Abstract
Background and Aims
Cholangiocytes are the target of a heterogeneous group of liver diseases, known as the cholangiopathies. An evolving understanding of the mechanisms driving biliary development provides the theoretical underpinnings for rational development of induced pluripotent stem cell (iPSC)-derived cholangiocytes (iDCs). Therefore, the aims of this study were to develop an approach to generate iDCs and to fully characterize the cells in vitro and in vivo.
Methods
Human iPSC lines were generated by forced expression of the Yamanaka pluripotency factors. We then pursued a step-wise differentiation strategy toward iDCs using precise temporal exposure to key biliary morphogens and we characterized the cells using a variety of morphologic, molecular, cell biologic, functional, and in vivo approaches.
Results
Morphology shows a stepwise phenotypic change toward an epithelial monolayer. Molecular analysis during differentiation shows appropriate enrichment in markers of iPSC, definitive endoderm, hepatic specification, hepatic progenitors, and ultimately cholangiocytes. Immunostaining, Western blotting, and flow cytometry demonstrate enrichment of multiple functionally relevant biliary proteins. RNA sequencing reveals that the transcriptome moves progressively toward that of human cholangiocytes. iDCs generate intracellular calcium signaling in response to ATP, form intact primary cilia, and self-assemble into duct-like structures in 3-dimensional culture. In vivo, the cells engraft within mouse liver following retrograde intra-biliary infusion.
Conclusions
In summary, we have developed a novel approach to generate mature cholangiocytes from iPSCs. In addition to providing a model of biliary differentiation, iDCs represent a platform for in vitro disease modelling, pharmacologic testing, and individualized, cell-based, regenerative therapies for the cholangiopathies.
Biliary diseases continue to be the cause of significant morbidity and mortality, in both children and adults(1). Cholangiocytes, the specialized epithelial cells lining the intra- and extra-hepatic bile ducts, are the target of a heterogeneous group of disorders known as the cholangiopathies(2). The obliterative cholangiopathies are a subset of these disorders that have, as a hallmark, progressive cholangiocyte destruction, culminating in ductopenia and cholestasis. Cholangiocytes also suffer damage during and after liver transplantation in the form of preservation injury, cellular rejection, disease recurrence, and ischemic cholangiopathy(3, 4). Most etiologies of the obliterative cholangiopathies result in progressive biliary fibrosis culminating in end-stage liver disease that is essentially untreatable without liver transplantation. However, an inadequate supply of donor organs limits the effectiveness of this surgical approach. Given the targeted cellular destruction typical of the cholangiopathies, these diverse disorders may be amenable to cell replacement strategies in these varying circumstances. Therefore, the biliary system is an attractive target for cell-based regenerative medicine approaches to study and potentially treat the disorders.
While the liver has remarkable intrinsic regenerative properties, this mechanism is impaired in the setting of chronic liver disease(5). Explosive growth in the field of liver regenerative medicine, including hepatic differentiation of induced pluripotent stem cells (iPSC), has the potential to provide a new platform for the study and treatment of liver disorders that could ultimately transform the care of patients with end-stage liver disease(6). The newly discovered ability of the Yamanaka factors to reprogram somatic cells to pluripotency has revealed remarkable cellular plasticity and indeed, it is now possible to generate iPSCs from virtually any tissue in the human body and to recapitulate developmental biology in vitro to generate diverse cellular phenotypes(7). Based on emerging details governing developmental biology of the liver(8), a number of groups have developed various methods for generating hepatocyte-like cells (HLCs) from iPSCs via stepwise differentiation strategies(9–19) or by direct differentiation from fibroblasts(20, 21). While some of these protocols described biliary elements, pluripotent stem cell-derived cholangiocytes had not been directly nor extensively studied until very recently when cholangiocytes were developed from embryonic stem cells and bipotent HepaRG cells(22), an approach that was also effective in iPSCs. Simultaneously, our group and others have begun to develop additional targeted approaches to create iPSC-derived cholangiocytes (iDCs). New understanding of the mechanisms driving biliary development(23–26) and cellular plasticity during liver regeneration / repair(27, 28) have provided the theoretical underpinnings for the rational development and use of iDCs as individualized disease models and potentially as regenerative therapeutics for biliary disease(29). Furthermore, this direction is conceptually appealing given the clinical access to the biliary tree in humans afforded by endoscopic retrograde cholangiopancreatography (ERCP), a technique readily available at every major academic medical center in the world.
Since hepatocytes and cholangiocytes share common precursors and since biliary differentiation pathways are now being more fully elucidated, we reasoned that targeted modifications to existing differentiation strategies should allow for generation of iDCs. This study provides technical and conceptual innovations by demonstrating that human myofibroblast-derived iPSCs can be reproducibly differentiated toward an adult bile duct epithelial fate, expressing numerous markers of functionally mature cholangiocytes. RNA sequencing at each phase of differentiation followed by principal component analysis and differential expression analysis confirms that the transcriptome is gradually altered from iPSC toward that of human cholangiocytes. In addition, the transcriptional profiles during the iPSC to iDC transition appear to recapitulate several aspects of biliary development. We go on to demonstrate that iDCs form primary cilia on their apical surface, have intact calcium signaling, and form duct-like structures in 3-dimensional (3D) culture. Furthermore, we show for the first time, that stem cell-derived cholangiocytes can engraft within the liver in vivo after retrograde intra-biliary infusion in a mouse model. The experimental results from this study provide new information regarding the feasibility iPSC-based differentiation strategies targeting cholangiocytes and produce the foundation for a detailed dissection of the mechanisms governing biliary differentiation. Furthermore, mature iDCs derived from patient-specific iPSCs could potentially be a limitless source of high-quality, individualized cells that could be: 1) studied in vitro as patient-specific models of biliary disease; 2) treated in vitro to test putative therapeutic compounds; 3) genetically modified to correct underlying disease-causing defects; and 4) transplanted (without the need for immunosuppression) as individualized, cell-based, regenerative therapeutics for the cholangiopathies.
Materials and Methods
Cholangiocyte culture
iDC, H69, and NHC were cultured in H69 media (DMEM/F12 supplemented with 10% fetal bovine serum, 1% penicillin/streptomycin, adenine, insulin, epinephrine, T3-T, hydrocortisone and epidermal growth factor).
Reprogramming / iPSC culture
Human myofibroblasts were cultured from biopsy specimens with appropriate IRB oversight and informed consent. These cells were induced to pluripotency by transient forced expression of OCT4, SOX2, KLF4, and c-MYC, using the Sendai system, as previously described (30). iPSC were seeded in culture plates pre-coated with 0.1% Matrigel (R&D systems) and cultured in nutristem XF/FF culture medium (Stemgent, Inc.) supplemented with 16% mTeSR1 basal medium (STEMCELL Technologies), mTeSR1 5X Supplement, 50X StemGS (ScienCell Research Laboratories) and 1% penicillin/streptomycin.
Generation of iDC
The step-wise differentiation toward iDCs was accomplished by a process of temporal exposure to biliary morphogens (Figure 1A). iPSC were induced to definitive endoderm (DE) for 4 days using Advanced RPMI medium changed and supplemented daily with addition of 50 ng/ml Activin A and 50 ng/ml Wnt3A in the presence of Matrigel. DE cells were treated daily with 10 ng/ml fibroblast growth factor 2 (FGF2), 20 ng/ml bone morphogenic protein 4 (BMP4) and 100μg/ml sonic hedgehog (SHH) for 4 days, for hepatic specification (HS). HS cells were induced to hepatic progenitor (HP) cells for 4 days with daily exposure to 50 μg/ml SHH and 100 μg/ml recombinant Jagged-1. To generate iDC, HP cells were treated for 4 days with H69 media changed and supplemented daily with 100 μg/ml transforming growth factor beta (TGF-β) and replacement of Matrigel with collagen. All growth factors were purchased from R&D Systems (Supplemental Table 1).
Figure 1. Protocol for generation of iDC from iPSC and morphologic analysis.
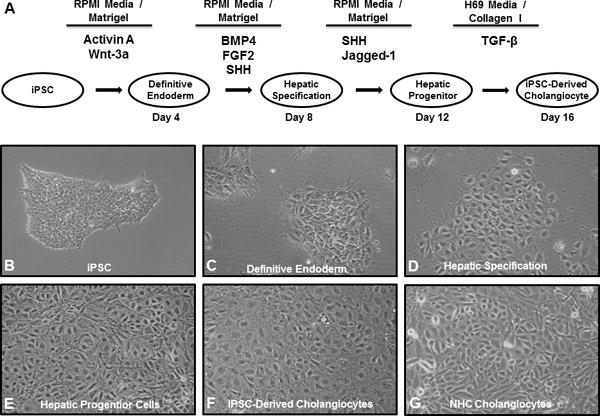
(A) The protocol proceeds through phases of DE, HS, and HP and incorporates defined culture media, precise extracellular components, and temporal exposure to key biliary morphogens. Light microscopy (10X) shows phenotypic changes at each stage of differentiation with the transformation of (B) the stem cell phenotype towards (C–F) an epithelial monolayer, similar to the (G) NHC cholangiocyte cell line.
Quantitative RT-PCR
Total RNA was extracted from human iPSC, DE, HS, HP and iDC cells using the RNeasy Plus Mini Kit (Qiagen). Reverse transcription was performed with 5 μg RNA using oligo (dT) primer and SuperScript III. Real-time PCR was performed in a volume of 25 μl using Sybr Green Master Mix and the 7500 Real-Time PCR System (Applied Biosystems). Primer sequences are shown in Supplemental Table 2.
Western blotting
Protein isolation was done with lysis buffer. Concentration was determined using a Spectrophotometer. 25 μg of protein was loaded onto 4–12% Tris-Glycine gels, electrophoresed, and transferred onto nitrocellulose membranes (Scientific Laboratory Supplies) for blotting. The membrane was blocked, incubated with primary antibodies, rinsed with TBST, and incubated with secondary antibodies (Supplemental Table 3). After washing, the membrane was exposed to luminol (Santa Cruz Biotechnology) and developed.
Immunofluorescence microscopy
Cells were seeded on glass coverslips coated with Matrigel. For observation of primary cilia, some cells were grown in monolayer for ~7 days followed by serum starvation to induce cilia. After fixation with paraformaldehyde and permeabilization with Triton X-100, cells were blocked with FBS, incubated with primary antibodies and then with fluorescence-conjugated secondary antibodies. Cells were washed and nuclei were stained with 4′6-diamidino-2-phenylindole dihydrochloride (DAPI, Sigma). Coverslips were mounted onto glass slides and observed under confocal microscopy (Carl Zeiss, Jena, Germany). Images were acquired using Zeiss LSM software.
Flow Cytometry
Fluorescence-activated cell sorting (FACS) was performed in the Mayo Flow Cytometry Laboratory, as previously described (31). Briefly, 1 million cells were treated with a Trypsin digestion solution and fixed in 4% paraformaldehyde for 20 minutes followed by 3 washes in FACS buffer (2% FBS + DPBS). Blocking was performed in 0.5% BSA in FACS buffer for 30 minutes at room temperature. The cell suspension was centrifuged at 300g for 5 minutes and the supernatant was removed by aspiration. Cells were incubated with primary antibody on ice for 30 minutes, washed three times, stained with fluorochrome-conjugated secondary antibodies, in the dark, on ice, for 60 minutes and washed another three times. Cells were analyzed on a FACSCalibur analytical flow cytometer in 3 independent sessions. Data files were analyzed using Cellquest software.
RNA sequencing
RNA sequencing was conducted in collaboration with the Mayo Medical Genomics Facility. From each phase of differentiation (iPSC, DE, HS, HP and iDC), from cholangiocyte cell lines (H69 and NHC), and from isolated human cholangiocytes (iHCs), total RNA was isolated. Quality of the RNA was assessed by the Mayo Gene Expression Core using Agilent Bioanalyzers. All samples had RNA Integrity Numbers greater than 7.0. RNA sequencing was performed as paired-end base reads on an Illumina HiSeq 2000 with three samples per lane, using the TruSeq SBS Sequencing Kit, Version 3. Base calling was performed using Illumina’s RTA version 1.12.4.2. Bioinformatics were performed with the assistance of the Mayo Division of Biostatistics and Informatics. Analysis of each sample (alignment statistics, in-depth quality control metrics, and gene and exon expression levels) was done using Mayo Clinic’s MAPRSeq v1.2(32). Reads were mapped using Tophat version 2.0.6(33) against the hg19 reference genome and gene counts were produced using htseq. Subsequent principal component analysis was performed using Partek® Genomics Suite™ version 6.6 (Partek Inc., St. Louis, MO). Differential expression analyses between samples were computed using an edgeR version 3.3.8 algorithm(34).
3D culture
Cells were suspended in a mixture of 1x DMEM, NaOH (10nM), type I collagen (2.0mg/ml), and 33% Matrigel. 1×104 cells were added to glass bottom culture dishes. The mixture solidified and was overlaid with media. Plates were cultured for 7 days and assessed by light and immunofluorescence microscopy.
Intracellular Ca2+-signaling
iDC and NHC cells were plated H69 media. After 24 hours, cells were serum-starved for 2 hours and incubated with Fluo-4AM (Invitrogen) in Hank’s Balanced Salt Solution supplemented with 20 mM HEPES and 2.5 mM Probenecid for 60 minutes. A FlexStation III (Molecular Devices) automated plate reader was used to measure calcium flux. After establishing baseline fluorescence for 30 seconds, ATP (ACROS Organics) or vehicle (10 μL) were added and fluorescence was further read for 2 minutes.
Electron microscopy
Scanning electron microscopy was performed, as previously described(35). iDC were grown in a monolayer for ~ 7 days followed by 24 hours of serum starvation, fixed in glutaraldehyde, post-fixed in osmium tetroxide, dehydrated, critical-point dried, sputter coated, and imaged with a scanning electron microscope (Hitachi) at 40,000x.
Retrograde intrabiliary infusion
Animals were anesthetized using a combination of Ketamine and xylazine. Using a dissecting microscope, the common bile duct was temporarily occluded (for a total of 2 hours) with a vascular clamp. The gallbladder was dissected from the liver (Supplemental Figure 1A) and two silk sutures were placed around the cystic duct (Supplemental Figure 1B). The gallbladder and cystic duct were cannulated with a 24-guage, ¾” angiocatheter (Supplemental Figure 1C) and sutured in place (Supplemental Figure 1D). 106cells were injected at a flow rate of 0.2 ml/min (Supplemental Figure 1E). Some cells were labelled with green fluorescent protein (GFP) using lentiviral transduction and selected with two weeks of exposure to puromycin. After infusion, a silk tie was placed between the gallbladder and the cystic duct and a cholecystectomy was completed. At 2 hours after clamping, the common bile duct was unclamped and the animals were closed using interrupted chromic gut sutures. At day 3 after cell transplantation, liver was collected, processed with parafin embedding and sectioning, and assessed for cell engraftment by immunostaining for the human-specific class 1 major histocompatibility complex (MHC1) and cytokeratin 7 (CK7), by confocal microscopy for GFP, and by immunofluorescence for CK7 and MHC1.
Statistical analysis
Data are presented as the mean ± standard error. Data represents typical experiments reproduced at least three times. Analysis was performed using Graph Stat Prizm software (GraphPad Software, Inc., La Jolla, CA). Statistical analyses used one-way analysis of variance (ANOVA) with a Tukey post-test.
Results
Generation of iDC
Human myofibroblasts were induced to pluripotency by the transient expression of the transcription factors (OCT4, SOX2, KLF4, and c-MYC) using the Sendai system. The iPSC had typical morphologic features of iPSC including discrete smooth-edged colonies and high nuclear-to-cytoplasmic ratio (Figure 1B). They also stained positive for numerous markers of pluripotency including TRA-1-60, OCT4, SOX2, NANOG, and alkaline phosphatase (Supplemental Figure 2A) and have gene expression profiles distinct form the parental fibroblasts but highly similar to a lentiviral-reprogrammed iPSC clone from keratinocytes and to a human embryonic stem cell line (Supplemental Figure 2B). Since hepatocytes and cholangiocytes share a common precursor, we postulated that subtle, but targeted, alterations to existing hepatocyte differentiation protocols should allow us to develop iDCs. We noticed that prior reports that had included SHH(13), Notch signaling(36), or TGF-β(37) had also reported some resulting biliary elements and thus these morphogens were included in our protocol. In addition, based on increasing fundamental knowledge of developmental biology of the biliary system(25) as well as liver regeneration(26), we further refined the novel stepwise protocol for the in vitro differentiation of iPSCs to iDCs (Figure 1A). We also avoided the hypoxia present in many hepatocyte differentiation protocols since cholangiocytes are sensitive to hypoxia (38). The differentiation protocol was performed in defined media (Advanced RPMI) with feeder-free culture conditions. Initially, iPSC on Matrigel were treated daily with Activin A and Wnt3a in RPMI for four days for induction of DE. DE cells were exposed daily to FGF2, BMP4 and to SHH for four days for hepatic specification. HP were induced via further daily exposure to SHH and Jagged-1 for four days. For the final stage of cholangiocyte maturation, the cells were exposed daily to TGF-β, RPMI media was replaced with H69 media (a hormonally-enriched media used to culture adult cholangiocytes), and Matrigel was replaced with collagen for the final four days.
iDC have biliary morphology
Morphologic analysis of the cells showed a step-wise phenotypic change toward an epithelial monolayer, consistent with biliary epithelium (Figure 1B–F). At the DE stage, the cells began to lose the characteristic features of iPSC (discrete smooth-edged colonies with high nuclear-to-cytoplasmic ratio). In the ensuing steps, the cells lost their ability to form clusters and began to grow in an epithelioid monolayer with features reminiscent of cultured cholangiocytes and similar to existing cholangiocyte cell lines, such as NHC (Figure 1G). iDCs typically survive in culture between 3 to 10 passages and can also be frozen in liquid nitrogen for storage and then thawed and recovered for subsequent analyses.
Molecular analysis of the iPSC to iDC transition
Quantitative real-time PCR analysis at the various stages of differentiation revealed appropriate enrichment of markers at each respective stage of differentiation (Figure 2). iPSC show enrichment in the pluripotency markers, SOX2 and NANOG (Figure 2A) that are then extinguished in the ensuing phases. However, iPSC lack significant expression of GATA4 and CXCR4, markers of the early endoderm lineage (Figure 2B), arguing against any spontaneous differentiation. In contrast, cells from the DE stage were highly enriched with these two markers. Markers of fetal liver and hepatic development such as HES1 and SALL4, were appropriately enriched in the hepatic specification phase (Figure 2C). Cells from the hepatic progenitor phase exhibited strong expression of hepatoblast genes, such as AFP and Prox-1 (Figure 2D). Notably, iDC lacked significant expression of these early hepatic markers, suggesting that these genes were down regulated during cholangiocyte maturation, something that has been challenging to achieve in hepatocyte differentiation protocols.
Figure 2. Molecular analysis.
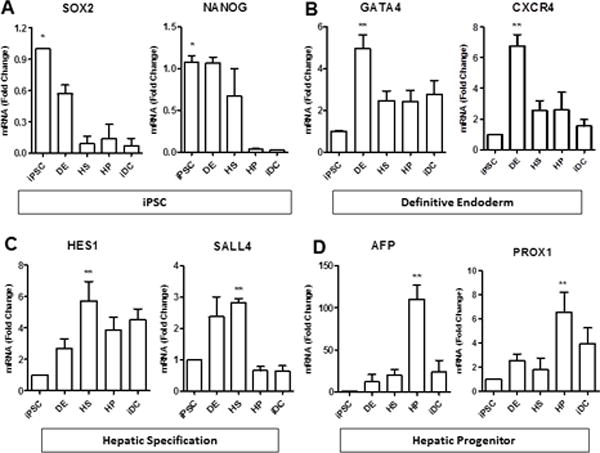
Real-time PCR showing expression of (A) pluripotency markers (Sox2 and NANOG) in iPSCs, followed by (B) expression of definitive endoderm markers (GATA4 and CXCR4), (C) hepatic specification markers (Hes1 and Sall4), and (D) hepatic progenitor markers (AFP and PROX1). *p≤0.05 compared to iDC, **p≤0.05 compared to iPSC. All data is normalized to GAPDH and iPSC expression is set to 1.0.
iDCs are enriched in biliary markers
Following the last step of differentiation, iDC showed high expression of multiple cholangiocyte markers including CK19, CK7, CFTR, PKD2, and AE2, by quantitative RT-PCR (Figure 3A). Since there is considerable variability within and among the existing cholangiocyte cell lines in terms of their expression of genes thought to be prototypical of cholangiocytes (Supplemental Figure 3A), there is no gold-standard control that expresses all of the cholangiocyte markers that are present in our iDCs. A significant increase in cholangiocyte markers begins to become apparent in the hepatic specification and hepatic progenitor phases, where SHH and Jagged-1 ligands, key biliary morphogens, were introduced to the protocol, suggesting that early priming events may already be initiating biliary programming at these phases. Importantly, iDC lack significant expression of pluripotency markers such as SOX2 and NANOG (Figure 2A), progenitor markers such as AFP, PROX-1 (Figure 2D), EpCAM and Tbx3 (Supplemental Figure 3B), as well as hepatocyte markers, such as ABCB4, albumin, α-1-antityrpsin, and tryptophan 2,3-dioxygenase (Figure 3B), indicating efficient differentiation and a relatively pure biliary fate. In fact, as long as Sonic Hedgehog and Notch ligands are present in the protocol, we do not detect hepatocyte differentiation, even in the presence of hepatocyte morphogens, such as hepatocyte growth factor, dexamethasone, and oncostatin.
Figure 3. Cholangiocyte gene expression.
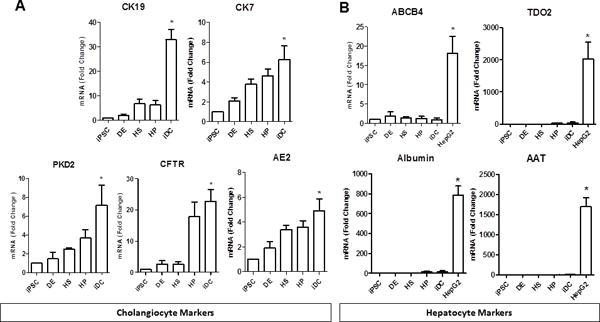
Real-time PCR shows that (A) iDCs are enriched in the cholangiocyte markers CK19, CK7, CFTR, PKD2, and AE2. iDCs lack expression of (B) hepatocyte markers, ABCB4, albumin, α-1-antitrypsin (A1AT), and tryptophan 2,3-dioxygenase (TDO2), *p≤0.05 compared to iPSC. All data is normalized to GAPDH and iPSC expression is set to 1.0.
iDCs are enriched in cholangiocyte proteins
Western blotting demonstrates significant and progressive enrichment of multiple, functionally relevant biliary proteins in iDCs (Figure 4A) including acetylated α-tubulin (a marker of primary cilia), AQP1 (a water channel), ASBT (a bile acid transporter), and SSTR-2 (a somatostatin receptor), along with CK7 and CK19 (the biliary cytokeratins). Notably, the iDC cells lack expression of the hepatocyte protein c-Met (the hepatocyte growth factor receptor). To demonstrate the homogeneity of cells following biliary differentiation, iDC were stained for CK7, which demonstrated robust and uniform staining. CK19 showed an appropriate intracellular and reticular pattern (Figure 4B), again suggesting efficient differentiation. To assess the overall enrichment of cholangiocyte proteins in iDCs, we performed flow cytometry for typical cholangiocyte markers. Flow cytometry analyses reveals that iDC cells were highly enriched in several cholangiocyte markers (Figure 4C) including CK7 (66.91%, compared to 73.25% in NHC and 2.04% in HepG2), CFTR (77.34%, compared to 22.29% in NHC and 10.47% in HepG2), and CK19 (85.3%, compared to 46.2% in NHC and 9.22% in HepG2). iDC cells also lack expression of EpCAM (2.03%, compared to 2.25% in NHC and 0.45% in HepG2).
Figure 4. Expression of cholangiocyte proteins.
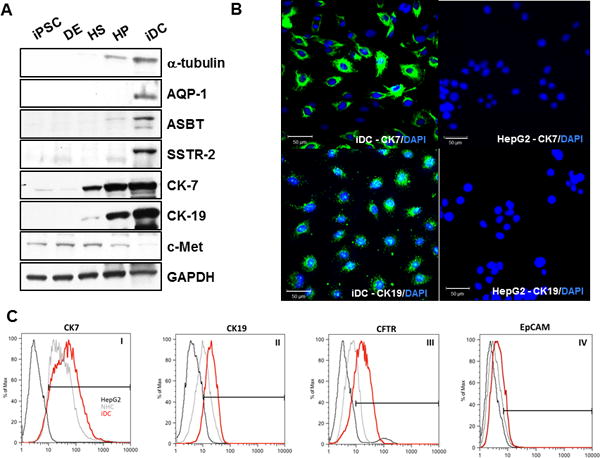
(A) Western blotting shows enrichment of multiple, functionally relevant biliary proteins in iDCs including acetylated α-tubulin, AQP-1, ASBT, SSTR-2, CK-7, and CK-19, while c-Met was not significantly detected. (B) iDCs were stained for CK7 and CK19 by immunofluorescence (left panels). HepG2 hepatocytes showed no staining for either marker (right panels). (C) Flow cytometry analyses of choalngiocyte markers, including CK7, CK19 and CFTR (panels I, II and III, respectively) and hepatic endoderm marker EpCAM (panel IV). Scale bars = 50 μm
RNA sequencing
In order to investigate the transcriptional changes during the iPSC to iDC transition, we performed an unbiased, genome-wide analysis of transcription using next generation sequencing. RNA sequencing was performed on three biological replicates from each of the five phases of differentiation (iPSC, DE, HS, HP, and iDC) as well as cholangiocyte cell lines (H69 and NHC), and iHCs (isolated from human surgical resection specimens). Principal component analysis shows a smooth transition in global transcriptional changes from iPSC (red) to iDC (gold) with a trajectory toward the transcriptional profiles of the human cholangiocytes (Figure 5A). The iDC cells remain in close proximity to the HP phase on the PCA plot since many progenitor markers are known to remain positive in cholangiocytes. However, they have also further differentiated from the progenitor phase toward cholangiocytes. Interestingly, iDCs appear to have an intermediate molecular phenotype between the freshly isolated human cholangiocytes and the cholangiocyte cell lines, which have been in long-term cell culture. Differential expression analysis confirms the similarities between iDCs and cholangiocyte cell lines. A Venn diagram (Figure 5B) showing similarly regulated genes (i.e. upregulated, unchanged, or down regulated, relative to iPSC) demonstrates that among the 17,725 genes analyzed, 11,031 are similarly regulated compared to H69 cells (62.2%) and 11,506 are similarly regulated compared to NHCs (65.0%). 9155 genes are similarly regulated relative to both groups (21.3%). This later group of genes (i.e. the center of the Venn Diagram) may represent a cholangiocyte signature of critical gene regulation that occurs during the iPSC to cholangiocyte transition. We also observed that as cells progress through differentiation, they become progressively less similar to iPSC, since the number of differentially expressed genes, relative to iPSC, increases (DE:1898; HS:3017; HP:6256; iDC:7941), as do the magnitude of the changes seen on Volcano plots (Supplemental Figure 3). Furthermore, the number (iDC:7941; H69:9724; NHC:9295) and magnitude (Supplemental Figure 3) of differentially expressed genes is ultimately similar to both cholangiocyte cell lines. A supervised interrogation of the expression profiles at each phase of differentiation reveals a heat map showing appropriate enrichment in prototypical developmental genes at each phase (Figure 5C). Genes associated with pluripotency are progressively down regulated (green) while characteristic cholangiocyte genes are increasingly activated (red). Overall, these data suggest that the transcriptome of iDCs has gradually diverged from that of iPSC and has become progressively more cholangiocyte-like and that our protocol may represent a legitimate in vitro model of biliary differentiation.
Figure 5. RNA sequencing.
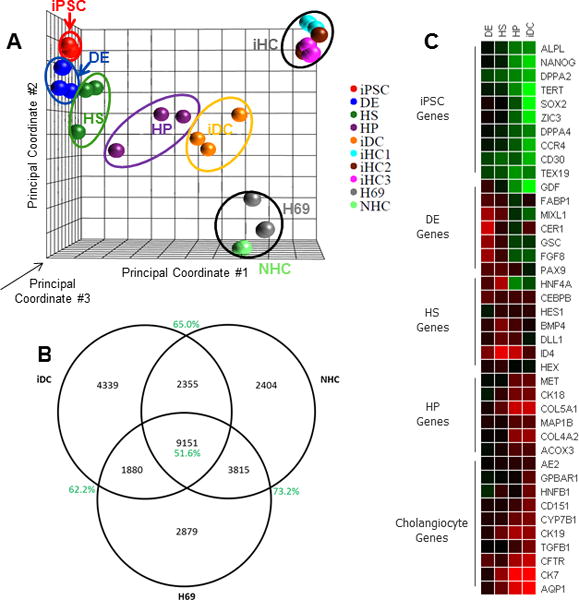
Sequencing of the entire transcriptome was performed at each phase of differentiation and compared to the transpcriptome of cholangiocyte cell lines and isolated human choloangiocytes. (A) Principal component analysis provided a 3-dimensional graphical representation of the gene expression clustering between groups based on three mathematically-defined principal components. Triplicate samples are contained within the ovals. We see a smooth transition from iPSC (red) to iDC (gold) with a trajectory toward the H69 (grey), NHC (green), and iHC (light blue, brown, and purple) cholangiocytes. (B) Differential expression analysis, performed relative to iPSC, displayed as a Venn diagram, confirms similarly regulated genes between iDCs, H69 cells, and NHCs. (C) A heatmap showing fold-change relative to iPSC demonstrates appropriate enrichment in developmental genes as differentiation progresses.
iDCs form primary cilia and have intact calcium signaling
In order to assess the functionality of iDCs, we first assessed their ability to form primary cilia. Primary cilia are important functional organelles known to be present on the apical membrane of cholangiocytes, but not on hepatocytes. Primary cilia act as osmoreceptors and mechanoreceptors that can detect and transmit multiple signals influencing cholangiocyte function. iDC cells cultured at confluence for 7 days and serum starved for 24 hours (conditions known to promote cilia formation in cholangiocytes) were able to form primary cilia as demonstrated by immunofluorescence for acetylated α-tubulin (Figure 6A). At the ultrastructural level, scanning electron microscopy also revealed the presence of mature primary cilia (Figure 6B). Since calcium signaling can be activated in cholangiocytes by purinergic receptors on primary cilia (39), we measured calcium signaling in response to ATP, as measured by fluorometric calcium assays. These demonstrated rapid and robust responses to the agonist, similar to a cholangiocyte cell line (Figure 6C) and consistent with other reports of stem cell-derived cholangiocytes(22).
Figure 6. Functional characteristics of iDCs.
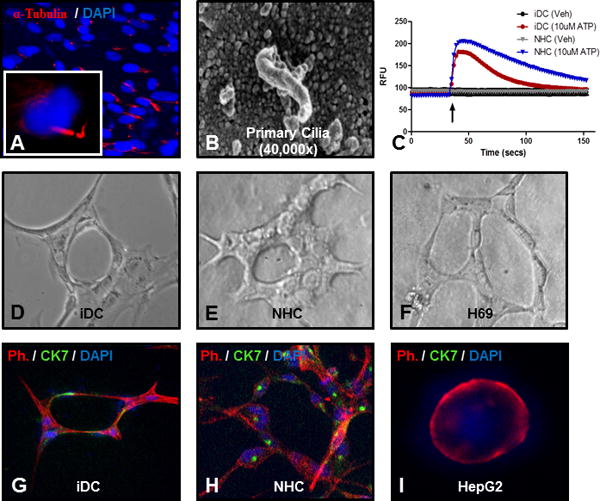
iDCs were able to form primary cilia as shown by (A) immunofluorescence for acetylated α-tubulin (red) and DAPI (blue) or by (B) scanning electron microscopy. (C) iDCs activate intracellular calcium signaling in response to ATP. (D) iDCs form round duct-like structures with a luminal space when cultured for 7 days in a 3D type I collagen / Matrigel culture system, similar to (E) NMC cells (F) H69 cells. (G) Co-staining with phalloidin (red), CK7 (green) and DAPI (blue) confirms CK7 positivity in the duct-like structures, similar to (H) NHC cells, but different from (I) HepG2 hepatocytes.
iDC form duct-like structures in 3D culture
When iDC were cultured for 7 days in a 3D collagen/Matrigel culture system, they were able to form round duct-like structures (Figure 6D) with a branching morphology and a central cavity, features that are typical of other cholangiocyte cells in culture (Figure 6E–F). Co-staining for phalloidin (to show cellular structure) and CK7 shows that the cells retain cholangiocyte markers in this configuration (Figure 6G), similar to NHC cells (Figure 6H), but distinct from HepG2 hepatocytes (Figure 6I). Together, the above data suggest that our differentiation method can be used to generate functional adult cholangiocytes from human iPSC.
iDCs engraft within mouse liver in vivo
We next sought to evaluate the functionality of iDC in vivo, by assessing their engraftment potential and behavior after delivery to the biliary system in a mouse model. We developed a microsurgical technique for retrograde intrabiliary infusion in the mouse as a means to deliver the cells from our last step of differentiation (Figure 7A). In this model, after clamping of the distal common bile duct, the gallbladder was dissected free from the liver and two sutures are placed around the cystic duct. Around 106 cells (some labelled with GFP) were then administered via a small angiocatheter followed by a completion of a cholecystectomy. Three days after cell transplantation, liver was collected and assessed for cell engraftment and morphology by immunohistochemical staining for human MHC1 (Figure 7B–C) and CK7 (Figure 7D), by dual label immunofluorescence microscopy for MHC1 and CK7 (Figure 7E), and by confocal microscopy for GFP expression (Figure 7F). The human iDCs engrafted within immune competent murine liver in vivo within existing portal tracts (Figure 7A) and were able to form peri-portal de novo duct-like structures (Figure 7C). Evidence of acute cellular rejection confimed that the cells were able to engraft and provoke an immune response, as expected. The cells maintain CK7 positivity in this configuration, similar to the native mouse bile ducts (Figure 7D), but can be definitvely distinguished from the native bile ducts using dual-label immunofluorescence which confirms co-localization of MHC1 and CK7 (Figure 7E). Developing cellular clusters without a fully-mature central lumen were also observed (Supplemental Figure 4). Furthermore, GFP-labelled iDCs can be followed using confocal microscopy (Figure 7F), which also demonstrated engraftment within and among existing bile ducts.
Figure 7. Retrograde intrabiliary infusion.
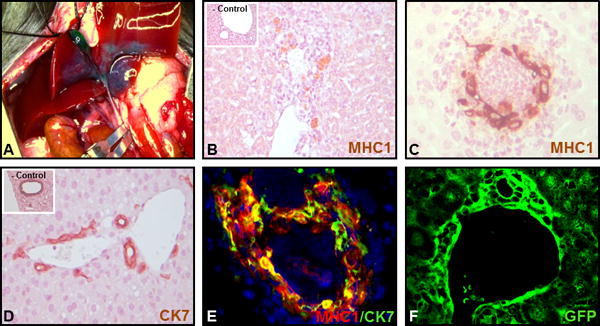
(A) Retrograde infusion of iDCs was accomplished by microsurgical cannulation of the cystic duct with temporary occlusion of the common bile duct. Engraftment was analyzed by (B–C) staining for human class 1 MHC (inset shows a cell-free control injection) or (D) CK7 (inset shows a cell-free control injection), by (E) dual label immunofluorescence for MHC1 (red) and CK7 (green), and if cells are pre-labelled with GFP, by (F) confocal microscopy for GFP.
Discussion
Cholangiocyte pathophysiology has been studied in earnest now for several decades. Early basic science investments to develop unique in vitro tools(40–42) as well as new animal models(43) have allowed the field to realize enormous advances in our understanding of normal and pathological cholangiocyte functions and has paid dividends in terms of mechanistic insights into various cholangiopathies. Despite these remarkable advances, most of the cholangiopathies remain biologically enigmatic and clinically problematic without definitive treatments strategies other than liver transplantation when the diseases reach their end-stage. In this context, the current study outlines a novel protocol for the reproducible differentiation of pluripotent stem cells to a mature and functional cholangiocyte phenotype. We have generated and fully characterized iDCs in vitro, analyzed the entire transcriptome by RNA sequencing, and for the first time, we demonstrate that stem cell-derived cholangiocytes can engraft within mouse liver in vivo. The iDC protocol represents an in vitro model of biliary differentiation that appears to recapitulate several aspects of biliary development and results in a scientific platform that may be useful to mechanistically test developmental hypotheses. The system should also allow us to model biliary disease in an individualized manner, to test therapeutic compounds in vitro, and to move toward cell-based therapies for the biliary system (Figure 8).
Figure 8. Development and potential uses for iDC.
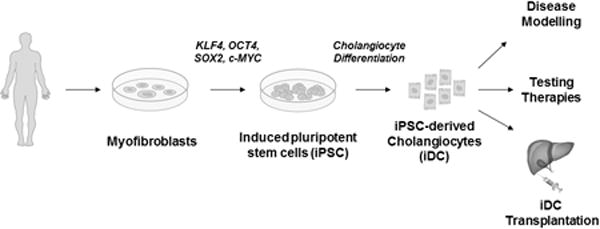
The flowchart outlines the development of iPSC from myofibroblasts, differentiation to iDCs, and potential downstream applications for the cells.
Research by several groups is now beginning to more fully elucidate many of the mechanisms that regulate and define biliary development(23–26). The foregut endoderm undergoes hepatic specification under the influence of FGF from the cardiac mesoderm and BMP from the septum transversum. Thereafter, the primitive hepatoblasts migrate into the septum transversum toward primitive portal venous structures. Hepatoblast proliferation and maturation follow as Wnt signaling is activated via interactions with endothelial cells and stellate cells. There appears to be a gradient of Notch ligands, TGF-β, and other molecules that results in the formation of a peri-portal ductal plate of immature cholangiocytes. Cholangiocyte maturation, proliferation, and morphogenesis into mature tubular structures then follows in this area while the remaining, non-peri-portal hepatoblasts move instead toward a hepatocyte fate. Accurate in vitro platforms for modelling the developmental biology of the biliary tree have been lacking and our system was designed to recapitulate several aspects of embryologic bile duct development. Since many cholangiopathies appear to have developmental defects or abnormal epigenetic patterning events that occur during embryogenesis, such a system may allow for a more detailed understanding of the pathogenesis of biliary disease as well as development of unique therapeutic targets and approaches. Whole transcriptome analysis by RNA sequencing suggests that the transcriptional profiles identified during the iPSC to iDC transition correlate with our current understanding of biliary development. As such, the system should be useful to mechanistically study how key transcriptional events may modulate hepatic lineage specification, either in the context of liver development or during progenitor cell activation in regeneration and remodeling during disease.
Rapid advances in liver regenerative medicine including differentiation of iPSC to hepatocytes has generated enthusiasm for individualized disease modelling(44–48) and cell-based therapy for hepatocellular disorders(29, 48, 49). The development of iDCs was an obvious and important extension of this work. In that regard, our group and others(22) have now begun to simultaneously develop similar, but unique, protocols to differentiate iPSC toward cholangiocytes. These disparate protocols each appear to result in cells with features of mature cholangiocytes. It is likely that each system will ultimately have strengths and limitations for various applications. Furthermore, head-to-head comparison at the transcriptomic, proteomic, and epigenetic levels should allow us to understand the subtleties that permit a multiplicity of developmental pathways to result in mature biliary cells while differentiation of iPSC to mature and functional hepatocytes has been notoriously difficult to achieve. One interpretation of this is that biliary differentiation may represent a “default” pathway in iPSC derivatives while differentiation to hepatocytes requires complex in vivo cues that are difficult to simulate in vitro. In this way, the biliary system may be an even more attractive target for regenerative medicine than parenchymal hepatocytes. There is recent evidence of biliary cells trans-differentiating to hepatocytes in the setting of massive parenchymal loss(50, 51). Furthermore, it is widely thought that hepatic progenitor cells residing in the terminal bile ductules may contribute to liver regeneration in the setting of disease(52), although this concept has also been recently questioned(53). While there are unresolved controversies, work by many groups is overall demonstrating remarkable plasticity and cellular trans-differentiation between hepatic epithelial cell types in various states of disease and regeneration and our work confirms that multiple parenchymal liver cell types can also be generated from iPSC.
In this study, we have developed a unique protocol to generate cholangiocytes from human iPSC. This process represents an in vitro model of biliary development and results in mature and functional cholangiocytes that are similar to normal human cholangiocytes. This system should allow us to test genetic and epigenetic mechanisms underlying biliary development and pathophysiology but also is a platform for individualized modelling of biliary disease and testing of pharmacotherapies. Importantly, this study is also the first to demonstrate engraftment of stem cell-derived cholangiocytes within liver in vivo, setting the stage for eventual cell-based therapeutic options for the cholangiopathies.
Supplementary Material
Acknowledgments
The authors acknowledge: Vijay Shah, Nicholas LaRusso, and Gregory Gores for valuable scientific discussions; Usman Yaqoob and Patrick Splinter for technical support; and Deb Hintz for secretarial support.
Financial Support: This work was supported by grants DK100575, DK084567, and the Loan Repayment Program from the National Institutes of Health; by the Department of Oncology and Hematology, the Second Hospital of Jilin University, China; and by the Mayo Clinic Center for Regenerative Medicine.
List of Abbreviations
- iPSC
induced pluripotent stem cells
- HLCs
hepatocyte-like cells
- iDC
iPSC-derived cholangiocytes
- ERCP
endoscopic retrograde cholangiopancreatography
- DE
definitive endoderm
- FGF2
fibroblast growth factor 2
- BMP4
bone morphogenic protein 4
- SHH
sonic hedgehog
- HS
hepatic specification
- HP
hepatic progenitor cells
- TGF-β
transforming growth factor beta
- DAPI
4′6-diamidino-2-phenylindole dihydrochloride
- MHC1
major histocompatibility complex 1
- ANOVA
analysis of variance
Footnotes
Disclosure/Duality of Interest
The authors have nothing to disclose.
Supplementary information is available at Laboratory Investigation’s website.
References
- 1.Lazaridis KN, Strazzabosco M, Larusso NF. The cholangiopathies: disorders of biliary epithelia. Gastroenterology. 2004;127(5):1565–1577. doi: 10.1053/j.gastro.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 2.O’Hara SP, Tabibian JH, Splinter PL, LaRusso NF. The dynamic biliary epithelia: molecules, pathways, and disease. J Hepatol. 2013;58(3):575–582. doi: 10.1016/j.jhep.2012.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kochhar G, Parungao JM, Hanouneh IA, Parsi MA. Biliary complications following liver transplantation. World J Gastroenterol. 2013;19(19):2841–2846. doi: 10.3748/wjg.v19.i19.2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mourad MM, Algarni A, Liossis C, Bramhall SR. Aetiology and risk factors of ischaemic cholangiopathy after liver transplantation. World J Gastroenterol. 2014;20(20):6159–6169. doi: 10.3748/wjg.v20.i20.6159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Williams MJ, Clouston AD, Forbes SJ. Links between hepatic fibrosis, ductular reaction, and progenitor cell expansion. Gastroenterology. 2014;146(2):349–356. doi: 10.1053/j.gastro.2013.11.034. [DOI] [PubMed] [Google Scholar]
- 6.Dianat N, Steichen C, Vallier L, Weber A, Dubart-Kupperschmitt A. Human pluripotent stem cells for modelling human liver diseases and cell therapy. Current gene therapy. 2013;13(2):120–132. doi: 10.2174/1566523211313020006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 8.Turner R, Lozoya O, Wang Y, et al. Human hepatic stem cell and maturational liver lineage biology. Hepatology. 2011;53(3):1035–1045. doi: 10.1002/hep.24157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Si-Tayeb K, Noto FK, Nagaoka M, et al. Highly efficient generation of human hepatocyte-like cells from induced pluripotent stem cells. Hepatology. 2010;51(1):297–305. doi: 10.1002/hep.23354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sancho-Bru P, Roelandt P, Narain N, et al. Directed differentiation of murine-induced pluripotent stem cells to functional hepatocyte-like cells. J Hepatol. 2011;54(1):98–107. doi: 10.1016/j.jhep.2010.06.014. [DOI] [PubMed] [Google Scholar]
- 11.Choi SM, Kim Y, Liu H, Chaudhari P, Ye Z, Jang YY. Liver engraftment potential of hepatic cells derived from patient-specific induced pluripotent stem cells. Cell cycle. 2011;10(15):2423–2427. doi: 10.4161/cc.10.15.16869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen YF, Tseng CY, Wang HW, Kuo HC, Yang VW, Lee OK. Rapid generation of mature hepatocyte-like cells from human induced pluripotent stem cells by an efficient three-step protocol. Hepatology. 2012;55(4):1193–1203. doi: 10.1002/hep.24790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakamura N, Saeki K, Mitsumoto M, et al. Feeder-free and serum-free production of hepatocytes, cholangiocytes, and their proliferating progenitors from human pluripotent stem cells: application to liver-specific functional and cytotoxic assays. Cell Reprogram. 2012;14(2):171–185. doi: 10.1089/cell.2011.0064. [DOI] [PubMed] [Google Scholar]
- 14.Yu Y, Liu H, Ikeda Y, et al. Hepatocyte-like cells differentiated from human induced pluripotent stem cells: relevance to cellular therapies. Stem Cell Res. 2012;9(3):196–207. doi: 10.1016/j.scr.2012.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aravalli RN, Cressman EN, Steer CJ. Hepatic differentiation of porcine induced pluripotent stem cells in vitro. Veterinary journal. 2012;194(3):369–374. doi: 10.1016/j.tvjl.2012.05.013. [DOI] [PubMed] [Google Scholar]
- 16.Zhu S, Rezvani M, Harbell J, et al. Mouse liver repopulation with hepatocytes generated from human fibroblasts. Nature. 2014;508(7494):93–97. doi: 10.1038/nature13020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Szkolnicka D, Farnworth SL, Lucendo-Villarin B, Hay DC. Deriving functional hepatocytes from pluripotent stem cells. Curr Protoc Stem Cell Biol. 2014;30:1G 5 1–1G 5 12. doi: 10.1002/9780470151808.sc01g05s30. [DOI] [PubMed] [Google Scholar]
- 18.Takayama K, Inamura M, Kawabata K, et al. Generation of metabolically functioning hepatocytes from human pluripotent stem cells by FOXA2 and HNF1alpha transduction. J Hepatol. 2012;57(3):628–636. doi: 10.1016/j.jhep.2012.04.038. [DOI] [PubMed] [Google Scholar]
- 19.Schwartz RE, Fleming HE, Khetani SR, Bhatia SN. Pluripotent stem cell-derived hepatocyte-like cells. Biotechnol Adv. 2014;32(2):504–513. doi: 10.1016/j.biotechadv.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang P, He Z, Ji S, et al. Induction of functional hepatocyte-like cells from mouse fibroblasts by defined factors. Nature. 2011;475(7356):386–389. doi: 10.1038/nature10116. [DOI] [PubMed] [Google Scholar]
- 21.Yu B, He ZY, You P, et al. Reprogramming fibroblasts into bipotential hepatic stem cells by defined factors. Cell Stem Cell. 2013;13(3):328–340. doi: 10.1016/j.stem.2013.06.017. [DOI] [PubMed] [Google Scholar]
- 22.Dianat N, Dubois-Pot-Schneider H, Steichen C, et al. Generation of functional cholangiocyte-like cells from human pluripotent stem cells and HepaRG cells. Hepatology. 2014;60(2):700–714. doi: 10.1002/hep.27165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zong Y, Stanger BZ. Molecular mechanisms of bile duct development. Int J Biochem Cell Biol. 2011;43(2):257–264. doi: 10.1016/j.biocel.2010.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Strazzabosco M, Fabris L. Development of the bile ducts: essentials for the clinical hepatologist. J Hepatol. 2012;56(5):1159–1170. doi: 10.1016/j.jhep.2011.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zong Y, Stanger BZ. Molecular mechanisms of liver and bile duct development. Wiley interdisciplinary reviews Developmental biology. 2012;1(5):643–655. doi: 10.1002/wdev.47. [DOI] [PubMed] [Google Scholar]
- 26.Tanimizu N, Miyajima A. Molecular mechanism of liver development and regeneration. Int Rev Cytol. 2007;259:1–48. doi: 10.1016/S0074-7696(06)59001-1. [DOI] [PubMed] [Google Scholar]
- 27.Greenbaum LE, Wells RG. The role of stem cells in liver repair and fibrosis. Int J Biochem Cell Biol. 2011;43(2):222–229. doi: 10.1016/j.biocel.2009.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Machado MV, Diehl AM. Liver renewal: detecting misrepair and optimizing regeneration. Mayo Clin Proc. 2014;89(1):120–130. doi: 10.1016/j.mayocp.2013.10.009. [DOI] [PubMed] [Google Scholar]
- 29.Huebert RC, Rakela J. Cellular therapy for liver disease. Mayo Clin Proc. 2014;89(3):414–424. doi: 10.1016/j.mayocp.2013.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kudva YC, Ohmine S, Greder LV, et al. Transgene-free disease-specific induced pluripotent stem cells from patients with type 1 and type 2 diabetes. Stem Cells Transl Med. 2012;1(6):451–461. doi: 10.5966/sctm.2011-0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang L, Kwon J, Popov Y, et al. Vascular endothelial growth factor promotes fibrosis resolution and repair in mice. Gastroenterology. 2014;146(5):1339–1350. e1331. doi: 10.1053/j.gastro.2014.01.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kalari KR, Nair AA, Bhavsar JD, et al. MAP-RSeq: Mayo Analysis Pipeline for RNA sequencing. BMC Bioinformatics. 2014;15:224. doi: 10.1186/1471-2105-15-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huebert RC, Jagavelu K, Liebl AF, et al. Immortalized liver endothelial cells: a cell culture model for studies of motility and angiogenesis. Lab Invest. 2010;90(12):1770–1781. doi: 10.1038/labinvest.2010.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jin LF, Ji SH, Yang JF, Ji WZ. Notch signaling dependent differentiation of cholangiocyte-like cells from rhesus monkey embryonic stem cells. Dongwuxue Yanjiu. 2011;32(4):391–395. doi: 10.3724/SP.J.1141.2011.04391. [DOI] [PubMed] [Google Scholar]
- 37.Akkari L, Haouzi D, Biname F, et al. Cell shape and TGF-beta signaling define the choice of lineage during in vitro differentiation of mouse primary hepatic precursors. J Cell Physiol. 2010;225(1):186–195. doi: 10.1002/jcp.22219. [DOI] [PubMed] [Google Scholar]
- 38.Feng L, Pang L, Guo Y, et al. Hypoxia/reoxygenation up-regulates death receptor expression and enhances apoptosis in human biliary epithelial cells. Life Sci. 2009;85(9–10):401–407. doi: 10.1016/j.lfs.2009.07.011. [DOI] [PubMed] [Google Scholar]
- 39.Yu J, Sheung N, Soliman EM, Spirli C, Dranoff JA. Transcriptional regulation of IL-6 in bile duct epithelia by extracellular ATP. Am J Physiol Gastrointest Liver Physiol. 2009;296(3):G563–571. doi: 10.1152/ajpgi.90502.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tietz PS, Chen XM, Gong AY, et al. Experimental models to study cholangiocyte biology. World J Gastroenterol. 2002;8(1):1–4. doi: 10.3748/wjg.v8.i1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Concepcion AR, Lopez M, Ardura-Fabregat A, Medina JF. Role of AE2 for pHi regulation in biliary epithelial cells. Front Physiol. 2013;4:413. doi: 10.3389/fphys.2013.00413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Walter TJ, Vanderpool C, Cast AE, Huppert SS. Intrahepatic bile duct regeneration in mice does not require Hnf6 or Notch signaling through Rbpj. Am J Pathol. 2014;184(5):1479–1488. doi: 10.1016/j.ajpath.2014.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fickert P, Pollheimer MJ, Beuers U, et al. Characterization of animal models for primary sclerosing cholangitis (PSC) J Hepatol. 2014;60(6):1290–1303. doi: 10.1016/j.jhep.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cayo MA, Cai J, DeLaForest A, et al. JD induced pluripotent stem cell-derived hepatocytes faithfully recapitulate the pathophysiology of familial hypercholesterolemia. Hepatology. 2012;56(6):2163–2171. doi: 10.1002/hep.25871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu D, Alipio Z, Fink LM, et al. Phenotypic correction of murine hemophilia A using an iPS cell-based therapy. Proc Natl Acad Sci U S A. 2009;106(3):808–813. doi: 10.1073/pnas.0812090106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yusa K, Rashid ST, Strick-Marchand H, et al. Targeted gene correction of alpha1-antitrypsin deficiency in induced pluripotent stem cells. Nature. 2011;478(7369):391–394. doi: 10.1038/nature10424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rashid ST, Corbineau S, Hannan N, et al. Modeling inherited metabolic disorders of the liver using human induced pluripotent stem cells. The Journal of clinical investigation. 2010;120(9):3127–3136. doi: 10.1172/JCI43122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yu Y, Fisher JE, Lillegard JB, Rodysill B, Amiot B, Nyberg SL. Cell therapies for liver diseases. Liver transplantation : official publication of the American Association for the Study of Liver Diseases and the International Liver Transplantation Society. 2012;18(1):9–21. doi: 10.1002/lt.22467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Subba Rao M, Sasikala M, Nageshwar Reddy D. Thinking outside the liver: induced pluripotent stem cells for hepatic applications. World J Gastroenterol. 2013;19(22):3385–3396. doi: 10.3748/wjg.v19.i22.3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Choi TY, Ninov N, Stainier DY, Shin D. Extensive conversion of hepatic biliary epithelial cells to hepatocytes after near total loss of hepatocytes in zebrafish. Gastroenterology. 2014;146(3):776–788. doi: 10.1053/j.gastro.2013.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.He J, Lu H, Zou Q, Luo L. Regeneration of liver after extreme hepatocyte loss occurs mainly via biliary transdifferentiation in zebrafish. Gastroenterology. 2014;146(3):789–800 e788. doi: 10.1053/j.gastro.2013.11.045. [DOI] [PubMed] [Google Scholar]
- 52.Miyajima A, Tanaka M, Itoh T. Stem/progenitor cells in liver development, homeostasis, regeneration, and reprogramming. Cell Stem Cell. 2014;14(5):561–574. doi: 10.1016/j.stem.2014.04.010. [DOI] [PubMed] [Google Scholar]
- 53.Yanger K, Knigin D, Zong Y, et al. Adult hepatocytes are generated by self-duplication rather than stem cell differentiation. Cell Stem Cell. 2014;15(3):340–349. doi: 10.1016/j.stem.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


