysis of total RNA from ATCC 58025, replacement mutants ΔBcaba1-2 and ΔBcaba1-27, and complemented mutant Bcaba1C5 also probed with the 5′ flank of pABA1Rep. Equal loading of the lanes was checked by hybridization with rDNA.
As a final proof for the role of bcaba1 in ABA biosynthesis, mutant ΔBcaba1-2 was transformed with vector pComABA1 containing a full-length copy of bcaba1, by using a nourseothricin-resistance selection cassette (see Materials and Methods). Altogether, 12 complemented mutants were screened for the presence of the complete gene by PCR (data not shown), and the bona fide complemented mutant Bcaba1C5 was further characterized. Since it contains probably multiple copies of the gene (see Fig. 8A), expression of bcaba1 in Bcaba1C5 is not only recovered but exceeds the wild-type level (see Fig. 8B). ABA production of the complemented mutant is comparable to the wild type (Table 1).
The LC-MS analyses showed that a peak at retention time 4.854 min with a major ion peak at m/z 235.172 was significantly increased in the mutants (Fig. 6C and D). The molecular mass corresponds to the postulated ABA intermediate α-(γ-)ionylidene acetic acid (see Fig. 1, component 1), although unequivocal identification of this metabolite would require further detailed analyses. Taken together, these data confirm that bcaba1 represents the first identified fungal ABA biosynthesis gene.
DISCUSSION
We have presented here conclusive evidence that the biosynthesis of ABA in B. cinerea follows—as postulated earlier on the basis of biochemical data—a direct pathway, including direct cyclization of farnesyl diphosphate and sequential oxidation. (i) DPA, which has been shown to be a potent inhibitor of phytoene dehydrogenase, i.e., of carotenogenesis, in several fungi, such as Phycomyces blakesleeanus (7) and Mucor circinelloides (11), did not affect ABA biosynthesis in B. cinerea as previously described for C. pini-densiflorae (21). (ii) Deletion of the bccpr1 gene probably encoding a cytochrome P450 oxidoreductase drastically reduced ABA biosynthesis, strongly suggesting the involvement of P450 monooxygenases in this fungal biosynthetic pathway (in contrast to the plant pathway). Most fungi investigated thus far seem to contain only one CPR encoding gene. However, ΔBccpr1 deletion mutants still seem to exhibit some residual ABA producing activity as determined by EIA. Furthermore, mutants are viable and even able to infect bean plants (data not shown), although P450 monooxygenases are also involved in primary metabolism, e.g., the formation of ergosterol (37). This leads to the assumption that at least one more electron-donating system has to be present in B. cinerea. In the zygomycete fungus Rhizopus nigricans, two putative CPR encoding genes have been identified sharing 66% identity (16). Therefore, the presence of a second CPR in Botrytis cannot be excluded completely. However, it is also possible that another electron donor, as described for cytochrome b5 in Saccharomyces cerevisiae (33), can (at least partially) overcome the defect and thus be responsible for residual ABA production. (iii) Deletion of a P450 monooxidase gene, bcaba1, completely abolished ABA biosynthesis, identifying the product of this gene as essential for ABA biosynthesis. These findings also rule out a minor role of the carotenoid pathway in this strain. This clearly demonstrates that the fungal ABA pathway, at least in B. cinerea, is different from that in higher plants (such as the pathways of GA3 and ethylene).
bcaba1 thus represents the first fungal ABA biosynthesis gene to be identified. The availability of this gene opens fascinating new opportunities for further investigations.
(i) The role of fungal ABA production in host-pathogen interactions can now be studied in detail. Expression of the gene in planta can be easily monitored and can give a clue about the timing of ABA synthesis; more importantly, a functional analysis by targeted inactivation in a pathogenic field isolate can give unequivocal evidence for the importance of fungal ABA in the pathogenic process (deletion of bcaba1 in the standard strain B05.10 is under way). However, in this context it has to be considered that also fungal ABA precursurs such as α-(γ-)ionylideneacetic acid can exhibit biological activity (reviewed by Oritani and Kiyota [23]). Therefore, overexpression experiments would also be helpful in this context. In addition, although the bcaba1 mutants do not show any defects in vegetative properties, an influence of ABA on other parts of the (nonpathogenic) life cycle, e.g., sexual propagation, cannot be ruled out thus far and can now be analyzed in detail.
(ii) Since genes of several secondary metabolic pathways in fungi are arranged in gene clusters, such as penicillin (9), sterigmatocystins (3), gibberellins (34), and ergot alkaloids (36), there is a high probability of identifying other genes encoding enzymes of the ABA pathway by a chromosome-walking approach. Indeed, preliminary analyses identified at least two genes downstream of bcaba1 that are candidates for further ABA biosynthetic genes (V. Siewers and P. Tudzynski, unpublished data).
(iii) The increasing number of fungal genomes available allows comparative evolutionary studies, since ABA biosynthesis seems to have been evolved in quite diverse groups of fungi, including ascomycetes such as Ceratocystis spp., zygomycetes such as Mucor spp., and basidiomycetes such as Agrocybe praecox (8, 10).
(iv) Detailed expression studies will make it possible to define more accurately the conditions for ABA production in axenic culture, an important prerequisite for biotechnological evaluation. In the same way that transformation of a G. fujikuroi wild-type strain with additional copies of the gibberellin gene cluster led to an up to threefold-enhanced gibberellin production (P. Linnemannstöns and B. Tudzynski, unpublished data), overexpression of bcaba1 (and other genes of the pathway) could be a helpful tool for the design of better ABA production strains.
We also demonstrated here the options provided by a “genomics” approach. Although only a part of the Botrytis genomic sequence is publicly available, a concerted approach with EST data, expression studies, and gene disruption was successful in identifying the first ABA biosynthesis gene. Conventional approaches, e.g., by differential cDNA screening had been unsuccessful. Thus, there is also a good chance for identification of genes involved in other phytohormone pathways in B. cinerea in the near future, especially for ethylene, which probably has significant impact on pathogenicity and might also affect other aspects of the fungal life cycle.
Acknowledgments
This study was supported by the Deutsche Forschungsgemeinschaft (Tu 50/9) and the European Community-Access to Research Infrastructure action of the Improving Human Potential Programme.
The technical support of Bayer Crop Science in establishing an EST library for strain ATCC 58025 is gratefully acknowledged. We thank Bettina Tudzynski and Peter Schreier (Bayer Crop Science) for helpful discussion, Brian Williamson (Dundee) for critical reading of the manuscript, Anke Bölting for excellent technical assistance, and E. Weiler (Bochum) for providing ABA-specific antisera and for help in establishment of the ABA-EIA.
REFERENCES
- 1.Assante, G., L. Merlini, and G. Nasini. 1977. (+)-Abscisic acid, a metabolite of the fungus Cercospora rosicola. Experimentia 33:1556-1557. [Google Scholar]
- 2.Audenaert, K., G. B. De Meyer, and M. M. Höfte. 2002. Abscisic acid determines basal susceptibility of tomato to Botrytis cinerea and suppresses salicylic acid-dependent signaling mechanisms. Plant Physiol. 128:491-501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown, D. W., J.-H. Yu, H. S. Kelkar, M. Fernandes, T. C. Nesbitt, N. P. Keller, T. H. Adams, and T. J. Leonard. 1996. Twenty-five co-regulated transcripts define a sterigmatocystin gene cluster in Aspergillus nidulans. Proc. Natl. Acad. Sci. USA 93:1418-1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cenis, J. L. 1993. Rapid extraction of fungal DNA for PCR amplification. Nucleic Acids Res. 20:2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chagué, V., Y. Elad, R. Barakat, P. Tudzynski, and A. Sharon. 2002. Ethylene biosynthesis in Botrytis cinerea. FEMS Microbiol. Ecol. 40:143-149. [DOI] [PubMed] [Google Scholar]
- 6.Chapman, D. J., and M. A. Regan. 1980. Evolution of a biochemical pathway: evidence from comparative biochemistry. Annu. Rev. Plant Physiol. 31:639-645. [Google Scholar]
- 7.Clarke, I. E., A. Delaconcha, F. J. Murillo, G. Sandmann, E. J. Skone, and P. M. Bramley. 1983. The effect of diphenylamine on carotenogenesis in Phycomyces blakesleeanus. Phytochemistry 22:435-439. [Google Scholar]
- 8.Crocoll, C., J. Kettner, and K. Dörffling. 1991. Abscisic acid in saprophytic and parasitic species of fungi. Phytochemistry 30:1059-1060. [Google Scholar]
- 9.Diez, B., S. Gutierrez, J. L. Barredo, P. van Solinger, L. H. M. van der Voort, and J. F. Martin. 1990. The cluster of penicillin biosynthetic genes. Identification and characterization of the pcbAB gene encoding the alpha-aminoadipyl-cysteinyl-valine synthetase and linkage to the pcbC and pcbDE genes. J. Biol. Chem. 265:16358-16365. [PubMed] [Google Scholar]
- 10.Dörffling, K., and W. Petersen. 1984. Abscisic acid in phytopathogenic fungi of the genera Botrytis, Ceratocystis, Fusarium, and Rhizoctonia. Z. Naturforsch. 39c:683-684. [Google Scholar]
- 11.Fraser, P. D., M. J. Ruiz-Hidalgo, M. A. Lopez-Matas, M. I. Alvarez, A. P. Eslava, and P. M. Bramley. 1996. Carotenoid biosynthesis in wild type and mutant strains of Mucor circinelloides. BBA Gen. Subjects 1289:203-208. [DOI] [PubMed] [Google Scholar]
- 12.Hedden, P., A. L. Phillips, M. C. Rojas, E. Carrera, and B. Tudzynski. 2002. Gibberellin biosynthesis in plants and fungi: a case of convergent evolution? J. Plant Growth Regul. 20:319-331. [DOI] [PubMed] [Google Scholar]
- 13.Hirai, N., M. Okamoto, and K. Koshimizu. 1986. The 1′,4′-trans-diol of abscisic acid, a possible precursor of abscisic acid in Botrytis cinerea. Phytochemistry 25:1865-1868. [Google Scholar]
- 14.Kettner, J. 1991. Untersuchungen zur Biosynthese von Abscisinsäure bei der Interaktion des pflanzenpathogenen Pilzes Botrytis cinerea Pers. mit der Kulturtomate Lycopersicon esculentum Mill. Ph.D. dissertation. Universität Hamburg, Hamburg, Germany.
- 15.Kettner, J., and K. Dörffling. 1995. Biosynthesis and metabolism of abscisic acid in tomato leaves infected with Botrytis cinerea. Planta 196:627-634. [Google Scholar]
- 16.Kunic, B., G. Truan, K. Breskvar, and D. Pompon. 2001. Functional cloning, based on azole resistance in Saccharomyces cerevisiae, and characterization of Rhizopus nigricans redox carriers that are differentially involved in the P450-dependent response to progesterone stress. Mol. Genet. Genomics 265:930-940. [DOI] [PubMed] [Google Scholar]
- 17.Malonek, S., M. Rojas, P. Hedden, P. Gaskin, and B. Tudzynski. The NADPH: cytochrome P450 reductase gene from Gibberella fujikuroi is essential for gibberellin biosynthesis. J. Biol. Chem., in press. [DOI] [PubMed]
- 18.Marumo, S., M. Katayama, E. Komori, Y. Ozaki, M. Natsume, and S. Kondo. 1982. Microbial production of abscisic acid by Botrytis cinerea. Agric. Biol. Chem. 46:1967-1968. [Google Scholar]
- 19.Milborrow, B. V. 2001. The pathway of biosynthesis of abscisic acid in vascular plants: a review of the present state of knowledge of ABA biosynthesis. J. Exp. Bot. 52:1145-1164. [PubMed] [Google Scholar]
- 20.Neill, S. J., R. Horgan, D. C. Walton, and C. A. M. Mercer. 1987. The metabolism of α-ionylidene compounds by Cercospora rosicola. Phytochemistry 26:2515-2519. [Google Scholar]
- 21.Okamoto, M., N. Hirai, and K. Koshimizu. 1988. Biosynthesis of abscisic acid. Memoirs College Agriculture Kyoto Univ. 132:79-115. [Google Scholar]
- 22.Okamoto, M., N. Hirai, and K. Koshimizu. 1988. Biosynthesis of abscisic acid from α-ionylideneethanol in Cercospora pini-densiflorae. Phytochemistry 27:3465-3469. [Google Scholar]
- 23.Oritani, T., and H. Kiyota. 2003. Biosynthesis and metabolism of abscisic acid and related compounds. Nat. Prod. Rep. 20:414-425. [DOI] [PubMed] [Google Scholar]
- 24.Pontecorvo, G. V., J. A. Poper, L. M. Hemmonns, K. D. Mac Donald, and A. W. J. Buften. 1953. The genetics of Aspergillus nidulans. Adv. Genet. 141:141-238. [DOI] [PubMed] [Google Scholar]
- 25.Quidde, T., P. Büttner, and P. Tudzynski, P. 1999. Evidence for three different specific saponin-detoxifying activities in Botrytis cinerea and cloning and functional analysis of a gene coding for a putative avenacinase. Eur. J. Plant Pathol. 105:273-283. [Google Scholar]
- 26.Reshetilova, T. A., T. F. Soloveva, B. P. Baskunov, and A. G. Kozlovskii. 1992. Investigation of alkaloid formation by certain species of fungi of the Penicillium genus. Microbiology 61:608-613. [Google Scholar]
- 27.Rolke, Y., S. Liu, T. Quidde, B. Williamson, A. Schouten, K.-M. Weltring, V. Siewers, K. B. Tenberge, B. Tudzynski, and P. Tudzynski. 2004. Functional analysis of H2O2-generating systems in Botrytis cinerea: the major Cu-Zn-superoxide dismutase (BCSOD1) contributes to virulence on French bean, whereas a glucose oxidase (BCGOD1) is dispensable. Mol. Plant Pathol. 5:17-27. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 29.Shaul, O., Y. Elad, and N. Zieslin. 1996. Suppression of Botrytis blight in cut rose flowers with gibberellic acid. Effects of exogenous application of abscisic acid and paclobutrazol. Postharvest Biol. Technol. 7:145-150. [Google Scholar]
- 30.Smedsgaard, J. 1996. Micro-scale extraction procedure for standardized screening of fungal metabolite production in cultures. J. Chromatogr. 760:264-270. [DOI] [PubMed] [Google Scholar]
- 31.Sprecher, E. 1959. Über die Guttation bei Pilzen. Planta 53:565-574. [Google Scholar]
- 32.ten Have, A., W. Mulder, J. Visser, and J. A. L. van Kan. 1998. The endopolygalacturonase gene Bcpg1 is required for full virulence of Botrytis cinerea. Mol. Plant-Microbe Interact. 11:1009-1016. [DOI] [PubMed] [Google Scholar]
- 33.Truan, G., J. C. Epinat, C. Rougeulle, C. Cullin, and D. Pompon. 1994. Cloning and characterization of a yeast cytochrome b(5)-encoding gene which suppresses ketoconazole hypersensitivity in a NADPH-P-450 reductase-deficient strain. Gene 142:123-127. [DOI] [PubMed] [Google Scholar]
- 34.Tudzynski, B., and K. Hölter. 1998. The gibberellin biosynthetic pathway in Gibberella fujikuroi: evidence for a gene cluster. Fungal Genet. Biol. 25:157-170. [DOI] [PubMed] [Google Scholar]
- 35.Tudzynski, B., and A. Sharon. 2002. Biosynthesis, biological role, and application of fungal hormones, p. 183-211. In H. D. Osiewacz (ed.), The Mycota X: industrial applications. Springer-Verlag, Berlin, Germany.
- 36.Tudzynski, P. 1999. Genetics of Claviceps purpurea, p. 79-93. In V. Kren and L. Cvak (ed.), Medicinal and aromatic plant-industrial profiles, vol. 6. Ergot, the genus Claviceps. Harwood Academic Publishers, Amsterdam, The Netherlands. [Google Scholar]
- 37.van den Brink, H. J. M., C. M. J. van Zeijl, J. F. Brons, and C. A. M. J. J. van den Hondel. 1995. Cloning and characterization of the NADPH-cytochrome P450 oxidoreductase gene from the filamentous fungus Aspergillus niger. DNA Cell Biol. 14:719-729. [DOI] [PubMed] [Google Scholar]
- 38.van den Brink, H. J. M., R. F. M. van Gorcom, C. A. M. J. J. van den Hondel, and P. J. Punt. 1998. Cytochrome P450 enzyme systems in fungi. Fungal Genet. Biol. 23:1-17. [DOI] [PubMed] [Google Scholar]
- 39.Weiler, E. W. 1982. An enzyme-immunoassay for cis-(+)-abscisic acid. Physiologia Plantarum 54:510-514. [Google Scholar]
- 40.Yamamoto, H., M. Inomata, S. Tsuchiya, M. Nakamura, T. Uchiyama, and T. Oritani. 2000. Early biosynthetic pathway to abscisic acid in Cercospora cruenta. Biosci. Biotechnol. Biochem. 64:2075-2082. [DOI] [PubMed] [Google Scholar]
- 41.Yanish-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strain: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 42.Yoder, O. C., and B. G. Turgeon. 2001. Fungal genomics and pathogenicity. Curr. Opin. Plant Biol. 4:315-321. [DOI] [PubMed] [Google Scholar]