Abstract
Although the Hispanic population is continuously growing in the United States, they are underrepresented in pharmacogenetic studies. This review addresses the need for compiling available pharmacogenetic data in US Hispanics, discussing the prevalence of clinically relevant polymorphisms in pharmacogenes encoding for drug-metabolizing enzymes. CYP3A5*3 (0.245–0.867) showed the largest frequency in a US Hispanic population. A higher prevalence of CYP2C9*3, CYP2C19*4, and UGT2B7 IVS1+985 A>Gwas observed in US Hispanic vs. non-Hispanic populations. We found interethnic and intraethnic variability in frequencies of genetic polymorphisms for metabolizing enzymes, which highlights the need to define the ancestries of participants in pharmacogenetic studies. New approaches should be integrated in experimental designs to gain knowledge about the clinical relevance of the unique combination of genetic variants occurring in this admixed population. Ethnic subgroups in the US Hispanic population may harbor variants that might be part of multiple causative loci or in linkage-disequilibrium with functional variants. Pharmacogenetic studies in Hispanics should not be limited to ascertain commonly studied polymorphisms that were originally identified in their parental populations. The success of the Personalized Medicine paradigm will depend on recognizing genetic diversity between and within US Hispanics and the uniqueness of their genetic backgrounds.
Keywords: admixture, CYP450, drug-metabolizing enzymes, genotypes, Hispanics, pharmacogenetics, Puerto Ricans
Introduction
On February 20, 2013, the Food and Drug Administration (FDA) expedited a new Boxed Warning – the strongest warning to restrict the use of codeine-containing medications in children as postoperative pain management. This warning resulted from reported adverse drug reactions such as life-threatening respiratory depression and three deaths of children that metabolized codeine faster, achieving lethal concentrations of morphine [1]. It is known that genetic polymorphisms affect a drug’s response by three mechanisms: (a) altering the metabolism of the drug, (b) producing unexpected effects (i.e., hemolysis), and (c) altering the drug target [2]. The mentioned reports of codeine use in children are an example of how genetic polymorphisms in drug-metabolizing enzymes result in serious and even lethal drug responses. Adverse drug events were found to account for 2.5% of emergency department visits [3]. A meta-analysis study found that 59% of drugs cited in reports of adverse drug events were metabolized by at least one enzyme with a known genetic polymorphism associated with lower enzyme activity [4]. Even when the FDA had labeled about 150 xenobiotics with pharmacogenetic information, the potential cost-effectiveness of genetic tests to avoid adverse drug events remains unclear and controversial [5]. The presence of individuals with different genetic backgrounds in clinical trials that attempted to determine the cost-effectiveness of genetic tests in pharmacotherapy might be an explanation for such controversial results.
Overrepresentation of European descendant populations in pharmacogenetic studies and the extrapolation of those findings to other populations are factors that undermine the strong evidence that supports the incorporation of precision medicine [6]. The majority of the case control studies that search for associations between a certain phenotype and genetic polymorphisms (i.e., Genome Wide Association Studies) are often conducted in Caucasians and are therefore limited to account for the effect on associations of multihybrid admixtures and unique stratification observed in Hispanics. Admixture is defined as “a common type of gene flow in human populations that occurs when individuals from two or more parental populations that have been isolated for several generations form a new hybrid population”. Admixture can be a confounding variable that may result in false associations due to differences in genetic backgrounds across individuals within a cohort [7]. Admixed populations may differ from other populations in frequency, distribution, and combination of allelic variants in the genome [8]. Hispanics in the United States are a case of admixture, with mainly a trichotomous ancestral contribution of the following distant parental populations: Europeans, Native Americans, and West-Africans [7, 9, 10]. Using principal component analysis of autosomal genotype data in Hispanics and other putative ancestral populations, a tri-hybrid admixture pattern was confirmed by Bryc and coworkers [11]. Indeed, the two principal components shown in Figure 1 illustrate that Native American, European, and African ancestries enriched the genomic diversity of Hispanics. The fitting of ellipses to the covariance matrix reveals different ancestry depictions (mixtures) per Hispanic/Latino population. The analysis on the X chromosome markers showed a similar pattern, although with greater variance (data not shown) [11].
Figure 1.
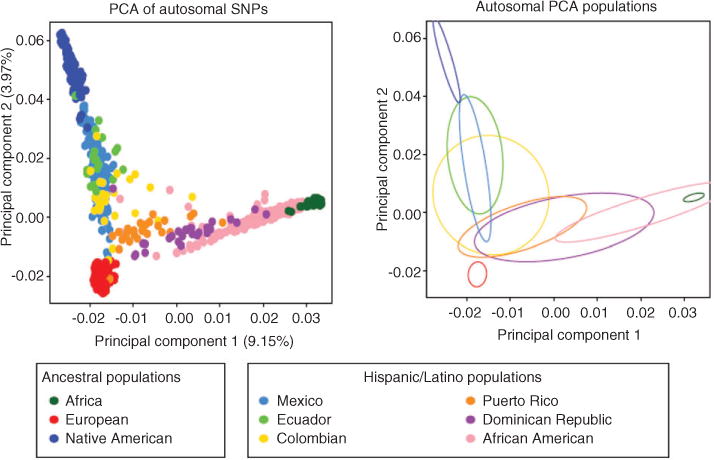
Principal component analysis (PCA) results of the Hispanic individuals with Europeans, Africans and Native Americans. PC 1 vs. PC2 scatter plots based on autosomal markers (left plot). Ellipses are fitted to the PCA results on the autosomes (right plot). Reprinted from Bryc et al. [11] with permission.
The relative contribution of each parental population is different among Hispanics: European and Native American contribution in autosomal genes is higher in Mexicans when compared to Caribbean Hispanics, while African contribution is higher in Caribbean Hispanics than in Mexicans. In admixed populations, the relative contribution of the parental populations will determine the distribution of the relevant variants [7]. As a result, Hispanics present a large variation in ancestry proportions among different ethnic groups and among individuals in a country [9, 11].
Another concern of extrapolating the findings of pharmacogenetic studies in European descendants to admixed populations is the role that linkage disequilibrium plays in population genetics [12]. Some polymorphisms associated to a certain phenotype in drug response may not be the causal variant but rather are in linkage disequilibrium with one or more causal or functional variants that occur specifically in a given population (i.e., rs12777823 in African-Americans and VKORC1-1639G>A in Asians, both associated to warfarin’s response). Since Hispanics are the result of three distant parental populations (including an enrichment of African genetic variants), a new combination of the haplotype block architecture is expected, giving rise to a unique linkage disequilibrium structure with new putative markers or association signals in this population. Linkage disequilibrium can also be affected by the ancestral contributions. Bryc and colleagues [11] found that linkage disequilibrium decays faster in Hispanic/Latinos with greater African ancestry.
The US Census Bureau reported that, by July 2012, 53 million Hispanics are living in the United States, corresponding to 16% of the total population in this country. The most represented ethnicities among Hispanics are Mexicans (65%), Puerto Ricans (9.4%), Salvadorans (3.8%), and Cubans (3.6%). Hispanics are the nation’s largest ethnic minority [13]. Although Hispanics are a continuing growing population in the United States, they are underrepresented in pharmacogenetic studies. Ongoing international efforts such as the 1000 Genomes Project and the HapMap Project are incorporating admixed populations due to the need of gaining knowledge of human genetic variations globally. These projects, in conjunction with the existing literature, are resources for studying genetic variations affecting human health and drug response across different populations in the world, including Hispanics. This review aims to evaluate the recent literature available about genetic polymorphism of drug-metabolizing enzymes (phase I and II) in Hispanics living in the United States, mainly in Mexicans, Puerto Ricans, and Cubans, and also to evaluate existing data available from the 1000 Genomes Project.
Phase I drug-metabolizing enzymes: cytochromes P450s
Cytochromes (CYP) P450 are a superfamily of proteins involved mainly in metabolism of xenobiotics (i.e., drugs) and endogenous compounds (including retinoic acid, prostaglandins, and eicosanoids) as well as the synthesis of endogenous compounds (steroid hormones) [14]. These enzymes have the capacity to interact with a large range of chemically different compounds. The CYP450s catalyze phase I reactions: oxidation, reduction, and hydrolysis, resulting in the introduction of functional groups (−OH, −COOH, −SH, −O−, or NH2) to the substrate. They are localized in the cytosolic face of the endoplasmic reticulum. CYP450s are expressed mainly in the liver, although some expression occurs in the gastrointestinal tract, lungs, kidney, and the central nervous system. The expression and function of CYPs are influenced by diet, environmental exposure, age, disease, genetic polymorphisms, and other factors [14, 15]. Mainly one or a few members of the CYP450s metabolize many drugs at pharmacologically relevant concentrations, even given the overlapping substrate specificity in these enzymes [15]. The superfamily of CYP450s is classified in families and subfamilies based on their amino acid sequence similarity. Twelve CYPs belonging to three families (families 1–3) have been identified to metabolize xenobiotics in humans, from which members of the subfamilies CYP2C, CY2D, and CYP3A are the most important for drug metabolism [14].
CYP2C9
CYP450, family 2, subfamily C, polypeptide 9 (CYP2C9) is a gene (ID:1559) within the CYP2C cluster of approximately 390 kilobases (Kb) in the chromosome 10q23.3 [16]. This locus also contains the CYP2C8, CYP2C18, and CYP2C19 genes [15]. CYP2C family members are responsible for 20% of the metabolism of clinically important drugs [17]. Among the members of the CYP2C family, CYP2C9 has the highest expression levels [15]. CYP2C9 is 50 Kb in size, codes for a 490-amino-acid protein (GI:527178914), and is thought to interact with weakly acidic ligands. It has been speculated that CYP2C9 protein may have more than one binding site [18]. CYP2C9 interacts with arachidonic acid to produce hydroxyeicosatetraenoic acid and epoxyeicosatrienoic acid [19]. This enzyme metabolizes drugs such as warfarin, phenytoin, losartan, glyburide, and most nonsteroidal anti-inflammatory drugs (NSAIDs) [15].
CYP2C9 functional and clinical significance of the most common variant alleles
CYP2C9 is one of the most studied CYPs in pharmacogenetics since it metabolizes a broad spectrum of commonly used drugs. Common or wild-type allele is known as CYP2C9*1 (http://www.cypalleles.ki.se/cyp2c9.htm) [20]. CYP2C9*2 (r s1799853; 430C>T at exon 3) results in a change in arginine for cysteine in residue 144, which had been associated with slower metabolism of S-warfarin (i.e., average 40% reduction in warfarin metabolism), diclofenac, and lauric acid (i.e., 60%–70% of normal enzyme activity) [21]. It is thought that this variant affects the interaction of the enzyme with NADPH CYP450 oxide-reductase [22, 23]. CYP2C9*2 has been associated with lower dose requirement of warfarin, phenytoin-induced neurotoxicity, and a greater decrease in blood pressure in patients treated with irbesartan [24–27]. CYP2C9*3 (rs1057910; 1075A>C at exon 7) produces an isoleucine to leucine, and a change in codon 359 results in lower enzyme activity (i.e., 5% of normal enzyme activity) [20, 22, 23, 28]. CYP2C9*5 (rs28371686; 1080C>G), *6 (rs9332131; 818delA), *8 (rs7900194; 449G>A), and *11 (rs28371685; 1003C>T) occur predominantly in individuals of African descent. Their allele frequencies range from 0.01 to 0.06 in African-Americans. They have been associated with lower warfarin dose requirements in African-Americans [29].
It is known that common variants in the CYP2C9 gene may decrease S-warfarin’s metabolic rate and therefore alter a patient’s sensitivity and dose requirements (i.e., approximately 16-fold interindividual variability in dose requirements). Notably, the CYP2C9*2 and *3 variants (most frequent among Caucasians) account for 15%–20% of the warfarin dose variability in various populations worldwide [30–33]. CYP2C9 genotypes are associated with decreased warfarin clearance, longer time to achieve stable anticoagulation therapy, 5–6 times higher likelihood of having initial International Normalized Ratio (INR) > 3 (indicative of overanticoagulation), and 4 times higher likelihood of developing major bleedings and intracranial hemorrhage during the initiation phase [34–36]. Likewise, patients carrying these single-nucleotide polymorphism (SNPs) may show a higher risk of developing acute gastrointestinal bleeding when using NSAIDs. On the basis of a meta-analysis of nine qualified studies, the FDA has approved reduced recommended warfarin dosage based on the presence of these variants [37].
We have recently found a significant association between CYP2C9 genotypes and warfarin dose requirements in Puerto Ricans. Patients carrying at least one copy of either the CYP2C9*2 or the *3 allele required a mean daily warfarin dose that was 26% less than that for patients with wild-type genotype. Moreover, carriers of functional CYP2C9 polymorphisms, along with the target of warfarin, vitamin K epoxide reductase complex subunit 1 (VKORC1)-1639G>A variant, demonstrated a higher incidence rate of multiple adverse events (i.e., 5.2 vs. 1.0 cases per 100 patient-months; RR=4.8) as compared to those with wild-type genotype in a cohort of Puerto Rican patients on warfarin. A significant association was observed between multiple adverse events and the carrier status (HR=2.5; 95% CI, 1.0–6.3; p=0.04). However, no significant associations between genotypes and individual outcomes over the first 90 days of therapy were found [38].
The 2010 Medco-Mayo Warfarin Effectiveness study suggested that genetic test in CYP2C9 and VKORC1 significantly reduced (by approximately 30%) the hospitalization rate for bleeding or thromboembolic events in patients starting anticoagulation therapy when compared with controls [39]. Both a report by the Centers for Medicare and Medicaid Services and the randomized clinical effectiveness CoumaGen-II trial (NCT00927862) concluded that pharmacogenetic guidance was superior to current dosing approaches (fixed and clinical) in achieving optimal anticoagulation (assessed by INR measurements) and also in reducing deaths, hemorrhages, and thromboembolisms [40, 41].
Warfarin ranks among the top 10 drugs with the largest number of serious adverse event reports in the FDA’s Adverse Event Reporting System and is also associated with about 29,000 emergency room visits per year for bleeding complications (50% major bleedings occurring within the first 90 days) [42]. If genotyping were known upon initiation of warfarin, 85,000 serious bleeding events and 17,000 strokes could be avoided annually, saving over $ 1 billion in healthcare [43]. Besides, if genotyping is performed after therapy begins, lesser but still worthwhile cost savings would result, in part due to reduced risk of hospitalization for bleeding or thromboembolism in outpatients initiating warfarin [39, 44].
Prevalence of the most common variants in US Hispanics
Currently, most of the pharmacogenetic studies conducted in Hispanics living in the United States have included mostly Mexican-Americans. The 1000 Genomes Project includes a Puerto Rican sample of 55 individuals [45]. Common CYP2C9 polymorphisms studied in the Puerto Rican population included in this project such as CYP2C9*2, *3, *5, and *8 were present in 0.173, 0.036, 0.009, and 0.009 of the population, respectively [45] (Table 1). Studies by Duconge and coworkers investigated minor allele frequencies (MAFs) in Puerto Ricans for CYP2C9*2, CYP2C9*3, CYP2C9*5, and CYP2C9*6, which are presented in Table 1 [51–53]. The CYP2C9*8 (rs7900194; 449G>A) variant has a frequency of 0.008 in a Puerto Rican population cohort of patients under warfarin therapy (unpublished data), not distant from the MAF reported for this population in the 1000 Genomes Project (0.009) [45].
Table 1.
Distribution of CYP2C9 allelic variants among Hispanics.
| Population | Na | *1 | *2 | *3 | *4 | *5 | *6 | *8 | *11 | *13 | Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Mexican-Americans | 98 | 0.86 | 0.08 | 0.06 | [46] | ||||||
| Mexican-Americans | 193 | 0.070 | 0.044 | 0.00 | 0.00 | 0.00 | [47] | ||||
| Hispanics (mostly Mexican-Americans) | 50 | 0.09 | 0.06 | [48] | |||||||
| Mexican-Americans | 38 | 0.86 | 0.08 | 0.06 | [49] | ||||||
| Mexican-Tapehuana | 99 | 0.01 | 0.015 | [50] | |||||||
| Mexican-Mestizos | 102 | 0.07 | 0.015 | [50] | |||||||
| Puerto Ricans | 100 | 0.701 | 0.0652 | 0.0543 | 0.000 | 0.000 | 0.0054 | [51] | |||
| Puerto Ricans | 163 | 0.840 | 0.107 | 0.043 | 0.01 | [52] | |||||
| Puerto Ricans | 103 | 0.748 | 0.189 | 0.053 | 0.01 | [53] | |||||
| Puerto Ricans | 55 | 0.173 | 0.036 | 0.009 | 0.009 | [45] | |||||
| Cuban-whites | 132 | 0.720 | 0.170 | 0.110 | 0.000 | 0.000 | 0.000 | 0.004 | [54] | ||
| Cuban-mestizos | 128 | 0.830 | 0.060 | 0.090 | 0.000 | 0.004 | 0.000 | 0.020 | [54] | ||
| Hispanics | 202 | 0.822 | 0.069 | 0.064 | 0.000 | 0.015 | 0.005 | 0.015 | 0.010 | 0.000 | [55] |
| Hispanics | 22 | 0.93 | 0.000 | 0.070 | [56] |
Number of individuals.
We have also observed a relationship between population substructure and warfarin-related genotypes in Puerto Ricans. Our group used physiogenomic (PG) markers to infer the structure and ancestry of participants, finding an absence of the *2 allele in the cluster of Puerto Ricans genetically resembling Amerindians, consistent with the prevalence reported in Asians by the HapMap Project [57]. This finding was not surprising since it is hypothesized that Amerindians originated from Asians who migrated from the Bering Strait (Bering Migration Theory). CYP2C9*2 was found in the cluster resembling Caucasians (7.7%), close to values reported for this ancestral population (10%) in the HapMap Project but different than expected in the West-African cluster (18.8% vs. 0% in Nigerians), showing significant admixture and the heterogeneity of the Puerto Rican population. CYP2C9*3 frequencies found in clusters genetically resembling Caucasians (7.6%) and West-Africans (0%) are comparable to reported values for the referred populations in the HapMap Project (6% and 0%, respectively). These findings suggest that interindividual variations in ancestral contribution may influence the response to warfarin of each Puerto Rican [58, 59].
The MAFs of *2 and *3, among other variants of CYP2C9, in Mexican-Americans are shown in Table 1 [46–50]. Both CYP2C9*2 and CYP2C9*3 are at lower frequency in Mexicans living in Mexico (Mexican-Mestizos and Mexican-Tapehuana) than in Mexican-Americans [46, 50]. CYP2C9 *4 (rs56165452; 1076T>C), *5, and *6 were not present in any of the individuals of Mexican-American descents examined [46, 47]. Reported literature in Cubans, classified as Cuban-whites (individuals descendants from four Caucasian grandparents) and Cuban-mestizos, showed that CYP2C9*2 and*3 MAFs were 0.06–0.17 and 0.09–0.11, respectively. CYP2C9*5 and *8 in Cubans were found in ranges of 0–0.004 and 0.004–0.02, respectively [54]. CYP2C9*6 was not found in any Cuban individual, while CYP2C9*4 was not found in any individual of Mexican-American, Puerto Rican, or Cuban origin [46–53].
Combined common allele variants in the CYP2C9 gene (*2, *3, *4, *5, *6, *8, *11, and *13) can be found with a frequency of 0.178 in the Hispanic population [55]. MAFs for the CYP2C9 *2 and *3 variants in Hispanics from unreported origin are available in Table 1 [55, 56]. The population of Hispanics in the study of Scott and colleagues are potentially mostly Caribbean Hispanics (Dominicans and Puerto Ricans), who constitute a large percentage of the Hispanic population in New York [60]. Frequencies of common variants for the CYP2C9 may fluctuate even among Hispanics, implicating special considerations to each population when designing dosing algorithms for drugs metabolized by CYP2C9. CYP2C9*4 was not found in Hispanics; this may implicate that this variant is too rare to be considered into genotyping panels for clinical decisions [55]. CYP2C9*8 is found at higher frequencies in African descendants, which may explain its presence in Cubans and Puerto Ricans due to their high African contribution when compared to other non-Caribbean Hispanic ethnicities [11, 61, 62]. The MAFs in the global population for CYP2C9*2, *3, *5, and *8 are 0.068, 0.042, 0.005, and 0.012, respectively [45].
CYP2C19
CYP450, family 2, subfamily C, polypeptide 19 (CYP2C19) is another member of the CYP2C family [63]. CYP2C19 has lower expression levels than CYP2C9 does [15]. The gene has an approximate size of 90.2 Kb (ID:1557) that code for a protein precursor of 490 amino acids (GI:4503219). This enzyme binds neutral or weak bases, and endogenously, it metabolizes progesterone and melatonin. CYP2C19 enzyme is involved in the metabolism of approximately 6.8% of drugs such as omeprazole, amitriptyline, carbamazepine, and clopidogrel [15]. Carriers of polymorphisms in the CYP2C19 gene that encodes for this enzyme may exhibit phenotypes classified as poor, intermediate, extensive, and ultrarapid metabolizers depending on the enzyme’s activity.
CYP2C19 functional and clinical significance of the most common variant alleles
At least 16 genetic variants have been associated with changes in enzyme’s activity, from which 7 result in an inactive enzyme [64]. As with other P450 genes, CYP2C19*1 identifies the wild-type allele. CYP2C19*2 (rs4244285; 681G>A) and CYP2C19*3 (rs4986893; 636G>A) null alleles produce an inactive enzyme (http://www.cypalleles.ki.se/cyp2c19.htm) [20]. CYP2C19*2 and CYP2C19*3 are both SNPs in exon 5 and exon 4, respectively. CYP2C19*2 leads to a splicing defect, while CYP2C19*3 results in a premature stop codon and a truncated protein that lacks enzymatic activity (Trp212X). Homozygotes are poor metabolizers who may benefit when using proton pump inhibitors since the plasma drug concentration is higher, resulting in better control of gastro-esophageal acidity [15]. CYP2C19*3 is associated with poor metabolism (PM) of proguanil (prophylactic antimalarial drug) [21].
Both CYP2C19*2 and *3 haplotypes are associated with diminished response to clopidogrel. Clopidogrel is an oral antiplatelet that needs activation by CYP2C19 in order to prevent blood clotting [15]. Compared to wild-types, carriers of these variants are at risk of major adverse cardiovascular events and get fewer benefits from clopidogrel treatment. The FDA added a black-box warning to the clopidogrel label, to alert patients and healthcare professionals about its lower effectiveness in individuals who have CYP2C19 variants associated with low enzymatic activity [65]. The gain-of-function CYP2C19*17 (rs12248560;-806C>T) variant increases transcription of the gene, which may result in increased drug’s metabolism.
Prevalence of the most common variants in US Hispanics
Polymorphisms in CYP2C19 have not been well described in the Hispanic population. In previous studies, the CYP2C19 *2 variant was present in 0.09–0.148 of the Puerto Rican population, while the *3 allele was found with a frequency of 0.035 in the studied group [66–68] (Table 2). Another study that used a PG array to interrogate 222 genes of cardiometabolic relevance in 71 genomic DNA specimens from Puerto Ricans found CYP2C19 *3 allele frequencies of 0.05 among individuals of higher Amerindian heritage, 0.037 among subjects of greater European contribution, and 0.021 among those showing high African ancestry compared to the expected HapMap values of 0.056, 0.017, and 0.003 for each ancestral population, respectively [68]. The higher-than-expected frequencies observed in individuals of either European or African descent might be a direct consequence of significant admixture. Concerning the 1000 Genomes Phase I selection, MAFs in Puerto Ricans for the CYP2C19 *2 and *3, among other variants, are shown in Table 2 [45].
Table 2.
Distribution of CYP2C19 allelic variants among Hispanics.
| Population | Na | *1 | *2 | *3 | *4 | *5 | *6 | *7 | *8 | *9 | *17 | Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Puerto Ricans | 100 | 0.09 | 0.000 | [66] | ||||||||
| Puerto Ricans | 122 | 0.139 | 0.000 | [67] | ||||||||
| Puerto Ricans | 55 | 0.127 | 0.000 | 0.009 | 0.009 | 0.145 | [45] | |||||
| Mexican-Americans | 346 | 0.902 | 0.097 | 0.001 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | [64] | ||
| Hispanics | 250 | 0.704 | 0.128 | 0.000 | 0.000–0.002 | 0.000 | 0.000 | 0.000 | 0.004 | 0.002 | 0.152 | [69] |
Number of individuals.
In a study with Mexican-Americans, a prevalence of 0.902, 0.097, and 0.001 for CYP2C19 *1, *2, and *3, respectively, was reported in 220 patients. This study did not observe *4 (rs28399504; 1A > G null), *5 (rs56337013; 1297 C>T null), *6 (rs72552267; 395G>A null), *7 (rs72558186; 19294T>A null), and *8 (rs41291556; 358T>C null) alleles in patients previously classified as poor metabolizers [64]. A single study conducted in 250 Hispanics living in New York of undetermined origin found that prevalence for CYP2C19 *1 was 0.704. This study genotyped *2 through *17 (rs12248560; −806C>T) and *22 (rs140278421; 557G>A) variants. Allele frequencies found in this study are presented in Table 2. Linkage disequilibrium with members of the CYP2C cluster (CYP2C9, CYP2C19, and CYP2C8) was found among this group of Hispanics [69]. Worldwide prevalence for CYP2C19 *2, *3, *4, *8, *9, *17, and *22 is 0.198, 0.014, 0.001, 0.002, 0.003, 0.152, and 0.000, respectively [45].
CYP2D6
CYP2D6 is the only member of the CYP2D subfamily that encodes for a functional protein [70]. This gene is approximately 4.4 Kb and is localized in chromosome 22q13.1 (ID:1565). The encoded protein is 497 amino acids long (GI:425706106) and is expressed in the endoplasmic reticulum of liver cells, gastrointestinal tract, and the brain. Interindividual variability in hepatic protein content might be explained, in part, by the presence of genetic polymorphisms. CYP2D6 in the brain is able to O-demethylate its substrate 5-methoxytriptamine, resulting in regeneration of serotonin [71]. CYP2D6 is also involved in progesterone hydroxylation [72]. Due to the role of CYP2D6 in the metabolism of endogenous compounds of the central nervous system, it was postulated that polymorphisms in the gene might have an effect on human behavior [73]. Furthermore, in Spanish and Cubans, one study found a significant association between CYP2D6 activity (measured by debrisoquine hydroxylation) and personality traits such as psychic anxiety and socialization [74]. CYP2D6 is the enzyme for metabolism of 15%–25% of drugs used in clinics [15]. Among drugs metabolized by CYP2D6 are tamoxifen, codeine, debrisoquine (antihypertensive), paroxetine, aripiprazole, and risperidone. Like other CYP450s, the metabolic capacity of CYP2D6 is measured using phenotyping tests and is classified as poor, intermediate, extensive, and ultrarapid metabolizers. CYP2D6 activity is measured using debrisoquine, sparteine, and dextromethorphan, as substrates [7].
CYP2D6 functional and clinical significance of the most common variant alleles
More than 100 variant alleles have been identified, including SNPs and structural variants [20]. The *1 and *2 variants have normal activity and are considered wild-type. CYP2D6*4 (rs3892097; 1846G > A) is a nonfunctional variant and results in a splicing defect. The CYP2D6*10 (rs1065852; 100C > T), *17 (rs16947; 1023C>T), and *41 (r s 28 3717 25; 2988G>A) variants result in decreased enzymatic activity, with *10 and *17 through coding SNPs in the primary protein structure (Pro34Ser in *10 and Thr107Ile and Arg296Cys in *17), while *41 causes splicing defects. Structural variants identified in CYP2D6 include deletions and amplifications of the gene. The CYP2D6*5 variant has a 11.5 Kb gene deletion. All of the alleles mentioned above are associated with lower or absent enzymatic activity, and therefore, drugs that need inactivation through CYP2D6 can accumulate in the plasma, and carriers might be at higher risk of intoxication. On the other hand, drugs that need activation may not be effective, as what occurs with tamoxifen. The anticancer drug tamoxifen needs bioconversion by CYP2D6 into the active metabolites: 4-hydroxy-tamoxifen and 4-hydroxy-N-desmethyl-tamoxifen (endoxifen). Risk of breast cancer mortality and relapse has been associated with the presence of the nonfunctional variant CYP2D6*4 [75].
Gene amplification commonly occurs in CYP2D6, resulting in higher-than-normal enzymatic activity if the amplified allele codes for a functional protein. Codeine is a prodrug that needs bioactivation to morphine by CYP2D6 in order to produce analgesia. Ultrarapid metabolizers are at greater risk of morphine-induced intoxication due to increased plasma concentration of morphine [76, 77]. In 2012, Kelly and colleagues [78] reported two fatal cases of children under codeine use: one had CYP2D6*1/*2AxN gene duplication and the other one presented higher-than-normal codeine concentration relative to morphine but unknown genotyping.
Prevalence of the most common variants in US Hispanics
González-Tejera et al. [79] performed CYP2D6 genotyping in a Puerto Rican cohort of psychiatric patients. The most prevalent allelic variants for this group of patients were normal activity alleles *1 and *2, the nonfunctional *4, and the reduced-function *41 (frequencies are shown in Table 3). Other variants such as *5, *9, *10, *17, *29, *31, *35, and *40 were also found at a range of 0.011–0.044. CYP2D6 *3 and *6 and duplications (*1XN, *2XN, *4XN, *10XN, *17XN, *35XN, and *41XN) were not found [79]. The CYP2D6 *10 variant was found with a frequency of 0.09 in 100 Puerto Rican newborn dried blood samples by Orengo-Mercado et al. [66], this frequency being higher than the one found in the patient cohort of 45 patients studied by González-Tejera (0.044) [79]. MAFs from 55 Puerto Ricans reported by the 1000 Genomes Project for CYP2D6 *4 and CYP2D6 *41, among others, are shown in Table 3. CYP2D6 *10 was found in 0.20 of the population; therefore, the *10 variant frequency and possibly those other frequencies in this population have not been consistent [45]. The study of González-Tejera and colleagues [79] mentioned previously included 45 psychiatric Puerto Ricans patients preselected based on suspected intolerance to drugs metabolized by the CYP2D6 Surprisingly, 2 out of 45 Puerto Rican patients were carriers of the rare *31 (4042G>A) variant with a MAF of 0.022 [79]. CYP2D6 *31 was also found in a group of Spanish and Hispanics with a lower frequency (0.0057 and 0.0033, respectively). Gaedigk and colleagues [84] validated the lack of activity of the *31 variant using dextromethorphan as a probe for phenotyping analysis in a Spanish patient with *4/*31 genotype. Interestingly, CYP2D6*31 was not found in Germans, Swiss, North-American Caucasians, African-Americans, Hispanics from South Florida, or Brazilians [84]. Therefore, it is suspected that the origin of CYP2D6*31, found in Spanish or Spanish descendants (Puerto Ricans and Hispanics), might be North African or Sephardic Jewish ancestry [84, 85].
Table 3.
Distribution of CYP2D6 allelic variants among Hispanics.
| Population | Na | *1 | *2 | *3 | *4 | *5 | *6 | *7 | *8 | *9 | *10 | *11 | *14 | *17 | *29 | *31 | *35 | *40 | *41 | *45 | *46 | Multi- plicationsb |
Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Puerto Ricans | 45 | 0.378 | 0.189 | 0.000 | 0.122 | 0.033 | 0.000 | 0.033 | 0.044 | 0.033 | 0.011 | 0.022 | 0.011 | 0.022 | 0.10 | 0.00 | [79] | ||||||
| Puerto Ricans | 100 | 0.09 | [66] | ||||||||||||||||||||
| Puerto Ricans | 55 | 0.173 | 0.200 | 0.409 | 0.155 | 0.000 | [45] | ||||||||||||||||
| Cubansc | 256 | 0.663–0.754 | 0.663–0.754 | 0.000 | 0.143–0.146 | 0.016–0.019 | 0.008–0.012 | 0.004–0.008 | 0.027–0.102 | 0.038–0.047 (wt) 0.00–0.004 (*4) |
[80] | ||||||||||||
| Mexican-Americans | 264 | 0.550 | 0.180 | 0.002 | 0.100 | 0.017 | 0.004 | 0.000 | 0.000 | 0.011 | 0.028 | 0.000 | 0.000 | 0.002 | 0.002 | 0.05 | 0.000 | 0.000 | 0.008 | [81] | |||
| Mexican-Americans | 50 | 0.000 | 0.170 | 0.020 | 0.000 | 0.000 | 0.000 | 0.010 | 0.020 | 0.030 | [82] | ||||||||||||
| Mexican-Americans | 349 | 0.228 | 0.003 | 0.103 | 0.023 | 0.000 | 0.074 | 0.007 | 0.010 | [83] |
Number of individuals.
Multiplications may refer to two copies or more. In Cubans, the frequency of multiplications studied by Llerena and Dorado was reported in conjunction with the presence of wild-type or *4. Frequency for multiplications with the wild-type allele was 0.038 to 0.047 and with *4 was 0 to 0.004. Gene duplications studied by Gonzâlez-Tejera et al. were *1XN, *2XN, *4XN, *10XN, *17XN, *35XN, and*41XN.
MAFs for the Cuban population were reported as Cuban-whites and Cuban-mestizos; therefore, values in the table are expressed in ranges that comprise values among all Cuban population.
An extensive study interrogating allelic variants of CYP2D6 in Mexican-Americans found that similar to Puerto Ricans, the most prevalent variants were *1, *2, *4, and *41. Variants *5, *9, and *10 were found at lower frequencies (Table 3), while variants *3, *6, *17, and *29 were very rare among Mexican-Americans. Gene duplications (*41XN, *2XN, and *4XN) were found at a frequency of 0.008, while alleles *7, *8, *11, *14, *45, and *46 were not found in this population. The combined allelic frequency of decreased and null alleles is 0.261. In this study, Luo and colleagues [81] were interested in identifying the SNPs at positions −1584 and 2988 G>A used as markers of *41 due to previous findings that these changes may predict metabolic activity of the carriers. These findings highlight the fact that genotype-phenotype associations found in a certain population should not be extrapolated to other ethnic groups.
Another study by Casner [82] genotyped the variants *4, *5, *10, and *17 and gene duplications (*XN, *XN/*4, *XN/*17) in Mexican-Americans, finding a MAF of 0.17, 0.02, 0.01, 0.02, and 0.03, respectively, and showing that the nonfunctional *4 was the most frequent of the investigated alleles [82]. The *6 and *3 variants were not found by Casner, while the *7 and *8 variants are apparently absent in Mexican-Americans according to published studies [81, 82]. Mendoza et al. [83] also studied a Mexican-American population and found that the most common variants were *2 and *4, followed by *10 and *5 and CYP2D6*XN (unidentified alleles) duplications (Table 3). This study also found that 63% of individuals with PM can be explained by the presence of *4. Mendoza et al. [83] explained that the detected allele frequencies for *4 and *10 in Mexican-Americans were more similar to the Spanish (0.12) than to the Asian (<0.001) population.
Currently, there is not much information available about genetic polymorphisms in the CYP2D6 gene for the Cuban population. Llerena and colleagues [80] studied CYP2D6 genetic variants in White-Cubans and Cuban mestizos and also included Nicaraguan-mestizos (Nicaraguans living in Nicaragua) for comparison purposes. In this study, the individuals were also phenotyped with debrisoquine to determine metabolic rate. The most frequent variants they found were *1 and *2, followed by *4 and *17 (Table 3). Among white Cubans, the MAFs for the *1 and *2 variants together were 0.754, for the *4 variant was 0.146, and for 17 variant was 0.027, while in Cuban-mestizos, the frequencies for the same CYP2D6 variants were 0.663 (*1 and *2 together), 0.143, and 0.102, respectively. CYP2D6 variants *5, *6, and *10 were also genotyped, and lower frequencies were found in both White-Cuban and Cuban-mestizos. Gene amplifications of normal alleles (*1XN or *2XN) were reported in frequencies within 0.038–0.047, while the −4XN variant was not present in Cuban-mestizos but was found with a frequency of 0.004 among White-Cubans. The CYP2D6*3 variant was not found in any individual [80].
It is important to highlight the differences in *17 prevalence even within the Cuban population; Cuban-Mestizos showed higher prevalence (0.102) when compared to White-Cubans (0.027) [80]. The CYP2D6*17 variant is found mostly in African populations, which might explain the higher allele frequency found among Cubans (0.027–0.102) and Puerto Ricans (0.033) when compared to Mexican-Americans (0.002–0.02) [79–83, 86]. This study suggested that differences in metabolism among extensive metabolizers could be explained by differential variations in defective or null alleles among populations and also to factors such as lifestyle and diet. Another study that evaluated the frequency of metabolic activities among Cubans (White-Cubans and Cuban-mestizos) and Spanish found similar frequencies of poor metabolizers between these ethnic groups (4.6% and 4.9%, respectively). But when comparisons were done among Cubans, the frequency of poor metabolizers was lower in Cuban-Mestizos (3.9%) when compared to White-Cubans (5.3%). In addition, the frequency of ultrarapid metabolizers in Spanish individuals (5.2%) was higher when compared to that in Cubans (3.8%) but almost identical when compared to that in White-Cubans (5.3%). The frequencies of the most common CYP2D6 allelic variants found in the global population for CYP2D6 *4, *9, *10, *17, and *41 are 0.106, 0.011, 0.256, 0.343, and 0.056, respectively [45].
CYP3A4/5
CYP3A4 and 3A5 belong to the CYP450s family 3 located in the 7q21.1 locus [15, 8 7, 88]. The CYP3A4 and CYP3A5 genes are about 27 Kb (ID:1576) and 32 Kb (1577) in size, respectively. The protein products of CYP3A4 (GI:13435386) and 3A5 (GI:4503231) are around 500 amino acids. Both are expressed in the endoplasmic reticulum of liver cells and intestines, but CYP3A5 is also expressed in lungs, prostate, and kidneys [89–91]. CYP3A4/5 interacts with large lipophilic substances and has overlapping substrate specificities. Endogenously, they are involved in the synthesis of sex hormones (progesterone and testosterone). CYP3A4/5 metabolizes a vast amount and wide spectrum of commonly used drugs in clinics, which include from antimicrobial and chemotherapeutic agents to psychoactive drugs (i.e., nevirapine, tamoxifen, and halo-peridol) [15].
CYP3A4/5 functional and clinical significance of the most common variant alleles
The most commonly studied variants in CYP3A4/5 are CYP3A4*1B (rs2740574; −392G>A) and CYP3A5*3 (rs776746; 6986A>G). The CYP3A4*1B variant is a base change in the promoter region of the gene that has been associated to prostate cancer, therapy-related acute myeloid leukemia, and higher antiviral exposure. The CYP3A4*1B variant is most frequent in African populations [92–94]. On the other hand, CYP3A *3 (6986 A>G) produces an enzyme without function due to errors in splicing, most frequent among Europeans [95]. CYP3A4 is found in kidneys and has the ability to produce hydroxyl-cortisol, a molecule involved in sodium and water retention. The presence of the wild-type allele (CYP3A5*1) was thought to predispose to hypertension, which was not supported by previous studies [96].
Prevalence of the most common variants in US Hispanics
A case control study conducted by Blanco and colleagues [93] looked for an association between CYP3A4*1B and the risk of developing therapy-induced myelodysplastic syndrome in children affected by acute lymphoblastic leukemia. Children classified as white, black, or Hispanic were genotyped for CYP3A4*1B and CYP3A5*3. Even though frequencies differed significantly among populations, the authors did not find an association between CYP3A4 or CYP3A5 polymorphisms and the risk of suffering therapy-induced myelodysplastic syndrome. CYP3A5 was also interrogated due to the fact that previous studies have identified the wild-type allele of this gene to be in linkage disequilibrium with CYP3A4*1B. Respective MAFs reported for CYP3A4*1B and CYP3A5*3 were 0.70 and 0.27 for blacks, 0.20 and 0.84 for Hispanics, and 0.036 and 0.91 for whites [93] (see Table 4). Another study that included Hispanics with HIV (Adult AIDS Clinical Trials Group) interrogated variants in CYP2B6 and CYP3A4/5 related to efavirenz (antiretroviral) exposure and central nervous system side effects, both potential causes of intolerance and eventual treatment failure. The CYP2B6 gene product is the main metabolic enzyme for efavirenz, while CYP3A gene products are minor contributors. The patients were recruited in the United States and Puerto Rico. Reported allelic frequencies in Hispanics for CYP3A4*1B (392 A>G) and CYP3A5*3 (6986 A>G) were 0.40 and 0.867, respectively. The CYP3A4*1B and C YP3A5*3 variants were weakly associated with higher exposure to efavirenz. Overall, CYP2B6 variants were associated with greater efavirenz exposure and increased central nervous system side effects, but the authors refrained from making conclusions about Hispanics due to “ small sample size ” (n=15 Hispanics) [94]. Six years, later the Adult AIDs Clinical Trials Group performed genotyping in a larger study cohort (n=149 Hispanics) and reported a MAF of 0.76 for CYP3A5*3 in Hispanics, but this time, they did not find an association between CYP3A5*3 and efavirenz exposure in any population [98]. Not far from the MAF found for CYP3A5*3 in Hispanics, Puerto Ricans present minor allele of 0.764 for this gene, while 0.20 was found for CYP3A4*1B according to data from the 1000 Genomes Project [45].
Table 4.
Distribution of CYP3A4/5 allelic variants among Hispanics of undetermined ethnicities and Puerto Ricans.
| Population | Na | CYP3A4*1B | CYP3A5*3 | CYP3A5*6 | Reference |
|---|---|---|---|---|---|
| Hispanics | 27 | 0.20 | 0.84 | [93] | |
| Hispanics | 15 | 0.40 | 0.867 | [94] | |
| Hispanics | ~149 | 0.760 | [95] | ||
| Hispanics | 55 | 0.164 | 0.245 | [96] | |
| Hispanics | 188 | 0.093 | [97] | ||
| Hispanics | 334 | 0.700 | 0.040 | [96] | |
| Puerto Ricans | 55 | 0.20 | 0.764 | [45] |
Number of individuals.
Establishing a potential association between CYP3A5*3 and other antiviral drugs is a subject of research in other studies as well. Zhang and colleagues [95] found that CYP3A5*3 did not affect the minimal concentration in plasma of the protease inhibitors lopinavir, amprenavir, and saquinavir. Global population MAF for CYP3A5*3 is 0.312 [45]. MAFs reported for CYP3A5*3 in Hispanics, blacks, and whites by Zhang et al. [95] were 0.755, 0.281, and 0.928, respectively. The mutant G allele is the most common in European descendants; therefore, the enzymatic activity of CYP3A5 is expected to be importantly reduced in this population [95]. As reported in previously mentioned studies, the G allele of C YP3A5 is also highly prevalent among Hispanics (Table 4) [93–96, 98]. Black-American and Hispanic patients who have two CYP3A5*1 functional alleles presented higher systolic blood pressure after verapamil antihypertensive therapy. Potential predisposition of CYP3A5*1 homozygotes to have higher systolic blood pressure might explain these results but is not evidenced in the study. CYP3A5*6 (14690G>A) frequency (0.04) was also investigated in verapamil-treated Hispanics, and a weak association of this variant with systolic blood pressure response to verapamil was found in blacks and Hispanics [96].
Paris et al. [92] found that CYP3A4*1B was significantly associated with prostate cancer in African-American men, and indeed, this polymorphism is most frequent in this population. This fact should be of concern for Caribbean Hispanics since they have a relatively large African ancestral contribution when compared to other non-Caribbean Hispanics. In fact, prostate cancer is the most prevalent cancer in Puerto Rican men [99]. CYP3A4*1B presents with wide variation among different populations: almost not found in Asians, prevalent among Hispanics, and more common in African-Americans than the wild-type (A) allele. The MAF of the CYP3A4*1B variant in the global population is 0.201 [45]. Paris [92] and colleagues selected a group of Hispanics from southern California and found that the frequency of the CYP3A4*1B variant in this population was 0.20, while Zhang et al. reported a frequency of 0.164 in Hispanics [92, 95]. Questions about whether the CYP3A4*1B variant is the causal polymorphism associated with prostate cancer and therapy-induced myelodysplastic syndrome merit further research. A large study conducted with about 800 individuals that included a broad range of racial groups aimed to genotype for the CYP3A4*1B variant and determine the related enzyme’s activity. Hispanic volunteers were recruited from Southern California and presented a frequency of 0.093 of CYP3A4*1B. Similarly to previously mentioned studies, this polymorphism was also highly prevalent in Black-Americans than in other racial groups but was not associated to lower metabolism of erythromycin [97].
Phase II metabolizing enzymes: N-acetyl transferases
N-acetyl transferase (NAT) 1 and 2, known as NAT1 (ID:9) and NAT2 (ID:10) genes, respectively, are located in chromosome 8p22 and comprehend a region of 180 Kb [100, 101]. NAT1 is present in all human tissues, whereas NAT2 is expressed mainly in liver and gastrointestinal tract [14]. NATs are responsible of the addition of an acetyl group derived from acetyl-coenzyme A to molecules containing an aromatic amine or hydrazine group, resulting in either activation or detoxification of these compounds [14, 102]. NAT enzymes metabolize isoniazid (used as treatment for tuberculosis), hydralazine (antihypertensive drug), and benzocaine (local anesthetic) [14]. Polymorphisms in NAT s are annotated in an online database (http://nat.mbg.duth.gr). Polymorphisms associated to slow acetylation may lead to accumulation of adducts, raising the question of a possible involvement in birth defects (clubfoot) and cancer (breast and bladder) [14, 100, 102]. A study conducted on Hispanics from New Mexico interrogated polymorphisms associated to NAT2 rapid acetylation: *4 (reference), *12 (rs1208 ; 803 A>G), and *13 (rs1041983 ; 282 C>T), whose frequencies are shown in Table 5. Polymorphisms associated with slow acetylation phenotype are *6 (rs1799930; 590G>A) and *7 (rs1799931 ; 857 G>A), and their prevalence, as well as for *5 alleles (rs1801280 ; 803 A>G; *5A, *5B, and 5C), is found in Table 5. In the same study, alleles associated with slow acetylation were more prevalent among non-Hispanic whites than among Hispanics [102]. Another study by Hecht and colleagues [100] found MAFs for NAT2*5, *6, *7, *11 (rs1799929 ; 481 C>T), and *13 (rs1041983; 282 C>T) equal to 0.3274, 0.1956, 0.1204, 0.3227, and 0.2996, respectively. In a different study, Lin et al. [103] genotyped only alleles *6, *7, and *11, finding minor allele frequencies corresponding to 0.23–0.32, 0.17–0.19, and 0.10–0.17, respectively, among two Hispanic populations living in California and Harbor. NAT1 variant alleles were found at frequencies of 0.4886 for NAT1*3 and 0.4972 for NAT1*10 [100]. Concerning Puerto Ricans, NAT1*3, NAT2*5, *6, *7, *11, *12, and *13 have MAFs as presented in Table 5. The worldwide frequencies for NAT1 and NAT2 alleles are 0.424, 0.296, 0.245, 0.072, 0.278, 0.315, and 0.356, respectively [45].
Table 5.
Distribution of NATI, NAT2, and GST allelic variants among Hispanics of undetermined ethnicities and Puerto Ricans.
| Population | Na | NAT1*3 | NAT1*4 | NAT*10 | NAT2*5 | NAT2*5A | NAT2*5B | NAT2*5C | NAT2*6 | NAT2*7 | NAT2*7B | NAT2*11 | NAT2*12A | NAT2*13 | GSTM1*0 | Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Hispanics | 157 | 0.489 | 0.497 | 0.327 | 0.196 | 0.120 | 0.323 | 0.300 | [100] | |||||||
| Hispanics | 496 | 0.289–0.314 | 0.004–0.006 | 0.355–0.359 | 0.002–0.012 | 0.076–0.092 | 0.014–0.024 | 0.002–0.010 | [102] | |||||||
| Hispanics | 296 | 0.170–0.190 | 0.100–0.170 | 0.230–0.320 | [103] | |||||||||||
| Puerto Ricans | 55 | 0.309 | 0.400 | 0.200 | 0.091 | 0.373 | 0.300 | [45] | ||||||||
| Hispanics | 146 | 0.400 | [104] |
Number of individuals.
Uridine diphosphate-glucuronosyl-transferases
Uridine diphosphate (UDP)-glucuronosyl-transferases (UGTs) are encoded by 19 genes, from which 9 located in chromosome 2 produce the enzymes known as UGT1 and 10 located in chromosome 4 produce UGT2. UGT enzymes transfer glucuronic acid from UDP-glucuronic acid to compounds in order to be excreted out of the body. The expression of these enzymes occurs mainly in the liver and gastrointestinal tract. UGT1A is an enzyme of great physiological relevance since it catalyzes the glucuronidation of bilirubin, a conversion needed for the excretion of this endogenous substance derived from breakdown of hemoglobin and proteins that contain heme-group [105]. Drugs metabolized by UGTs are irinotecan, acetaminophen, morphine, and oxazepam [14]. Pharmacogenetic studies of UGT s in Hispanics are not abundant. Li and colleagues reported MAFs for seven SNPs in UGT2B7 in Hispanics. The database for UGT allelic variants is available at http://www.pharmacogenomics.pha.ulaval.ca/cms/ugt_alleles/ [106]. Frequencies reported by Li et al. [107] were retrieved from the 1000 Genomes Project until 2012, which included 17 Mexican-Americans and 5 Puerto Ricans. In order to provide updated information, the same SNPs reported by Li et al. [107] were accessed from the 1000 Genomes Project for this review (Table 6). The −327 G>A (rs7662029), −138 G>A (rs73823859), −125 T>C (rs7668282), 211 G>T (rs12233719), IV1S +985 A>G (rs62298861), 802 T>C (rs7439366), and 1191 C>T (rs57913007) mutated alleles are present in Puerto Ricans with a frequency of 0.600, 0.055, 0.973, 0.009, 0.236, 0.409, and 0.055, respectively, and for Mexicans-Americans, these SNP frequencies were 0.265, 0.038, 0.008, 0.008, 0.432, 0.311, and 0.038, respectively. In the global population, these allelic variants are present with a frequency of 0.356, 0.030, 0.032, 0.039, 0.185, 0.397 and 0.031, respectively [45].
Table 6.
Distribution of UGT2B7 allelic variants among Puerto Ricans and Mexican-Americans.
| Population | Na | UGT2B7 (rs7662029) |
UGT2B7 (rs73823859) |
UGT2B7 (rs7668282) |
UGT2B7 (rs12233719) |
UGT2B7 (rs62298861) |
UGT2B7 (rs7439366) |
UGT2B7 (rs57913007) |
Reference |
|---|---|---|---|---|---|---|---|---|---|
| Puerto Ricans | 55 | 0.400 | 0.055 | 0.027 | 0.009 | 0.236 | 0.409 | 0.055 | [45] |
| Mexican-Americans | 66 | 0.265 | 0.038 | 0.008 | 0.008 | 0.432 | 0.311 | 0.038 | [45] |
Number of individuals.
Glutathione S-transferases
Glutathione S-transferases (GSTs) are classified as cytosolic and microsomal. The cytosolic type is divided in classes termed as alpha (GSTA), mu (GSTM), omega, pi, sigma, theta (GSTT), and zeta. The genes for the different classes are dispersed among different human chromosomes. The main function of GSTs is to transfer glutathione to reactive electrophilic molecules, preventing the interaction of the latter with macromolecules (proteins, lipids, DNA, and carbohydrates). Endogenous ligands of GSTs are hydrophobic molecules such as prostaglandins, leukotriene, heme, bilirubin, and bile acids [14, 108]. GSTs are involved in the inactivation of anticancer drugs. GSTs are also polymorphic, where GSTM1 and GSTT1 are common among all populations [109, 110]. Both GSTM1*0 and GSTT1*0 present a gene deletion that produces a null phenotype, which may result in drug-induced toxicity of chemotherapeutic agents [14]. GSTM1*0 frequency has a prevalence of 0.40 among Mexican-Americans, quite close to that of Mexican-Mestizos (0.426) (Table 5) [104, 111]. A study by Woo and colleagues [109] aimed to determine the association of GST null alleles with the predisposition to present treatment-related myeloid malignancies in children with acute lymphoblastic leukemia (ALL). Among Hispanics, 36% of study controls (patients with ALL but not myeloid malignancies) and 50% of study cases (patients with ALL and myeloid malignancies) had GSTM1 null allele, while 11% of controls and 17% of cases had the GSTT1 null allele. The percentage of controls and cases presenting both null alleles (GSTM1 and GSTT1) was 7% and 17% of Hispanics, respectively. Woo et al. [109] found that carriers of GSTM1 and GSTT1 null alleles are not predisposed to develop treatment-induced myeloid malignancies.
Clinical implementations in US Hispanics
Currently, multiple sites in the US mainland have been operationally implementing a prospective (often preemptive) genotyping program by interrogating many of the pharmacogenes herein discussed. Such comprehensive programs (e.g., Vanderbilt PREDICT project, Mayo Clinic) are linked to advanced, computer-aided, clinical (point-of-care) decision-support systems and algorithms and have been deployed at medical and academic health institutions to guide individualized healthcare (i.e., the choice of prescription medicines and dosages for each patient) in clinical settings. Although these initiatives are also available for people of Hispanic heritage residing in the United States, cultural, socioeconomic, and legal barriers limit their access to these programs. Clinical Pharmacogenetics Implementation Consortium guidelines for drugs metabolized through CYP2C9, CYP2C19, CYP2D6, and TPMT are currently available, but their validity and clinical utility have not yet been fully investigated in US Hispanics [112 115]. To the best of our knowledge, only a limited number of studies have been conducted in US Hispanics to validate and ascertain the benefits of genotyping warfarin pharmacogenes (CYP2C9 and VKORC1) in this population, but not for the other genetic markers.
The corresponding derivation or replication cohorts for some of the available multiethnic pharmacogenetic algorithms (e.g., International Warfarin Pharmacogenetics Consortium [IWPC] derived, Gage et al. algorithm, and Wu et al. algorithm) have included a few individuals of Hispanic descent; however, there are reports of pharmacogenetic-guided algorithms developed to predict effective warfarin dosing in US Hispanic populations (i.e., Mexican-Americans and Puerto Ricans). These algorithms include the clinically actionable CYP2C9 and VKORC1 polymorphisms commonly interrogated in other populations (e.g., CYP2C9*2, *3 and VKORC1-1639G>A) [48, 52, 116].
The algorithm for warfarin dose prediction in Puerto Ricans explained two-thirds (R2=0.68) of variability in dose requirements and provided better dose estimations than the clinical algorithms did [52]. In addition, a pharmacogenetic algorithm for 3-day initial dosing that includes genotypes, admixture, and clinical covariates explained 48% of the dose variability in Puerto Ricans. A pharmacogenetic dose-refinement algorithm after the third day accounted for 76% of observed variability, after incorporating natural logarithm-transformed INR at day 4 and initial doses for days 1–3 [116]. The algorithms provided consistently better dose predictions in Puerto Ricans than the IWPC algorithm did, particularly for patients who required low or intermediate doses (≤ 7 mg/day).
Another study by Cavallari et al. [48] investigated the performance of pharmacogenetic-driven dosing algorithms in 50 US Hispanics from Chicago, most of whom were of Mexican descent. The authors developed a pharmacogenetic regression model to predict warfarin dosing in the study population and found that patients with CYP2C9*2 or *3 required 42% less of the warfarin dose than noncarriers did. Polymorphisms in CYP2C9 and VKORC1 explained 55% of the interpatient variability in warfarin dose among Hispanics. Their Hispanic cohort had a similar prevalence of CYP2C9 and VKORC1 alleles when compared to European Caucasians, which explains the effectiveness of previously published dosing algorithms [48]. On the other hand, the Clarification of Optimal Anticoagulation Through Genetics trial selected a group of 65 Hispanics among the studied cohort to compare the pharmacogenetic vs. clinical algorithms, but the data analysis of this subgroup was not performed separately from other studied populations [5].
Conclusions
This review article discussed the prevalence of some clinically relevant polymorphisms in pharmacogenes encoding for drug-metabolizing enzymes (phase I and II), addressing a need for compiling available pharmacogenetic data in the admixed Hispanic populations living within the Unites States. Reviewed data for this publication consider mainly allele frequencies reported for the biggest Hispanic populations in the United States: Mexican-Americans, Puerto Ricans, and Cubans (Supplemental Material, Table 1, that accompanies the article, http://www.degruyter.com/view/j/dmdi.2015.30.issue-2/dmdi-2015-0023/dmdi-2015-0023.xml?format=INT) but also considers data from the 1000 Genomes Project, further including data of Colombians. From the alleles reviewed, the most prevalent alleles found in Hispanics were CYP2C9*3 (rs 1057910; 1075A>C at exon 7) that results in decreased CYP2C9 activity and the loss-of-function CYP2C19*4 (rs 28399504; 1A>G null) identified in mephenytoin poor-metabolizers (Supplemental Material, Table 2) [117]. CYP2C9*3 frequency in Hispanics is close to the allele frequency reported in Europeans genotyped in the 1000 Genomes Project. CYP2C19*4 is a rare variant not found in Africans or Europeans (as reported by the 1000 Genomes Project) but was reported in 0.002 of Asians, which is a lower prevalence when compared to Hispanics (ranges from 0.000 to 0.006). UGT2B7 IVS1+985 A>G (rs62298861) is a change in intron 1 associated with an increase in morphine glucuronidation and mRNA levels, with higher frequency in Hispanics than in other populations [118]. CYP2C9*13; CYP2C19 *5, *6, *7, *10, *12, *16, and *22; and CYP2D6 *7, *8, *11, *14, and *45 were not found in Hispanics. There are no reports about novel polymorphisms found in Hispanics as far as we know.
Currently, a few ongoing international efforts in this field (e.g., the 1,000 Genomes Project) are recruiting individuals of Hispanic heritage (i.e., Colombians, Mexican-Americans, and Puerto Ricans), becoming an invaluable resource for gaining knowledge about the frequency and distribution of common genetic variants in these often marginally represented minority populations, thus bridging the gap of information between white Caucasians and these ethnically diverse groups. In addition, some independent studies of relatively small sample sizes have been conducted by a limited number of researchers. However, there are still quite a few issues that need to be addressed before making significant progress in the field with regard to the inclusion of Hispanics for a proper pharmacogenetic ascertainment. Some of these concerns are highlighted below.
The first and most important point to consider is the pharmacogenetic variability and the wide range of genetic diversity (i.e., mosaic of ancestries) observed within the Hispanic population. We investigated the population structure of 35 Puerto Ricans using the Hap1M Duo Bead-Chip™ on Illumina iScan™ by a high-resolution total genome-driven pairwise allelic dissimilarity analysis and compared the result to a randomly selected subset of a US reference population from Kentucky (30 Caucasians and 10 African-Americans). It can be seen clearly that the reference population is segregated into two distinct groups (Figure 2). As suspected, all the dark stripes (no. 31–40) in the upper panel belong to individuals who self-identified as African-Americans. It is remarkable how clearly the whole genome array is able to distinguish ethnic origin here. The gradation in the black bands indicates the range of Caucasian admixture in the African-American group. There appears to be little corresponding admixture in the Caucasians (stripes, no. 1–30). The same procedure on Puerto Ricans yields a much different image (bottom). The population is much more admixed and there is no clear distinction between separate groups. However, the average distance between individuals is clearly larger than within the same ethnic group in the reference population, as indicated by the darker background (gray) color. It is easy to use ancestry as a covariate in the United States and Europe because the grouping is so distinctive and categorical. For Hispanics, the admixture is continuous and is best quantified by a vector.
Figure 2.
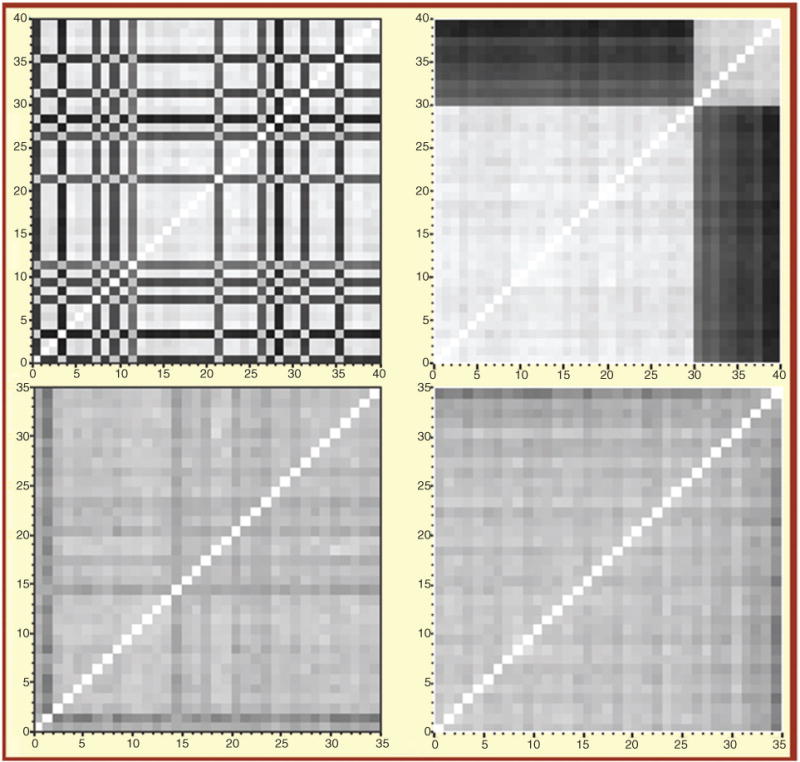
Genome-wide allelic dissimilarity and genomic admixture of a Puerto Rican population (n=35), as compared to a reference population from Kentucky (n=40, including European- and African-Americans).
Plot depicts allelic dissimilarities as a distance matrix for reference (top) and Puerto Ricans (bottom). Each square represents a pair of individuals. Data are shown with samples reordered according to nearest neighbor clustering (right panels) and random order (left panels). The darker the spot, the more genetically distant the individuals are.
In this review article, we reported results from several published studies on Hispanics without a clear definition of their ethnic or geographic origin, which makes it difficult to draw valid conclusions on the relevance of the findings to a certain ethnic group (e.g., Caribbean Hispanics). However, cultural barriers to recruit and retain individuals of Hispanic origin in clinical studies also need further consideration. The label “ Hispanic or Latino ” comprises multiple countries of origin that share the same language and culture and similar habits and history, but they should not be considered as a single monolithic population for pharmacogenetic purposes. Intergroup differences, cryptic population structures, and stratification within the Hispanic population are expected due in part to a differential pattern of ancestral contributions and admixture, but intraethnic differences have also been observed. For example, CYP2C9*3 prevalence is higher in Mexican-Americans than in Mexican-Mestizos and Mexican indigenous Tapehuana [46, 47, 50]. CYP2C9 polymorphisms also showed intraethnic variations among Cuban-Mestizos and Cuban-Whites [54]. Therefore, given the great genetic diversity within Hispanics, we should treat the person as an individual rather than a member of an ethnic group while applying the DNA-guided personalized medicine paradigm in this population [8, 80].
Considering the high level of admixture and heterogeneity of Hispanics, new methodological approaches should be considered as part of the experimental designs to gain more insight into the clinical relevance of genetic polymorphisms involved in drug responses among these groups. Some of these methods are admixture mapping, pathway analyses, and population-based sequencing. Changes in linkage disequilibrium need to be considered given a distinct population structure that characterizes Hispanics as a consequence of a differential contribution from multiple ancestries; new putative markers or association signals may arise from a unique combination of haplotype blocks.
At present, the majority of the commercially available genotyping platforms include only the most common variants, failing to detect those “rare” polymorphisms showing higher prevalence in admixed minority populations [79]. González-Tejera and coworkers [79] interrogate the CYP2D6 gene for polymorphisms on this locus in Puerto Rican patients who are intolerant to drugs metabolized by this enzyme. Strikingly, two patients carried the rare variant CYP2D6*31, and one has the CYP2D6*40 variant, both of them not included in commercial platforms. The authors also found uncommon combinations of alleles and suggested that the Puerto Rican population is unique and may harbor other alleles less common to other populations [79].
Pharmacogenetic studies in the admixed Hispanic population should not be limited to ascertain commonly occurring allelic variants. Instead, they should also encompass other less frequent variants that might be either part of multiple causative loci or in extended linkage disequilibrium with functional alleles relevant for drug responses in a certain population. Further studies are warranted to accomplish this major goal. The success of the Personalized Medicine paradigm will depend on our capabilities to recognize the genetic diversity existing between and within different ethnic groups, as well as the uniqueness of their genetic backgrounds [8, 80].
Supplementary Material
Acknowledgments
The material in this review is partially the result of work supported with resources and the use of facilities at the Veteran Affairs Caribbean Health System in San Juan, Puerto Rico. We thank the UPR-MSC RCMI Center for Genomics in Health Disparities and Rare Disorders and the Laboratory of Personalized Medicine, Hartford, CT, for support. We also thank Dr. Susan Corey, Dr. Andrea Gaedigk, and Mr. Freddie Hernandez for their help in this review. Finally, we thank all patients for their participation in these studies.
Research funding: This work was supported in part by a grant from the National Heart, Lung and Blood Institute (SC2HL110393) and Research Center in Minority Institutions (RCMI) grants from the National Center for Research Resources (2G12-RR003051) and the National Institute on Minority Health and Health Disparities (8G12-MD007600) of the National Institutes of Health (NIH). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH or the US Government.
Footnotes
Author contributions: All the authors have accepted responsibility for the entire content of this submitted manuscript and approved submission. They have no other relevant affiliation or financial involvement with any organization or entity with a financial interest in or conflicts of interest with the subject matter or materials discussed in the article that need to be disclosed. No writing assistance was utilized in the production of this manuscript.
Employment or leadership: G. Ruaño is founder and president of Genomas Inc. Jorge Duconge and Karla Claudio-Campos also held a without compensation (WOC) employment status with the VA Caribbean Healthcare Systems (VACHS), Pharmacy Service, in San Juan, Puerto Rico.
Honorarium: None declared.
Competing interests: The funding organization(s) played no role in the study design; in the collection, analysis, and interpretation of data; in the writing of the report; or in the decision to submit the report for publication.
Supplemental Material: The online version of this article (DOI: 10.1515/dmdi-2014-0023) offers supplementary material, available to authorized users.
Contributor Information
Karla Claudio-Campos, Email: karla.claudio2@upr.edu, School of Medicine, Medical Sciences Campus, Department of Pharmacology and Toxicology, University of Puerto Rico, PO Box 365067, San Juan, Puerto Rico, PR 00936-5067, Phone: (787)-758-2525 ext. 1638, Fax: (787)-767-2796.
Jorge Duconge, School of Pharmacy, Medical Sciences Campus, Pharmaceutical Sciences Department, University of Puerto Rico, San Juan, Puerto Rico.
Carmen L. Cadilla, Molecular Genetics Lab, School of Medicine, Medical Sciences Campus, Department of Biochemistry, University of Puerto Rico, San Juan, Puerto Rico
Gualberto Ruaño, Genetics Research Center, Hartford Hospital, Hartford, CT, USA.
References
- 1.U.S. Food and Drug Administration (FDA) Codeine Use in Certain Children after tonsillectomy and/or adenoidectomy: Drug Safety Communication-risk of rare, but life-threatening adverse events or death. Available at: www.fda.gov/safety/MedWatch/SafetyInformation/SafetyAlertsforHumanMedicalProducts/ucm315627.htm (accessed April 18, 2014)
- 2.Meyer UA. Adverse drug reactions. Pharmacogenetics and adverse drug reactions. Lancet. 2000;356:1667–71. doi: 10.1016/S0140-6736(00)03167-6. [DOI] [PubMed] [Google Scholar]
- 3.Budnitz DS, Pollock DA, Weidenbach KN, Mendelsohn AB, Schroeder TJ, Annest JL. National surveillance of emergency department visits for outpatient adverse drug events. J Am Med Assoc. 2006;296:1858–66. doi: 10.1001/jama.296.15.1858. [DOI] [PubMed] [Google Scholar]
- 4.Phillips KA, Veenstra DL, Oren E, Lee JK. Potential role of pharmacogenomics: a systematic review. J Am Med Assoc. 2001;286:2270–9. doi: 10.1001/jama.286.18.2270. [DOI] [PubMed] [Google Scholar]
- 5.Kimmel SE, French B, Kasner SE, Johnson JA, Anderson JL, Gage BF, et al. A pharmacogenetic versus a clinical algorithm for warfarin dosing. N Engl J Med. 2013;369:2283–93. doi: 10.1056/NEJMoa1310669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ramos E, Callier SL, Rotimi CN. Why personalized medicine will fail if we stay the course. Per Med. 2013;9:839–47. doi: 10.2217/PME.12.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Suarez-Kurtz G. Pharmacogenomics in admixed populations. Austin, Texas: Landes Bioscience; 2007. [Google Scholar]
- 8.Suarez-Kurtz G, Pena SD. Pharmacogenomics in the Americas: the impact of genetic admixture. Curr Drug Targets. 2006;7:1649–58. doi: 10.2174/138945006779025392. [DOI] [PubMed] [Google Scholar]
- 9.Via M, Gignoux CR, Roth LA, Fejerman L, Galanter J, Choudhry S, et al. History shaped the geographic distribution of genomic admixture on the island of Puerto Rico. PLoS One. 2011;6:e16513. doi: 10.1371/journal.pone.0016513. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3031579&tool=pmcentrez&rendertype=abstract. Cited May 4, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ruaño G, Duconge J, Windemuth A, Cadilla CL, Villagra D, Renta J, et al. Physiogenomic analysis of the Puerto Rican population. Pharmacogenomics. 2010;10:565–77. doi: 10.2217/pgs.09.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bryc K, Velez C, Karafet T, Moreno-Estrada A, Reynolds A, Auton A, et al. Colloquium paper: genome-wide patterns of population structure and admixture among Hispanic/Latino populations. Proc Natl Acad Sci USA. 2010;107:8954–61. doi: 10.1073/pnas.0914618107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ong RT, Teo Y-Y. varLD: a program for quantifying variation in linkage disequilibrium patterns between populations. Bioinformatics. 2010;26:1269–70. doi: 10.1093/bioinformatics/btq125. [DOI] [PubMed] [Google Scholar]
- 13.U.S. Census Bureau. Profile America facts for features. Hispanic Heritage Month 2013: Sept. 15–Oct. 15. Available at: https://www.census.gov/content/dam/Census/newsroom/facts-for-features/2013/cb13ff-19_hispanicheritage.pdf (accessed April 18, 2014)
- 14.Goodman LS, Gilman A, Brunton LL, Lazo JS, Parker KL. In: Goodman & Gilman’s the pharmacological basis of therapeutics. 11. Brunton LL, Lazo JS, Parker KL, editors. New York: McGraw-Hill; 2005. [Google Scholar]
- 15.Zanger UM, Schwab M. Cytochrome P450 enzymes in drug metabolism: regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol Ther. 2013;138:103–41. doi: 10.1016/j.pharmthera.2012.12.007. [DOI] [PubMed] [Google Scholar]
- 16.Van Booven D, Marsh S, McLeod H, Carrillo MW, Sangkuhl K, Klein TE, et al. Cytochrome P450 2C9-CYP2C9. Pharmacogenet Genomics. 2010;20:277–81. doi: 10.1097/FPC.0b013e3283349e84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goldstein JA. Clinical relevance of genetic polymorphisms in the human CYP2C subfamily. Br J Clin Pharmacol. 2001;52:349–55. doi: 10.1046/j.0306-5251.2001.01499.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Williams PA, Cosme J, Ward A, Angove HC, Matak Vinković D, Jhoti H. Crystal structure of human cytochrome P450 2C9 with bound warfarin. Nature. 2003;424:464–8. doi: 10.1038/nature01862. [DOI] [PubMed] [Google Scholar]
- 19.Rifkind A, Lee C, Chang TK, Waxman DJ. Arachidonic acid metabolism by human cytochrome oxygenation and evidence for a role for CYP2C enzymes in arachidonic acid epoxygenation in human liver microsomes. Arch Biochem Biophys. 1995;380:380–9. doi: 10.1016/0003-9861(95)90023-3. [DOI] [PubMed] [Google Scholar]
- 20.Human Cythochrome P450 (CYP) Allele Nomenclature Committee. The Human Cytochrome P450 (CYP) Allele Nomenclature Database. Available at: http://www.cypalleles.ki.se/index.htm. Cited April 19, 2014.
- 21.Cariaso M, Lennon G, SNPedia SNPedia: a wiki supporting personal genome annotation, interpretation and analysis. 2012:D1308–D1312. doi: 10.1093/nar/gkr798. Available at: http://www.snpedia.com/index.php/Rs4986893. Cited June 23, 2014. [DOI] [PMC free article] [PubMed]
- 22.Rettie AE, Wienkers LC, Gonzalez FJ, Trager WF, Korzekwa KR. Impaired (S)-warfarin metabolism catalysed by the R144C allelic variant of CYP2C9. Pharmacogenetics. 1994;4:39–42. doi: 10.1097/00008571-199402000-00005. [DOI] [PubMed] [Google Scholar]
- 23.Crespi C, Miller V. The R144C change in the CYP2C9*2 allele alters interaction of the cytochrome P450 with NADPH: cytochrome P450 oxidoreductase. Pharmacogenetics. 1997;7:203–10. doi: 10.1097/00008571-199706000-00005. [DOI] [PubMed] [Google Scholar]
- 24.van der Weide J, Steijns LS, van Weelden MJ. The effect of genetic polymorphism of cytochrome P450 CYP2C9 on phenytoin dose requirement. Pharmacogenetics. 2001;11:287–91. doi: 10.1097/00008571-200106000-00002. [DOI] [PubMed] [Google Scholar]
- 25.Dorado P, Lopez-Torres E, Penas-Lledo E, Martinez-Anton J, Llerena A. Neurological toxicity after phenytoin infusion in a pediatric patient with epilepsy: influence of CYP2C9, CYP2C19, and ABCB1 genetic polymorphisms. Pharm J. 2013;13:359–61. doi: 10.1038/tpj.2012.19. [DOI] [PubMed] [Google Scholar]
- 26.Hallberg P, Karlsson J, Kurland L, Lind L, Kahan T, Malmqvist K, et al. The CYP2C9 genotype predicts the blood pressure response to irbesartan: results from the Swedish Irbesartan Left Ventricular Hypertrophy Investigation vs Atenolol (SILVHIA) trial. J Hypertens. 2002;20:2089–93. doi: 10.1097/00004872-200210000-00030. [DOI] [PubMed] [Google Scholar]
- 27.King BP, Khan TI, Aithal GP, Kamali F, Daly AK. Upstream and coding region CYP2C9 polymorphisms: correlation with warfarin dose and metabolism. Pharmacogenetics. 2004;14:813–22. doi: 10.1097/00008571-200412000-00004. [DOI] [PubMed] [Google Scholar]
- 28.Steward DJ, Haining RL, Henne KR, Davis G, Rushmore TH, Trager WF, et al. Genetic association between sensitivity to warfarin and expression of CYP2C9*3. Pharmacogenetics. 1997;7:361–7. doi: 10.1097/00008571-199710000-00004. [DOI] [PubMed] [Google Scholar]
- 29.Cavallari L, Langaee TY, Momary K, Shapiro N, Nutescu E, Coty W, et al. Genetic and clinical predictors of warfarin dose requirements in African Americans. Clin Pharmacol Ther. 2010;87:459–64. doi: 10.1038/clpt.2009.223. [DOI] [PubMed] [Google Scholar]
- 30.Seip RL, Duconge J, Ruaño G. Implementing genotype-guided antithrombotic therapy. Future Cardiol. 2010;6:409–24. doi: 10.2217/fca.10.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taube J, Halsall D, Baglin T. Influence of cytochrome P-450 CYP2C9 polymorphisms on warfarin sensitivity and risk of over-anticoagulation in patients on long-term treatment. Blood. 2000;96:1816–9. [PubMed] [Google Scholar]
- 32.Wu AH. Use of genetic and nongenetic factors in warfarin dosing algorithms. Pharmacogenomics. 2007;8:851–61. doi: 10.2217/14622416.8.7.851. [DOI] [PubMed] [Google Scholar]
- 33.Hillman MA, Wilke RA, Caldwell MD, Berg RL, Glurich I, Burmester JK. Relative impact of covariates in prescribing warfarin according to CYP2C9 genotype. Pharmacogenetics. 2004;14:539–47. doi: 10.1097/01.fpc.0000114760.08559.dc. [DOI] [PubMed] [Google Scholar]
- 34.Scordo MG, Pengo V, Spina E, Dahl ML, Gusella M, Padrini R. Influence of CYP2C9 and CYP2C19 genetic polymorphisms on warfarin maintenance dose and metabolic clearance. Clin Pharmacol Ther. 2002;72:702–10. doi: 10.1067/mcp.2002.129321. [DOI] [PubMed] [Google Scholar]
- 35.Higashi MK, Veenstra DL, Kondo LM, Wittkowsky AK, Srinouanprachanh S, Farin FM, et al. Association Between CYP2C9 Genetic Variants and Anticoagulation-related outcomes during warfarin therapy. J Am Med Assoc. 2002;287:1690–8. doi: 10.1001/jama.287.13.1690. [DOI] [PubMed] [Google Scholar]
- 36.Schwarz UI, Ritchie MD, Bradford Y, Li C, Dudek SM, Frye-Anderson A, et al. Genetic determinants of response to warfarin during initial anticoagulation. N Engl J Med. 2008;358:999–1008. doi: 10.1056/NEJMoa0708078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.U.S. Food and Drug Administration (FDA) Medication Guide Coumadin (warfarin sodium) Available at: http://www.fda.gov/downloads/Drugs/DrugSafety/ucm088578.pdf (accessed June 17, 2014). Reference id: 3022954.
- 38.Valentin II, Rivera G, Nieves-Plaza M, Cruz I, Renta JY, Cadilla CL, et al. Pharmacogenetic association study of warfarin safety endpoints in Caribbean Hispanics. P R Health Sci J. 2014;33:97–104. [PMC free article] [PubMed] [Google Scholar]
- 39.Epstein RS, Moyer TP, Aubert RE, O Kane DJ, Xia F, Verbrugge RR, et al. Warfarin genotyping reduces hospitalization rates results from the MM-WES (Medco-Mayo Warfarin Effectiveness study) J Am Coll Cardiol. 2010;55:2804–12. doi: 10.1016/j.jacc.2010.03.009. [DOI] [PubMed] [Google Scholar]
- 40.Centers for Medicare and Medicaid Services. Decision memo for pharmacogenomic testing for warfarin response (CAG-00400N) Centers for Medicare and Medicaid Services; 2009. Available at: http://www.cms.hhs.gov/mcd/search.asp. Cited June 21, 2014. [PubMed] [Google Scholar]
- 41.Anderson JL, Horne BD, Stevens SM, Woller SC, Samuelson KM, Mansfield JW, et al. A randomized and clinical effectiveness trial comparing two pharmacogenetic algorithms and standard care for individualizing warfarin dosing (CoumaGen-II) Circulation. 2012;125:1997–2005. doi: 10.1161/CIRCULATIONAHA.111.070920. [DOI] [PubMed] [Google Scholar]
- 42.Wysowski DK, Nourjah P, Swartz L. Bleeding complications with warfarin use: a prevalent adverse effect resulting in regulatory action. J Am Med Assoc. 2007;167:1414–9. doi: 10.1001/archinte.167.13.1414. [DOI] [PubMed] [Google Scholar]
- 43.McWilliam A, Lutter R, Nardinelli C. (Working Paper 06–23, AE1-Brookings Joint Center for Regulatory Studies).Health care savings from personalized medicine using genetic testing: The case of warfarin. 2006 Available at: www.aei-brookings.org/admin/authorpdfs/page.php?id=1337&PHPsEssID=7b3a3ae4b-30d77cb76223e29535e7590. Cited October 18, 2012.
- 44.Eckman M, Rosand J, Greenberg S, Gage B. Cost-effectiveness of using pharmacogenetic information in warfarin dosing for patients with nonvalvular atrial fibrillation. Ann Intern Med. 2009;150:73–83. doi: 10.7326/0003-4819-150-2-200901200-00005. [DOI] [PubMed] [Google Scholar]
- 45.Amigo J, Salas A, Phillips C, Carracedo A. SPSmart: adapting population based SNP genotype databases for fast and comprehensive web access. BMC Bioinf. 2008;9:428. doi: 10.1186/1471-2105-9-428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Llerena A, Dorado P, O’Kirwan F, Jepson R, Licinio J, Wong M-L. Lower frequency of CYP2C9*2 in Mexican-Americans compared to Spaniards. Pharm J. 2004;4:403–6. doi: 10.1038/sj.tpj.6500278. [DOI] [PubMed] [Google Scholar]
- 47.Kramer MA, Rettie AE, Rieder MJ, Cabacungan ET, Hines RN. Novel CYP2C9 Promoter variants and assessment of their impact on gene expression. Mol Pharmacol. 2008;73:1751–60. doi: 10.1124/mol.107.044149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cavallari LH, Momary KM, Patel SR, Shapiro NL, Nutescu E, Viana MA. Pharmacogenomics of warfarin dose requirements in Hispanics. Blood Cells Mol Dis. 2011;46:147–50. doi: 10.1016/j.bcmd.2010.11.005. [DOI] [PubMed] [Google Scholar]
- 49.Rosemary J, Adithan C. The pharmacogenetics of CYP2C9 and CYP2C19: ethnic variation and clinical significance. Curr Clin Pharmacol. 2007;2:93–109. doi: 10.2174/157488407779422302. [DOI] [PubMed] [Google Scholar]
- 50.Dorado P, Sosa-Macias MG, Peñas-Lledó EM, Alanis-Bañuelos RE, Wong M-L, Licinio J, et al. CYP2C9 allele frequency differences between populations of Mexican-Mestizo, Mexican-Tepehuano, and Spaniards. Pharm J. 2011;11:108–12. doi: 10.1038/tpj.2010.29. [DOI] [PubMed] [Google Scholar]
- 51.Duconge J, Cadilla C, Windemuth A, Kocherla M, Gorowski K, Seip RL, et al. Prevalence of combinatorial CYP2C9 and VKORC1 genotypes in Puerto Ricans: implications for warfarin management in Hispanics. Ethn Dis. 2009;19:390–5. [PMC free article] [PubMed] [Google Scholar]
- 52.Ramos AS, Seip RL, Rivera-Miranda G, Felici-Giovanini ME, Garcia-Berdecia R, Alejandro-Cowan Y, et al. Development of a pharmacogenetic-guided warfarin dosing algorithm for Puerto Rican patients. Pharmacogenomics. 2012;13:1937–50. doi: 10.2217/pgs.12.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Valentin II, Vazquez J, Rivera-Miranda G, Seip RL, Velez M, Kocherla M, et al. Prediction of warfarin dose reductions in Puerto Rican patients, based on combinatorial CYP2C9 and VKORC1 genotypes. Ann Pharmacother. 2012;46:208–18. doi: 10.1345/aph.1Q190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Llerena A, Alvarez M, Dorado P, Gonzalez I, Peñas-Lledó E, Perez B, et al. Interethnic differences in the relevance of CYP2C9 genotype and environmental factors for diclofenac metabolism in Hispanics from Cuba and Spain. Pharm J. 2013;14:229–34. doi: 10.1038/tpj.2013.28. [DOI] [PubMed] [Google Scholar]
- 55.Scott SA, Khasawneh R, Peter I, Kornreich R, Desnick RJ. Combined CYP2C9, VKORC1 and CYP2F4 frequencies among racial and ethnic groups. Pharmacogenomics. 2010;11:781–91. doi: 10.2217/pgs.10.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu AH, Wang P, Smith A, Haller C, Drake K, Linder M, et al. Dosing algorithm for warfarin using CYP2C9 and VKORC1 genotyping from a multi-ethnic population: comparison with other equations. Pharmacogenomics. 2008;9:169–78. doi: 10.2217/14622416.9.2.169. [DOI] [PubMed] [Google Scholar]
- 57.International HapMap Consortium. (HapMap project dataset release no. 28, Phase I, II & III).International HapMap Project. 2010 Available at: http://hapmap.ncbi.nlm.nih.gov/. Cited September 1, 2010.
- 58.Villagra D, Duconge J, Windemuth A, Cadilla CL, Kocherla M, Gorowski K, et al. CYP2C9 and VKORC1 genotypes in Puerto Ricans: a case for admixture-matching in clinical pharmacogenetics studies. Clin Chim Acta. 2010;411:1306–11. doi: 10.1016/j.cca.2010.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Duconge J, Ruaño G. The emerging role of admixture in the pharmacogenetics of Puerto Rican Hispanics. J Pharm Pharmacoproteomics. 2010;1:1–9. doi: 10.4172/2153-0645.1000101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tayo BO, Teil M, Tong L, Qin H, Khitrov G, Zhang W, et al. Genetic background of patients from a university medical center in Manhattan: implications for personalized medicine. PLoS One. 2011;6:e19166. doi: 10.1371/journal.pone.0019166. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3087725&tool=pmcentrez&rendertype=abstract. Cited May 6, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Scott SA, Jaremko M, Lubitz SA, Kornreich R, Halperin JL, Desnick RJ. CYP2C9*8 is prevalent among African-Americans: implications for pharmacogenetic dosing. Pharmacogenet Genomics. 2009;10:1243–55. doi: 10.2217/pgs.09.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yong L, Jeong H-Y, Takahashi H, Drozda K, Patel SR, Shapiro NL, et al. Decreased warfarin clearance with CYP2C9 R150H (*8) polymorphism. Clin Pharmacol Ther. 2012;91:660–5. doi: 10.1038/clpt.2011.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Scott SA, Sangkuhl K, Shuldiner AR, Hulot J-S, Thorn CF, Altman RB, et al. PharmGKB summary: very important pharmacogene information for cytochrome P450, family 2, subfamily C, polypeptide 19. Pharmacogenet Genomics. 2012;22:159–65. doi: 10.1097/FPC.0b013e32834d4962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Luo H-R, Poland RE, Lin K-M, Wan Y-JY. Genetic polymorphism of cytochrome P450 2C19 in Mexican Americans: a cross-ethnic comparative study. Clin Pharmacol Ther. 2006;80:33–40. doi: 10.1016/j.clpt.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 65.FDA. FDA Announces new boxed warning on Plavix alerts patients, health care professionals to potential for reduced effectiveness. FDA News Release. 2010 Available at: http://www.fda.gov/newsevents/newsroom/pressannouncements/ucm204253.htm. Cited June 17, 2014.
- 66.Orengo-Mercado C, Nieves B, López L, Vallés-Ortiz N, Renta JY, Santiago-Borrero PJ, et al. Frequencies of functional polymorphisms in three pharmacokinetic genes of clinical interest within the admixed Puerto Rican population. J Pharm Pharmacoproteomics. 2013;4:1–6. doi: 10.4172/2153-0645.1000113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Duconge J, Cadilla CL, Renta JY, Silen-Rivera P, Piovanetti P, Garcia-Berdecia R, et al. Prevalence of CYP2C19 gene polymorphisms in the Puerto Rican population: a preliminary report. P R Health Sci J. 2008;27:2–3. [PMC free article] [PubMed] [Google Scholar]
- 68.Duconge J, Escalera O, Korchela M, Ruaño G. Clinical Implications of Genetic Admixture in Hispanic Puerto Ricans: Impact on the Pharmacogenetics of CYP2C19 and PON1. In: Sanodou D, editor. Clinical Applications of Pharmacogenetics. Rijeka: InTech; 2012. pp. 151–63. [Google Scholar]
- 69.Martis S, Peter I, Hulot J-S, Kornreich R, Desnick RJ, Scott SA. Multi-ethnic distribution of clinically relevant CYP2C genotypes and haplotypes. Pharmacogenomics. 2013;13:369–77. doi: 10.1038/tpj.2012.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Owen RP, Sangkuhl K, Klein TE, Altman RB. Cytochrome P450 2D6. Pharmacogenet Genomics. 2009;19:559–62. doi: 10.1097/FPC.0b013e32832e0e97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yu A-M, Idle JR, Herraiz T, Küpfer A, Gonzalez FJ. Screening for endogenous substrates reveals that CYP2D6 is a 5-methoxyindolethylamine O-demethylase. Pharmacogenetics. 2003;13:307–19. doi: 10.1097/01.fpc.0000054094.48725.b7. [DOI] [PubMed] [Google Scholar]
- 72.Hiroi T, Kishimoto W, Chow T, Imaoka S, Igarashi T, Funae Y. Progesterone oxidation by cytochrome P450 2D isoforms in the brain. Endocrinology. 2001;142:3901–8. doi: 10.1210/endo.142.9.8363. [DOI] [PubMed] [Google Scholar]
- 73.Niwa T, Hiroi T, Tsuzuki D, Yamamoto S, Narimatsu S, Fukuda T, et al. Effect of genetic polymorphism on the metabolism of endogenous neuroactive substances, progesterone and p-tyramine, catalyzed by CYP2D6. Brain Res Mol Brain Res. 2004;129:117–23. doi: 10.1016/j.molbrainres.2004.06.030. [DOI] [PubMed] [Google Scholar]
- 74.González I, Peñas-Lledó EM, Pérez B, Dorado P, Alvarez M, LLerena A. Relation between CYP2D6 phenotype and genotype and personality in healthy volunteers. Pharmacogenomics. 2008;9:833–40. doi: 10.2217/14622416.9.7.833. [DOI] [PubMed] [Google Scholar]
- 75.Bijl MJ, van Schaik RH, Lammers LA, Hofman A, Vulto AG, van Gelder T, et al. The CYP2D6*4 polymorphism affects breast cancer survival in tamoxifen users. Breast Cancer Res Treat. 2009;118:125–30. doi: 10.1007/s10549-008-0272-2. [DOI] [PubMed] [Google Scholar]
- 76.Kirchheiner J, Schmidt H, Tzvetkov M, Keulen J-T, Lötsch J, Roots I, et al. Pharmacokinetics of codeine and its metabolite morphine in ultra-rapid metabolizers due to CYP2D6 duplication. Pharm J. 2007;7:257–65. doi: 10.1038/sj.tpj.6500406. [DOI] [PubMed] [Google Scholar]
- 77.Gasche Y, Daali Y, Fathi M, Chiappe A, Cottini S, Dayer P, et al. Codeine intoxication associated with ultrarapid CYP2D6 metabolism. N Engl J Med. 2004;351:2827–31. doi: 10.1056/NEJMoa041888. [DOI] [PubMed] [Google Scholar]
- 78.Kelly LE, Rieder M, van den Anker J, Malkin B, Ross C, Neely MN, et al. More codeine fatalities after tonsillectomy in North American children. Pediatrics. 2012;129:e1343–7. doi: 10.1542/peds.2011-2538. [DOI] [PubMed] [Google Scholar]
- 79.González-Tejera G, Gaedigk A, Corey S. Genetic variants of the drug-metabolizing enzyme CYP2D6 in Puerto Rican psychiatry patients: a preliminary report and potential implications for breast cancer patients. P R Health Sci J. 2010;29:299–304. [PubMed] [Google Scholar]
- 80.Llerena A, Dorado P, Ramírez R, González I, Alvarez M, Peñas-Lledó EM, et al. CYP2D6 genotype and debrisoquine hydroxylation phenotype in Cubans and Nicaraguans. Pharm J. 2012;12:176–83. doi: 10.1038/tpj.2010.85. [DOI] [PubMed] [Google Scholar]
- 81.Luo H-R, Gaedigk A, Aloumanis V, Wan Y-JY. Identification of CYP2D6 impaired functional alleles in Mexican Americans. Eur J Clin Pharmacol. 2005;61:797–802. doi: 10.1007/s00228-005-0044-4. [DOI] [PubMed] [Google Scholar]
- 82.Casner PR. The effect of CYP2D6 polymorphisms on dextromethorphan metabolism in Mexican Americans. J Clin Pharmacol. 2005;45:1230–5. doi: 10.1177/0091270005280755. [DOI] [PubMed] [Google Scholar]
- 83.Mendoza R, Wan Y-JY, Poland RE, Smith M, Zheng Y, Berman N, et al. CYP2D6 polymorphism in a Mexican American population. Clin Pharmacol Ther. 2001;70:552–60. doi: 10.1067/mcp.2001.120675. [DOI] [PubMed] [Google Scholar]
- 84.Gaedigk A, Isidoro-García M, Pearce RE, Sánchez S, García-Solaesa V, Lorenzo-Romo C, et al. Discovery of the nonfunctional CYP2D6 31 allele in Spanish, Puerto Rican, and US Hispanic populations. Eur J Clin Pharmacol. 2010;66:859–64. doi: 10.1007/s00228-010-0831-4. [DOI] [PubMed] [Google Scholar]
- 85.Adams SM, Bosch E, Balaresque PL, Ballereau SJ, Lee AC, Arroyo E, et al. The genetic legacy of religious diversity and intolerance: paternal lineages of Christians, Jews, and Muslims in the Iberian Peninsula. Am J Hum Genet. 2008;83:725–36. doi: 10.1016/j.ajhg.2008.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bradford LD, Gaedigk A, Leeder JS. High frequency of CYP2D6 poor and “intermediate” metabolizers in black populations: a review and preliminary data. Psychopharmacol Bull. 1998;34:797–804. [PubMed] [Google Scholar]
- 87.Whirl-Carrillo M, McDonagh E, Hebert J, Gong L, Sangkuhl K, Thorn CF, et al. Pharmacogenomics knowledge for Personalized Medicine. Clin Pharmacol Ther. 2013;92:414–7. doi: 10.1038/clpt.2012.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lamba J, Hebert JM, Schuetz EG, Klein TE, Altman RB. PharmGKB summary: very important pharmacogene information for CYP3A5. Pharmacogenet Genomics. 2012;22:555–8. doi: 10.1097/FPC.0b013e328351d47f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Paine MF, Hart HL, Ludington SS, Haining RL, Rettie AE, Zeldin DC. The human intestinal cytochrome P450 “PIE.”. Drug Metab Dispos. 2006;34:880–6. doi: 10.1124/dmd.105.008672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Macé K, Bowman ED, Vautravers P, Shields PG, Harris CC, Pfeifer AM. Characterisation of xenobiotic-metabolising enzyme xxpression in human bronchial mucosa and peripheral lung issues. Eur J Cancer. 1998;34:914–20. doi: 10.1016/s0959-8049(98)00034-3. [DOI] [PubMed] [Google Scholar]
- 91.Agúndez JA, Martínez C, Olivera M, Gallardo L, Ladero JM, Rosado C, et al. Expression in human prostate of drug- and carcinogen-metabolizing enzymes: association with prostate cancer risk. Br J Cancer. 1998;78:1361–7. doi: 10.1038/bjc.1998.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Paris PL, Kupelian PA, Hall JM, Williams TL, Levin H, Klein EA, et al. Association between a CYP3A4 genetic variant and clinical presentation in African-American prostate cancer patients. Cancer Epidemiol Biomarkers Prev. 1999;8:901–5. [PubMed] [Google Scholar]
- 93.Blanco JG, Edick MJ, Hancock ML, Winick NJ, Dervieux T, Amylon MD, et al. Genetic polymorphisms in CYP3A5, CYP3A4 and NQO1 in children who developed therapy-related myeloid malignancies. Pharmacogenetics. 2002;12:605–11. doi: 10.1097/00008571-200211000-00004. [DOI] [PubMed] [Google Scholar]
- 94.Haas DW, Ribaudo HJ, Kim RB, Tierney C, Wilkinson GR, Gulick RM, et al. Pharmacogenetics of efavirenz and central nervous system side effects: an Adult AIDS Clinical Trials Group study. AIDS. 2004;18:2391–400. [PubMed] [Google Scholar]
- 95.Zhang X, Tierney C, Albrecht M, Demeter LM, Morse G, DiFrancesco R, et al. Discordant associations between SLCO1B1 521T→C and plasma levels of ritonavir-boosted protease inhibitors in AIDS clinical trials group study A5146. Ther Drug Monit. 2013;35:209–16. doi: 10.1097/FTD.0b013e318280d0ad. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Langaee TY, Gong Y, Yarandi HN, Katz DA, Cooper-DeHoff RM, Pepine CJ, et al. Association of CYP3A5 polymorphisms with hypertension and antihypertensive response to verapamil. Clin Pharmacol Ther. 2007;81:386–91. doi: 10.1038/sj.clpt.6100090. [DOI] [PubMed] [Google Scholar]
- 97.Ball SE, Scatina J, Kao J, Ferron GM, Fruncillo R, Mayer P, et al. Population distribution and effects on drug metabolism of a genetic variant in the 5′ promoter region of CYP3A4. Clin Pharmacol Ther. 1999;66:288–94. doi: 10.1016/S0009-9236(99)70037-8. [DOI] [PubMed] [Google Scholar]
- 98.Ribaudo HJ, Liu H, Schwab M, Schaeffeler E, Eichelbaum M, Motsinger-Reif AA, et al. Effect of CYP2B6, ABCB1, and CYP3A5 polymorphisms on efavirenz pharmacokinetics and treatment response: an AIDS Clinical Trials Group study. J Infect Dis. 2010;202:717–22. doi: 10.1086/655470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Figueroa-Vallés N, Ortiz-Ortiz K. Cancer in Puerto Rico, 2004–2009. Puerto Rico Central Cancer Registry; 2012. Available at: http://scholar.google.com/scholar?hl=en&btnG=Search&q=intitle:Cancer+in+Puerto+Rico+2004-2009#9. [Google Scholar]
- 100.Hecht JT, Ester A, Scott A, Wise CA, Iovannisci DM, Lammer EJ, et al. NAT2 variation and idiopathic talipes equinovarus (Club-foot) Am J Med Genet. 2007;143:2285–91. doi: 10.1002/ajmg.a.31927. [DOI] [PubMed] [Google Scholar]
- 101.McDonagh EM, Boukouvala S, Aklillu E, Hein DW, Altman RB, Klein TE. PharmGKB summary: very important pharmacogene information for N-acetyltransferase 2. Pharmacogenet Genomics. 2014;24:409–25. doi: 10.1097/FPC.0000000000000062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Baumgartner KB, Schlierf TJ, Yang D, Doll MA, Hein DW. N-acetyltransferase 2 genotype modif cation of active cigarette smoking on breast cancer risk among Hispanic and non-Hispanic white women. Toxicol Sci. 2009;112:211–20. doi: 10.1093/toxsci/kfp199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lin HJ, Han CY, Lin BK, Hardy S. Slow acetylator mutations in the human polymorphic N-acetyltransferase gene in 786 Asians, blacks, Hispanics, and whites: application to metabolic epidemiology. Am J Hum Genet. 1993;52:827–34. [PMC free article] [PubMed] [Google Scholar]
- 104.Kelsey K, Spitz M, Zuo Z, JK W. Polymorphisms in the glutathione S-transferase class mu and theta genes interact and increase susceptibility to lung cancer in minority population (Texas, United States) Cancer Causes Control. 1997;8:554–9. doi: 10.1023/a:1018434027502. [DOI] [PubMed] [Google Scholar]
- 105.Barbarino JM, Haidar CE, Klein TE, Altman RB. PharmGKB summary: very important pharmacogene information for UGT1A1. Pharmacogenet Genomics. 2014;24:177–83. doi: 10.1097/FPC.0000000000000024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.UGT Nomenclature Committee. UGT alleles nomenclature home page. 2005 Available at: http://www.ugtalleles.ulaval.ca. Cited June 5, 2014.
- 107.Li J, Menard V, Benish RL, Jurevic RJ, Guillemette C, Stoneking M, et al. Worldwide variation in human drug-metabolism enzyme genes CYP2B6 and UGT2B7: implications for HIV/AIDS treatment. Pharmacogenomics. 2012;13:555–70. doi: 10.2217/pgs.11.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Salinas AE, Wong MG. Glutathione S-transferases – a review. In: Atta-ur-Rahman, Craik DJ, Dax SL, Rees DC, editors. Current medicinal chemistry. Vol. 61. Amsterdam: Bentham Science Publishers; 1999. pp. 279–309. [PubMed] [Google Scholar]
- 109.Woo MH, Shuster JJ, Chen C, Bash RO, Behm FG, Camitta B, et al. Glutathione S-transferase genotypes in children who develop treatment-related acute myeloid malignancies. Leukemia. 2000;14:232–7. doi: 10.1038/sj.leu.2401660. [DOI] [PubMed] [Google Scholar]
- 110.Thorn CF, Ji Y, Weinshilboum RM, Altman RB, Klein TE. PharmGKB summary: very important pharmacogene information for GSTT1. Pharmacogenet Genomics. 2012;22:646–51. doi: 10.1097/FPC.0b013e3283527c02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Montero R, Araujo A, Carranza P, Mejía-Loza V, Serrano L, Albores A, et al. Genotype frequencies of polymorphic GSTM1, GSTT1, and cytochrome P450 CYP1A1 in Mexicans. Hum Biol. 2007;79:299–312. doi: 10.1353/hub.2007.0037. [DOI] [PubMed] [Google Scholar]
- 112.Johnson JA, Gong L, Whirl-Carrillo M, Gage BF, Scott SA, Stein CM, et al. Clinical Pharmacogenetics Implementation Consortium guidelines for CYP2C9 and VKORC1 genotypes and warfarin dosing. Clin Pharmacol Ther. 2011;90:625–9. doi: 10.1038/clpt.2011.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Scott SA, Sangkuhl K, Gardner EE, Stein CM, Hulot J-S, Johnson J, et al. Clinical Pharmacogenetics Implementation Consortium guidelines for cytochrome P450-2C19 (CYP2C19) genotype and clopidogrel therapy. Clin Pharmacol Ther. 2011;90:328–32. doi: 10.1038/clpt.2011.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Crews KR, Gaedigk A, Dunnenberger HM, Leeder JS, Klein TE, Caudle KE, et al. Clinical Pharmacogenetics Implementation Consortium guidelines for cytochrome P450 2D6 genotype and codeine therapy: 2014 update. Clin Pharmacol Ther. 2014;95:376–82. doi: 10.1038/clpt.2013.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Relling MV, Gardner EE, Sandborn WJ, Schmiegelow K, Pui C-H, Yee SW, et al. Clinical Pharmacogenetics Implementation Consortium guidelines for thiopurine methyltransferase genotype and thiopurine dosing: 2013 update. Clin Pharmacol Ther. 2013;93:324–5. doi: 10.1038/clpt.2013.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bermudez A. A proposal for an individualized pharmacogenetic-guided warfarin dosage regimen for Puerto Rican patients commencing anticoagulation therapy. J Pharm Pharmacoproteomics. 2014;T1:1–11. doi: 10.4172/2153-0645.T-001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Scott SA, Martis S, Peter I, Kasai Y, Kornreich R, Desnick RJ. Identification of CYP2C19*4B: pharmacogenetic implications for drug metabolism including clopidogrel responsiveness. Pharm J. 2012;12:297–305. doi: 10.1038/tpj.2011.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Innocenti F, Liu W, Fackenthal D, Ramírez J, Ye X, Wu X, et al. SNP discovery and functional assessment of variation in the UDP-glucuronosyltransferase 2B7 (UGT2B7) gene. Pharmacogenet Genomics. 2009;18:683–97. doi: 10.1097/FPC.0b013e3283037fe4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


