Abstract
CL-20 (2,4,6,8,10,12-hexanitro-2,4,6,8,10,12-hexaazaisowurtzitane) (C6H6N12O12), a future-generation high-energy explosive, is biodegradable by Pseudomonas sp. strain FA1 and Agrobacterium sp. strain JS71; however, the nature of the enzyme(s) involved in the process was not understood. In the present study, salicylate 1-monooxygenase, a flavin adenine dinucleotide (FAD)-containing purified enzyme from Pseudomonas sp. strain ATCC 29352, biotransformed CL-20 at rates of 0.256 ± 0.011 and 0.043 ± 0.003 nmol min−1 mg of protein−1 under anaerobic and aerobic conditions, respectively. The disappearance of CL-20 was accompanied by the release of nitrite ions. Using liquid chromatography/mass spectrometry in the negative electrospray ionization mode, we detected a metabolite with a deprotonated mass ion [M − H]− at 345 Da, corresponding to an empirical formula of C6H6N10O8, produced as a result of two sequential N denitration steps on the CL- 20 molecule. We also detected two isomeric metabolites with [M − H]− at 381 Da corresponding to an empirical formula of C6H10N10O10. The latter was a hydrated product of the metabolite C6H6N10O8 with addition of two H2O molecules, as confirmed by tests using 18O-labeled water. The product stoichiometry showed that each reacted CL-20 molecule produced about 1.7 nitrite ions, 3.2 molecules of nitrous oxide, 1.5 molecules of formic acid, and 0.6 ammonium ion. Diphenyliodonium-mediated inhibition of salicylate 1-monooxygenase and a comparative study between native, deflavo, and reconstituted enzyme(s) showed that FAD site of the enzyme was involved in the biotransformation of CL-20 catalyzed by salicylate 1-monooxygenase. The data suggested that salicylate 1-monooxygenase catalyzed two oxygen-sensitive single-electron transfer steps necessary to release two nitrite ions from CL-20 and that this was followed by the secondary decomposition of this energetic chemical.
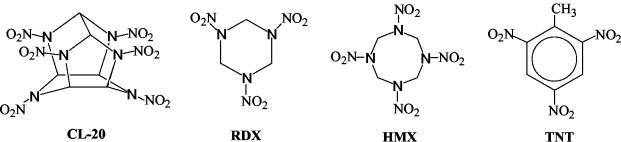
CL-20 (2,4,6,8,10,12-hexanitro-2,4,6,8,10,12-hexaazaiso-wurtzitane) is a high-energy polycyclic nitramine compound with a rigid caged structure (25) (Fig. 1). Due to its superior explosive properties, it may replace the conventionally usedexplosives such as hexahydro-1,3,5-trinitro-1,3,5-triazine(RDX), octahydro-1,3,5,7-tetranitro-1,3,5,7-tetrazocine (HMX) and 2,4,6-trinitrotoluene (TNT) (Fig. 1) in the future. The environmental, biological, and health impacts of CL-20 and its metabolic products are not known. Severe environmental contamination (7, 12, 24) and biological toxicity (21, 30,34, 36) as a result of the widespread use of monocyclic nitramine explosives such as RDX and HMX have been well documented. The U.S. Environmental Protection Agency has recommended a lifetime health advisory for RDX (9) and HMX (23). It is likely that due to its structural similarity to RDX and HMX, CL-20 (Fig. 1) may also cause similar environmental problems in soil, sediment, and groundwater. Therefore, the biodegradation of CL-20 must be understood in order to determine and predict its environmental fate and impact.
FIG. 1.
Molecular structure of the explosives CL-20, RDX, HMX, and TNT.
Previous reports of biotransformation and biodegradation of RDX and HMX by a variety of microorganisms and enzymes have shown that initial N denitration led to ring cleavage and decomposition (2, 3, 5, 10, 14, 28). Trott et al. (31) reported aerobic biodegradation of CL-20 by the soil isolate Agrobacterium sp. strain JS71, which utilized CL-20 as a sole nitrogen source and assimilated 3 mol of nitrogen per mol of CL-20. However, no information was provided about the initial reactions involved in the biodegradation of CL-20. In a recent study, we reported that a flavoenzyme(s) from Pseudomonas sp. strain FA1 might have been responsible for the biotransformation of CL-20 via an initial N-denitration mechanism (4); however, we did not detect initial metabolite(s) to support our hypothesis. We also reported the presence of CL-20 biotransformation products such as nitrite, nitrous oxide, and formate (4).
Salicylate 1-monooxygenase (EC 1.14.13.1), a flavin adenine dinucleotide (FAD)-containing enzyme from Pseudomonas sp. strain ATCC 29352, is capable of catalyzing a variety of biochemical reactions (19). The physiological role of salicylate 1-monooxygenase is to biotransform salicylate to catechol (20). However, it demonstrates activity against many other substrates such as o-halogenophenol and o-nitrophenol (29), benzoates, and variety of other compounds (19). We hypothesized that CL-20, being an oxidized chemical, might act as a substrate of salicylate 1-monooxygenase by accepting an electron(s).
In the present study, we used salicylate 1-monooxygenase as a model flavoenzyme in experiments to determine the initial enzymatic reaction(s) involved in the biodegradation of CL-20 and to gain insights into how flavoenzyme-producing bacteria biotransform CL-20. In liquid chromatography/mass spectrometry (LC/MS) studies, uniformly ring-labeled [15N]CL-20 and H218O were used to identify the intermediate(s) produced during the course of reaction. Additionally, we studied the involvement of the FAD-site of salicylate 1-monooxygenase in CL-20 biotransformation.
MATERIALS AND METHODS
Chemicals.
CL-20 (ɛ-form and 99.3% purity) and uniformly ring-labeled [15N]CL-20 (ɛ-form and 90.0% purity) were provided by ATK Thiokol Propulsion, Brigham City, Utah. NADH, diphenyliodonium chloride (DPI), and flavin adenine dinucleotide (FAD) were purchased from Sigma Chemicals, Oakville, Ontario, Canada. Nitrous oxide (N2O) was purchased from Scott Specialty Gases, Sarnia, Ontario, Canada. H218O (95% normalized 18O atoms) was purchased from Aldrich Chem. Co., Milwaukee, Wis. All other chemicals were of the highest purity grade.
Enzyme preparation and modification.
Salicylate 1-monooxygenase from Pseudomonas sp. strain ATCC 29352 was purchased as a lyophilized powder (protein approximately 45% by the Biuret method) from Sigma Chemicals. Native enzyme activity against salicylate was determined as specified by the manufacturer. The enzyme was washed with phosphate buffer (pH 7.0) at 4°C using Biomax-5K membrane centrifuge filter units (Sigma Chemicals) to remove preservatives and then resuspended in the same buffer. The protein content was determined by using the bicinchoninic acid protein assay kit from Pierce Chemical Co., Rockford, Ill.
Apoenzyme (deflavo form) was prepared by removing FAD from salicylate 1-monooxygenase by using a previously reported method (32). Reconstitution was carried out by incubating the apoenzyme with 100 μM FAD in 50 mM potassium phosphate buffer (pH 7.0) for 1 h at 4°C. The unbound FAD was removed by washing the enzyme with the same buffer, using Biomax-5K membrane centrifuge filter units.
Biotransformation assays.
Enzyme-catalyzed biotransformation assays were performed under aerobic as well as anaerobic conditions by using 6-ml glass vials. Anaerobic conditions were created by purging the reaction mixture with argon gas for 20 min and by replacing the headspace air with argon in sealed vials. Each assay vial contained, in 1 ml of assay mixture, CL-20 or uniformly ring-labeled [15N]CL-20 (25 μM or 11 mg liter−1), NADH (100 μM), enzyme preparation (250 μg), and potassium phosphate buffer (50 mM; pH 7.0). Higher CL-20 concentrations than its aqueous solubility of 3.6 mg liter−1 (11) were used to allow detection and quantification of the intermediate(s). Reactions were performed at 30°C. Three different controls were prepared by omitting enzyme, CL-20, or NADH from the assay mixture. Boiled enzyme was also used as a negative control. NADH oxidation was measured spectrophotometrically at 340 nm as described previously (2). Samples from the liquid and gas phases in the vials were analyzed for residual CL-20 and biotransformed products.
To determine the residual CL-20 concentrations during biotransformation studies, the reaction was performed with multiple identical vials. At each time point, the total CL-20 content in one reaction vial was solubilized in 50% aqueous acetonitrile and analyzed by high-pressure liquid chromatography (HPLC) (see below). The CL-20 biotransformation activity of salicylate 1-monooxygenase was expressed as nanomoles per minute per milligram of protein unless otherwise stated.
To demonstrate the effect of enzyme concentration on CL-20 biotransformation, a progress curve was generated for reactions under anaerobic conditions by incubating salicylate 1-monooxygenase at increasing concentrations (0.25 to 2.0 mg/ml) with 45 μM CL-20 and 100 μM NADH. The reactions conditions were the same as these described above.
To determine the incorporation of water into the CL-20 metabolite(s), H218O was mixed with 100 mM potassium phosphate buffer (pH 7.0) at a ratio of 8:2. All other reaction ingredients and conditions were the same as those described above for the biotransformation of CL-20.
Enzyme inhibition studies.
Inhibition with DPI, an inhibitor of flavoenzymes that acts by forming a flavin-phenyl adduct (6), was assessed by incubating salicylate 1-monooxygenase (1 mg) with DPI (0 to 0.5 mM) at room temperature for 15 to 20 min. After incubation, CL-20 biotransformation activities of the enzyme were determined by monitoring the residual CL-20 as described above.
Analytical procedures.
CL-20 and its intermediates were detected and analyzed with a Bruker bench top ion trap mass detector attached to a Hewlett-Packard 1100 series HPLC system equipped with a photodiode array detector. The samples were injected into a 5-μm-pore-size Zorbax SB-C18 capillary column (0.5-mm internal diameter by 150 mm; Agilent) at 25°C. The solvent system was an acetonitrile/water gradient (30 to 70% [vol/vol]) at a flow rate of 15 μl/min. For mass analysis, ionization was performed in the negative electrospray ionization mode (ES−), producing mainly deprotonated molecular mass ions [M − H]−. The mass range was scanned from 40 to 550 Da.
The CL-20 concentration was quantified with an HPLC apparatus connected to a photodiode array detector (λ230) as described previously (4). Nitrous oxide (N2O) and formaldehyde (HCHO) were analyzed as previously reported (2, 3, 18). Ammonium cations were analyzed by an SP 8100 HPLC system equipped with a Waters 431 conductivity detector and a Hamilton PRP-X200 (250 mm by 4.6 mm by 10 μm) analytical column as described previously (17). Formic acid (HCOOH) and nitrite (NO2) were quantified using a Waters HPLC system equipped with a conductivity detector as reported previously (4).
15N-labeled N2O (i.e., 15N14NO) was analyzed by a 6890 gas chromatograph (Hewlett-Packard Mississauga, Ontario, Canada) coupled with a 5973 quadrupole mass spectrometer. A GS-Gas Pro capillary column (30 m by 0.32 mm) (J & W Scientific, Folsom, Calif.) was used under splitless conditions. The injector and mass spectrometer interface (MSD) were maintained at 150 and 250°C, respectively. The column was set at −25°C for 1.5 min and then raised to −10°C at a rate of 10°C/min, which was held for 5 min. The MSD was used in the scan mode (electron impact) between 10 and 50 Da. The total run time was 8 min, and 15N14NO was detected at a retention time of 5.5 min with a molecular mass of 45 Da.
RESULTS AND DISCUSSION
Salicylate 1-monooxygenase-catalyzed biotransformation of CL-20.
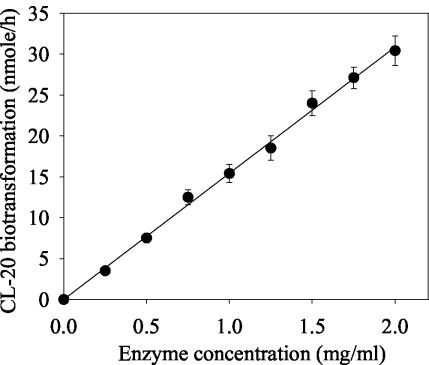
Biotransformation of CL-20 with a purified salicylate 1-monooxygenase from Pseudomonas sp. strain ATCC 29352 was found to be NADH dependent and optimal at pH 7.0 and 30°C under dark conditions. A progress curve demonstrated a linear increase in CL-20 biotransformation as a function of enzyme concentration (Fig. 2). The rates of CL-20 biotransformation were 0.256 ± 0.011 and 0.043 ± 0.003 nmol min−1 mg of protein−1 under anaerobic and aerobic conditions, respectively, indicating the involvement of an initial oxygen-sensitive process. On the other hand, salicylate 1-monooxygenase activity against the physiological substrate salicylate was 580 nmol min−1 mg of protein−1 under aerobic conditions. The low activity of enzyme against CL-20 might be due to three probable reasons. First, CL-20 is a hydrophobic compound with very little aqueous solubility (3.6 mg/liter at 25°C) (11), hence, its biotransformation rate is limited by the rate of mass transfer from solid to aqueous phase. In comparison, the aqueous solubility of salicylate is about 2,000 mg/liter at 20°C (Merck Index, 13th ed.) and would be higher at 30°C. Second, CL-20 is not a physiological substrate of salicylate 1-monooxygenase; hence, it is expected that its interaction with this enzyme would be rather slow. Third, CL-20 was biotransformed under anaerobic conditions, in contrast to the aerobic biotransformation of salicylate.
FIG. 2.
Progress curve demonstrating CL-20 biotransformation as a function of salicylate 1-monooxygenase concentration. The linear-regression curve has a gradient of 15.42 and an r2 of 0.99. Data are means of results from triplicate experiments, and error bars indicate standard errors. Some error bars are not visible due to their small size.
In a previous study, the rates of CL-20 biotransformation with a flavoenzyme(s) from Pseudomonas sp. strain FA1, under anaerobic and aerobic conditions, were 0.191 ± 0.006 and 0.041 ± 0.001 nmol min−1 mg of protein−1, respectively (4), which appear to be similar to those which we observed in the present study. The total flavin content in salicylate 1-monooxygenase and the flavoenzyme(s) from Pseudomonas sp. strain FA1 were 20.5 and 12.6 nmol mg of protein−1, respectively. Hence, under anaerobic conditions, CL-20 biotransformation by the two enzymes in terms of their flavin contents were 0.012 and 0.015 nmol min−1 nmol of flavin moiety−1, respectively. We found that in all controls (see Materials and Methods), abiotic degradation of CL-20 was negligible after 1 h of reaction time. The maximum abiotic degradation of CL-20 (0.010 ± 0.001 nmol min−1) was seen in a control with NADH under anaerobic conditions, which was only 4% of the CL-20 degradation catalyzed by salicylate 1-monooxygenase.
Molecular oxygen (O2) inhibits CL-20 biotransformation in two possible ways: (i) by competing with CL-20 for accepting electrons at the FAD site, since O2, in the absence of substrate, is known to accept electrons from the reduced salicylate 1-monooxygenase to produce H2O2 (19), and (ii) by quenching an electron from the CL-20 anion radical (described below), converting it back to the parent molecule (CL-20) and thus enforcing a futile redox cycling. Analogously, O2-mediated inhibition of RDX anion radical formation was observed during biotransformation of RDX catalyzed by diaphorase (2). Due to the inhibitory effect of oxygen, the subsequent experiments were carried out under anaerobic conditions.
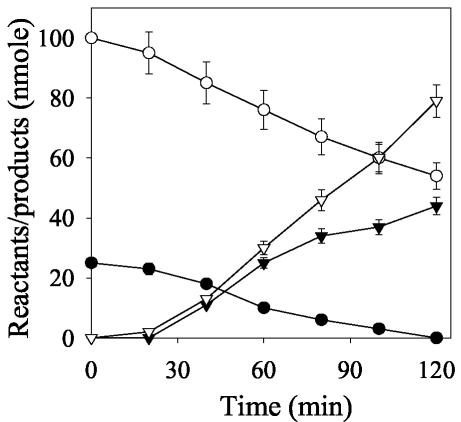
Time course studies showed a gradual disappearance of CL-20 at the expense of the electron donor NADH with concomitant release of nitrite (NO2−) and nitrous oxide (N2O) (Fig. 3). We found that N2O, although produced during later steps of CL-20 biotransformation (described below), appeared before NO2− in the assay medium, as shown in Fig. 3. This could be explained by two facts: (i) there was a large difference in the stoichiometries of nitrite (1.7) and nitrous oxide (3.2), and (ii) the nitrous oxide detection method (gas chromatography [GC] with electron capture detector) was much more sensitive (lowest-detection limit, 0.022 nmol) than the nitrite detection method (HPLC conductivity detector) (lowest detection limit, 5.434 nmol/ml). Hence, nitrous oxide was detected as early as 20 min into the reaction whereas nitrite was detected only after 30 min (Fig. 3). After 2 h of reaction, each reacted CL-20 molecule consumed about 1.9 NADH molecules and produced 1.7 nitrite ions, 3.2 molecules of nitrous oxide, 1.5 molecules of formic acid, and 0.6 molecule of ammonium (Table 1). Of the total 12 N atoms and 6 C atoms per reacted CL-20 molecule, we recovered approximately 9 N (as NO2−, N2O, and NH4+) and 2 C (as HCOOH) atoms, respectively. The remaining 3 N and 4 C atoms were probably present in an unidentified product(s). In comparison, a membrane-associated flavoenzyme(s) from Pseudomonas sp. strain FA1 produced 2.3 nitrite ions, 1.5 molecules of nitrous oxide, and 1.7 molecules of formic acid from each reacted CL-20 molecule (Table 1). Furthermore, during photodegradation of CL-20 at 300 nm in acetonitrile aqueous solution, which was also initiated by N denitration, the product distribution was similar to that in the present study but the stoichiometry was different; i.e., each reacted molecule of CL-20 produced 5.0, 5.3, 1.4, and 0.3 molecules of NO2−, HCOOH, NH3, and N2O, respectively (15). The probable reason for the higher yields of NO2− and HCOOH in photolysis of CL-20 was attributed to the intense action of the high-energy wavelength λ300 (energy, 400 kJ/mol), which caused rapid cleavage of N-NO2 and -HC-NNO2 bonds in CL-20 (15).
FIG. 3.
Time course study of NADH-dependent biotransformation of CL-20 by salicylate 1-monooxygenase (1 mg) under anaerobic conditions. Data for residual CL-20 (•), NADH (○), nitrite (▾), and nitrous oxide (▿) are shown. The data are means of results from triplicate experiments, and error bars indicate standard errors. Some error bars are not visible due to their small size.
TABLE 1.
Comparative stoichiometries of reactants and products during biotransformation of CL-20 by the salicylate 1-monooxygenase from Pseudomonas sp. strain ATCC 29352 and the membrane-associated enzyme(s) from Pseudomonas sp. strain FA1
| Reactant or product |
Pseudomonas sp. strain ATCC 29352a,c
|
Pseudomonas sp. strain FA1b,c
|
||
|---|---|---|---|---|
| Amt (nmol) | Molar ratio of reactant to product per reacted CL-20 molecule | Amt (nmol) | Molar ratio of reactant to product per reacted CL-20 molecule | |
| Reactants | ||||
| CL-20 | 25 ± 1.4 | 1.0 ± 0.05 | 20 ± 1.2 | 1.0 ± 0.06 |
| NADH | 48 ± 2.7 | 1.9 ± 0.10 | 90 ± 7.1 | 4.5 ± 0.35 |
| Products | ||||
| Nitrite (NO2−) | 43 ± 2.5 | 1.7 ± 0.09 | 46 ± 3.2 | 2.3 ± 0.16 |
| Nitrous oxide (N2O) | 79 ± 5.3 | 3.2 ± 0.21 | 29 ± 1.6 | 1.5 ± 0.08 |
| Formate (HCOOH) | 37 ± 2.4 | 1.5 ± 0.09 | 34 ± 2.8 | 1.7 ± 0.13 |
| Ammonium (NH4+) | 15 ± 0.9 | 0.6 ± 0.04 | NDd | ND |
Reaction was performed at pH 7.0 and 30°C for 2 h under anaerobic conditions.
Data are from reference 4.
Data are mean ± standard error (n = 3).
ND, not determined.
Involvement of the flavin-moiety (FAD) in CL-20 biotransformation.
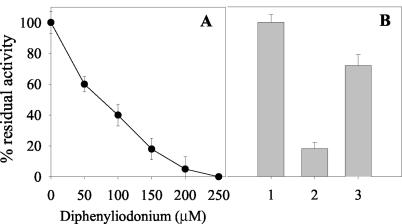
Salicylate 1-monooxygenase from Pseudomonas sp. strain ATCC 29352 is a dimeric protein with two identical subunits, each of which has an approximate molecular mass of 45.5 kDa and contains one molecule of FAD (33). DPI inhibited the biotransformation of CL-20 in a concentration-dependent manner (Fig. 4A). It is known that DPI inhibits flavoenzymes by the formation of flavin-phenyl adduct (6), which indicates the involvement of FAD in CL-20 biotransformation. Furthermore, DPI targets flavin-containing enzymes that catalyze one-electron transfer reactions (6, 26), which provided strong circumstantial evidence that a one-electron transfer process was involved in the initial reaction that might have caused N denitration of CL-20.
FIG. 4.
(A) Concentration-dependent inhibition of salicylate 1-monooxygenase-catalyzed biotransformation of CL-20 by DPI. (B) Biotransformation of CL-20 by the native (bar 1), deflavo (bar 2), and reconstituted (bar 3) salicylate 1-monooxygenase (3). A 100% CL-20 biotransformation activity was equivalent to 0.256 ± 0.011 nmol min−1 mg of protein−1. Data are mean percentages of CL-20 biotransformation activity ± standard errors (n = 3).
The involvement of FAD was additionally confirmed by assaying the deflavo and reconstituted forms of salicylate 1-monooxygenase against CL-20. The specific activities of the native, deflavo, and reconstituted forms of salicylate 1-monooxygenase against CL-20 were 0.256 ± 0.011, 0.045 ± 0.003, and 0.183 ± 0.008 nmol min−1 mg of protein−1, respectively, revealing that deflavo enzyme had lost about 82% of its activity compared to the native enzyme (Fig. 4B). The remaining 18% activity observed in the deflavo form was due to incomplete removal of FAD (data not shown), whereas the reconstituted enzyme, prepared by reconstitution of the deflavo enzyme with FAD, restored the CL-20 biotransformation activity up to 72% (Fig. 4B). The above results indicate the direct involvement of FAD in biotransformation of CL-20.
Free FAD (100 μM) also transformed CL-20 in the presence of NADH (100 μM) at a rate of 0.035 ± 0.003 nmol min−1; however, the biotransformation rate was only 14% of that of the bound FAD present in native salicylate 1-monooxygenase. The above finding additionally supported the involvement of FAD in CL-20 biotransformation and suggested that the flavin moiety has to be enzyme bound in order to function efficiently. The involvement of a flavoenzyme(s) in biotransformation of RDX (2) and HMX (3) via a one-electron transfer process has previously been reported. In a study with flavin mononucleotide containing diaphorase from Clostridium kluyveri, we reported an oxygen-sensitive one-electron transfer reaction that caused an initial N denitration of RDX followed by spontaneous decomposition (2). On the other hand, a FAD-containing xanthine oxidase also catalyzed a similar reaction with HMX (3).
Detection and identification of metabolites.
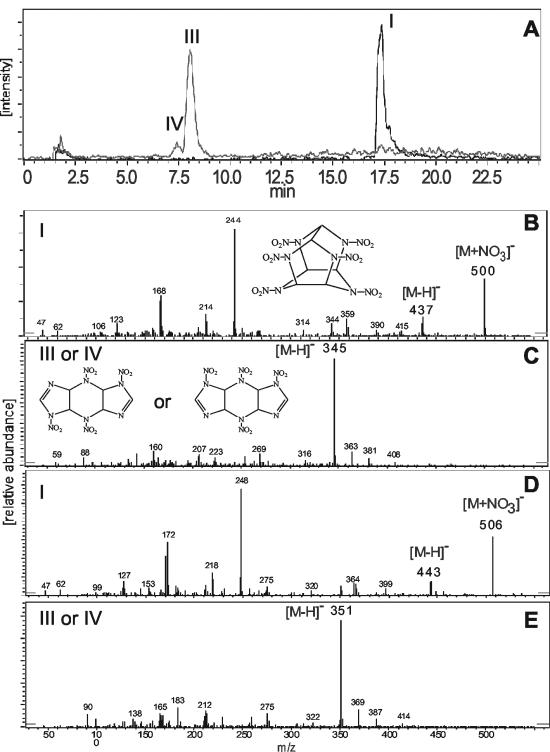
The metabolites, obtained by a reaction between salicylate 1-monooxygenase and CL-20 (metabolite I), were detected by their deprotonated molecular mass ion [M − H]− in LC/MS (ES−) studies and are listed in Table 2. Intermediates III and IV appeared simultaneously, with retention times (Rt) of 8.2 and 7.5 min, respectively (Fig. 5A), but with the same [M − H]− at 345 Da (Fig. 5C), corresponding to an empirical formula C6H6N10O8. When ring-labeled [15N]CL-20 was used in the reaction, both intermediates III and IV showed a deprotonated molecular ion [M − H]− at 351 Da (Table 2; Fig. 5E), indicating that each intermediate included the six nitrogen atoms from the CL-20 ring. Moreover, the use of H218O did not affect the mass of intermediates III and IV. Intermediates III and IV, previously observed during photolysis of CL-20 (15), were tentatively identified as isomers resulting from the loss of two -NO2 groups during the initial reaction(s) between CL-20 and salicylate 1-monooxygenase. In the present study, we described the secondary decomposition of intermediate III, (Fig. 6). Intermediate IV, being an isomer of III, might also decompose like intermediate III.
TABLE 2.
Properties of metabolites detected and identified by LC/MS (ES−) during biotransformation of CL-20 catalyzed by salicylate 1-monooxygenase from Pseudomonas sp. strain ATCC 29352
| Metabolitea | Retention time (Rt) (min) | Mass area in LC/MS (ES−) extracted ion- chromatogramb | [M − H]− (Da)c | No. of nitrogen atoms from [15N]CL-20 ring | No. of oxygen atoms from H218O | Proposed empirical formula |
|---|---|---|---|---|---|---|
| III | 8.2 | 207,980 | 345 | 6 | 0 | C6H6N10O8 |
| IVd | 7.5 | 26,089 | 345 | 6 | 0 | C6H6N10O8 |
| V | 12.5 | 11,092 | 381 | 6 | 2 | C6H10N10O10 |
| VI | 1.9 | 71,672 | 381 | 6 | 2 | C6H10N10O10 |
| VII | 1.7 | 7,637 | 293 | 4 | 4 | C6H10N6O8 |
FIG. 5.
(A) LC/MS (ES−) extracted ion chromatogram of CL-20 (I) and its metabolite (III or IV) produced by the reaction of CL-20 with salicylate 1-monooxygenase; the extracted mass ion of CL-20 was an adduct mass ion [M + NO3] at 500 Da, and the deprotonated mass ions [M − H]− of CL-20 (I) and its metabolite (III or IV) were 437 and 345 Da, respectively. (B to E) LC/MS (ES−) spectra of nonlabeled CL-20 (B), its metabolite III or IV (C), uniformly ring-labeled [15N]CL-20 (D), and its metabolite III or IV (E) showed deprotonated mass ions [M − H]− at 437, 345, 443, and 351 Da, respectively.
FIG. 6.
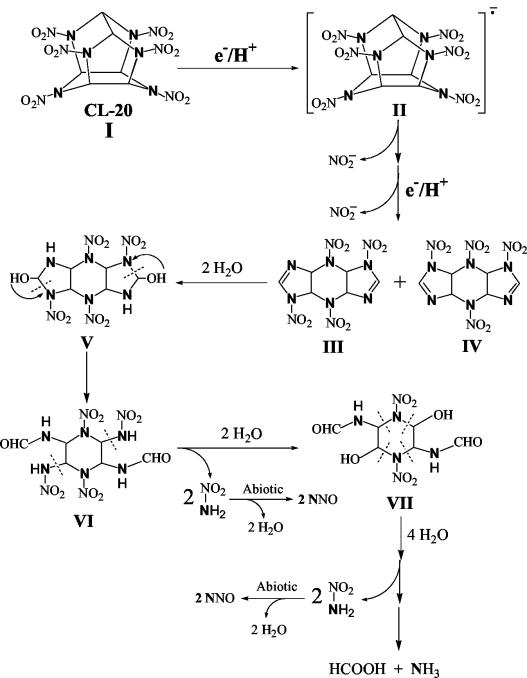
Proposed pathway of the initial biotransformation of CL-20 catalyzed by salicylate 1-monooxygenase followed by secondary decomposition. Nitrogen atoms shown in bold were actually ring- nitrogens and were uniformly labeled in [15N] CL-20. Secondary decomposition of intermediate III is shown; intermediate IV might also decompose like intermediate III. The intermediate shown in brackets was not detected.
Several other products, including V, VI, and VII, were also detected with Rt at 12.5, 1.9, and 1.7 min, respectively, and with [M − H]− at 381, 381, and 293 Da corresponding to empirical formulae of C6H10N10O10, C6H10N10O10, and C6H10N6O8, respectively (Table 2). Likewise, when uniformly ring-labeled [15N]CL-20 was used, the [M − H]− of products V, VI, and VII were observed at 387, 387, and 297 Da, respectively, indicating the incorporation of six 15N atoms into products V and VI and only four 15N atoms into product VII (Table 2). When the experiment was repeated with H218O, the [M − H]− of products V, VI, and VII were observed at 385, 385, and 301 Da, respectively, indicating the incorporation of two 18O atoms into products V and VI and four 18O atoms into product VII (Table 2). Based on the above data, intermediate V was tentatively identified as a carbinol adduct (C6H10N10O10) whereas intermediates VI (C6H10N10O10) and VII (C6H10N6O8) were identified as sequential ring cleavage products (Table 2, Fig. 6). All the above metabolites were transient and completely disappeared after 2.5 h of reaction.
To determine the source of N2O, experiments were performed with uniformly ring-labeled [15N]CL-20. 15N14NO was detected with a molecular mass of 45 Da by GC-MS analysis, confirming that of the two nitrogen atoms in N2O, one was from the labeled [15N]CL-20 ring and the second was from the unlabeled peripheral nitro (-NO2) group. We found that all of the N2O in the present study was labeled and produced from N-NO2 groups released from the CL-20, as previously suggested by Patil and Brill (27). Analogously, N2O generation from N-NO2 has also been reported during biodegradation of RDX (13) and HMX (3). In a previous study, we reported that N2O, besides being produced from N-NO2, was also produced via nitrite reduction catalyzed by an enzyme preparation from Pseudomonas sp. strain FA1 (4). In contrast, the present study did not show N2O production through nitrite reduction. For instance, when we incubated NaNO2 (1 mM) with salicylate 1-monooxygenase and NADH under similar reaction conditions to those used for CL-20 biotransformation, we did not detect N2O during 2 h of reaction, which additionally supported the idea that N-NO2 was the only source of N2O in the present study.
Proposed mechanism(s) of the initial reaction(s) followed by secondary decomposition of CL-20.
Based on the gradual appearance of nitrite (Fig. 3), reaction inhibition by oxygen and DPI, detection of an initial intermediate(s) (Fig. 5), and analogy to other systems (2, 3), we propose that salicylate 1-monooxygenase catalyzes a single-electron transfer to the CL-20 molecule (metabolite I) to produce an anion-radical (intermediate II) (Fig. 6). Spontaneous N denitration of intermediate II (i.e., release of the first NO2−) would produce a transient N-centered free radical, as previously proposed during biotransformation (4), thermolysis (27) and photodegradation (15) of CL-20. The N-centered free-radical, being unstable, must undergo rapid rearrangement by cleavage at the weaker C—C bond bridging the two cyclopentanes in CL-20 (35). The rearranged molecule would accept a second electron in order to release a second NO2− to produce two isomeric intermediates, III and IV (Table 2; Fig. 5 and 6). Stoichiometrically, two N- denitration steps would require an obligatory transfer of two electrons (equivalent to one NADH molecule); however, we found that 1.9 NADH molecules were consumed for each reacted CL-20 molecule (Table 1). The excess of NADH (i.e., the remaining 0.9 molecule) utilization could be due to two possible reasons: (i) some NADH could bind to the enzyme and go undetected, or (ii) it could be utilized by a yet unidentified CL-20 metabolite(s).
Intermediate III, produced as a result of two N denitration steps, underwent hydrolysis by the addition of two H2O molecules across the two imine bonds (—C=N—) to produce an unstable carbinol derivative V, as confirmed by the experiment with H218O (discussed above). Imine bonds are known to be unstable and to decompose rapidly in water (22). Hence, V might cleave at the O2NN—CH(OH) bond following rearrangement to produce intermediate VI, which has a similar [M − H]− of 381 Da to that of intermediate V (Table 2; Fig. 6). Addition of a water molecule across an imine bond followed by ring opening has previously been detected during photolysis of CL-20 (15) and RDX (16) and during cytochrome P450-catalyzed biotransformation of RDX (5). Stoichiometric addition of two H2O molecules to intermediate VI, confirmed by the experiment with H218O, produced intermediate VII, with concomitant release of two nitramide molecules (NH2-NO2) (Table 2; Fig. 6). Intermediate VII, being an α-hydroxyalkyl nitramine, was unstable in water (8) and therefore decomposed to finally produce nitrous oxide, formic acid, and ammonia, as quantified in Table 1 and shown in Fig. 6.
In conclusion, we have provided the first biochemical evidence of the initial reaction(s) involved in the transformation of CL-20 catalyzed by salicylate 1-monooxygenase under anaerobic conditions. The mechanism described here is consistent with our previous enzymatic studies with RDX and HMX (2, 3), which also suggested that one-electron transfer is necessary and sufficient to cause N- denitration of RDX and HMX, leading to their spontaneous decomposition. Some of the CL-20 products observed in the present study are consistent with those reported previously with respect to alkaline hydrolysis (1), biotransformation (4), and photolysis (15). So far, very few reports are available with regard to microbial degradation of CL-20. For instance, Agrobacterium sp. strain JS71, a soil isolate, degraded CL-20 and assimilated 3 mol of nitrogen per mol of CL-20 (31). In a recent study, Pseudomonas sp. strain FA1 degraded CL-20 and assimilated about 4 mol of nitrogen per mol of CL-20 (4). However, the two previous studies (4, 31) detected no initial metabolite(s) and did not describe the involvement of any specific enzyme(s). The present study thus advances our understanding of the initial steps involved in biotransformation of CL-20 by flavoenzyme(s)-producing bacteria. Pseudomonas, being the source of salicylate 1-monooxygenase and other flavoenzymes, seems to be a promising degrader of CL-20. The ubiquitous presence of Pseudomonas and similar bacteria in environments such as soil, marine and freshwater sediments, and estuaries would therefore help in understanding the fate of CL-20 in such environments.
Acknowledgments
B. Bhushan thanks the Natural Sciences and Engineering Research Council (NSERC) and the National Research Council (NRC) of Canada for a visiting fellowship. We sincerely acknowledge the U.S. Strategic Environmental Research and Development Program (SERDP) for financial support (CP 1256). We also thank the Department of National Defense, Val Bélair, Canada, for their support.
We gratefully acknowledge the analytical and technical support of A. Corriveau and C. Beaulieu. Finally, the critical suggestions of reviewers helped in improving the manuscript.
REFERENCES
- 1.Balakrishnan, V. K., A. Halasz, and J. Hawari. 2003. Alkaline hydrolysis of the cyclic nitramine explosives RDX, HMX, and CL-20: new insights into degradation pathways obtained by the observation of novel intermediates. Environ. Sci. Technol. 37:1838-1843. [DOI] [PubMed] [Google Scholar]
- 2.Bhushan, B., A. Halasz, J. C. Spain, and J. Hawari. 2002. Diaphorase catalyzed biotransformation of RDX via N-denitration mechanism. Biochem. Biophys. Res. Commun. 296:779-784. [DOI] [PubMed] [Google Scholar]
- 3.Bhushan, B., L. Paquet, A. Halasz, J. C. Spain, and J. Hawari. 2003. Mechanism of xanthine oxidase catalyzed biotransformation of HMX under anaerobic conditions. Biochem. Biophys. Res. Commun. 306:509-515. [DOI] [PubMed] [Google Scholar]
- 4.Bhushan, B., L. Paquet, J. C. Spain, and J. Hawari. 2003. Biotransformation of 2,4,6,8,10,12-hexanitro-2,4,6,8,10,12-hexaazaisowurtzitane (CL-20) by denitrifying Pseudomonas sp. strain FA 1. Appl. Environ. Microbiol. 69:5216-5221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhushan, B., S. Trott, J. C. Spain, A. Halasz, L. Paquet, and J. Hawari. 2003. Biotransformation of hexahydro-1,3,5-trinitro-1,3,5-triazine (RDX) by a rabbit liver cytochrome P450: insight into the mechanism of RDX biodegradation by Rhodococcus sp. strain DN22. Appl. Environ. Microbiol. 69:1347-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chakraborty, S., and V. Massey. 2002. Reaction of reduced flavins and flavoproteins with diphenyliodonium chloride. J. Biol. Chem. 277:41507-41516. [DOI] [PubMed] [Google Scholar]
- 7.Clausen, J., J. Robb, D. Curry, and N. Korte. 2004. A case study of contaminants on military ranges: Camp Edwards, Massachusetts, USA. Environ. Pollut. 129:13-21. [DOI] [PubMed] [Google Scholar]
- 8.Druckrey, H. 1973. Specific carcinogenic and teratogenic effects of ‘indirect’ alkylating methyl and ethyl compounds, and their dependency on stages of ontogenic development. Xenobiotica 3:271-303. [Google Scholar]
- 9.Etnier, E. L., and W. R. Hartley. 1990. Comparison of water quality criterion and lifetime health advisory for hexahydro-1,3,5-trinitro-1,3,5-triazine (RDX). Regul. Toxicol. Pharmacol. 11:118-122. [DOI] [PubMed] [Google Scholar]
- 10.Fournier, D., A. Halasz, J. C. Spain, P. Fiurasek, and J. Hawari. 2002. Determination of key metabolites during biodegradation of hexahydro-1,3,5-trinitro-1,3,5-triazine (RDX) with Rhodococcus sp. strain DN22. Appl. Environ. Microbiol. 68:166-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Groom, C. A., A. Halasz, L. Paquet, P. D'Cruz, and J. Hawari. 2003. Cyclodextrin-assisted capillary electrophoresis for determination of the cyclic nitramine explosives RDX, HMX and CL-20: comparison with high-performance liquid chromatography. J. Chromatogr. A 999:17-22. [DOI] [PubMed] [Google Scholar]
- 12.Haas, R., E. V. Löw, I. Schreiber, and G. Stork. 1990. Conception for the investigation of contaminated munitions plants. 2. Investigation of former RDX-plants and filling stations. Fresenius J. Anal. Chem. 338:41-45. [Google Scholar]
- 13.Halasz, A., J. Spain, L. Paquet, C. Beaulieu, and J. Hawari. 2002. Insights into the formation and degradation of methylenedinitramine during the incubation of RDX with anaerobic sludge. Environ. Sci. Technol. 36:633-638. [DOI] [PubMed] [Google Scholar]
- 14.Hawari, J. 2000. Biodegradation of RDX and HMX: from basic research to field application, p. 277-310. In J. C. Spain, J. B. Hughes, and H.-J. Knackmuss (ed.), Biodegradation of nitroaromatic compounds and explosives. CRC Press, Inc., Boca Raton, Fla.
- 15.Hawari, J. 2003. Environmental fate and transport of a new energetic material, CL-20. Annual Project Report, December 2003 (CP 1256). Strategic Environmental Research and Development Program, Arlington, Va.
- 16.Hawari. J., A. Halasz, C. Groom, S. Deschamps, L. Paquet, C. Beaulieu, and A. Corriveau. 2002. Photodegradation of RDX in aqueous solution: a mechanistic probe for biodegradation with Rhodococcus sp. Environ. Sci. Technol. 36:5117-5123. [DOI] [PubMed] [Google Scholar]
- 17.Hawari, J., A. Halasz, S. Beaudet, L. Paquet, G. Ampleman, and S. Thiboutot. 2001. Biotransformation routes of octahydro-1,3,5,7-tetranitro-1,3,5,7-tetrazocine by municipal anaerobic sludge. Environ. Sci. Technol. 35:70-75 [DOI] [PubMed] [Google Scholar]
- 18.Hawari, J., A. Halasz, T. Sheremata, S. Beaudet, C. Groom, L. Paquet, C. Rhofir, G. Ampleman, and S. Thiboutot. 2000. Characterization of metabolites during biodegradation of hexahydro-1,3,5-trinitro-1,3,5-triazine (RDX) with municipal sludge. Appl. Environ. Microbiol. 66:2652-2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kamin, H., R. H. White-Stevens, and R. P. Presswood. 1978. Salicylate hydroxylase. Methods Enzymol. 53:527-543. [DOI] [PubMed] [Google Scholar]
- 20.Katagiri, M., H. Maeno, S. Yamamoto, O. Hayaishi, T. Kitao, and S. Oae. 1965. Salicylate hydroxylase, a monooxygenase requiring flavin adenine dinucleotide. II. The mechanism of salicylate hydroxylation to catechol. J. Biol. Chem. 240:3414-3417. [PubMed] [Google Scholar]
- 21.Levine, B. S., E. M. Furedi, D. E. Gordon, J. J. Barkley, and P. M. Lish. 1990. Toxic interactions of the munitions compounds TNT and RDX in F344 rats. Fundam. Appl. Toxicol. 15:373-380. [DOI] [PubMed] [Google Scholar]
- 22.March, J. 1985. Advanced organic chemistry, 3rd ed., p. 784-785. Wiley-Interscience, New York, N.Y.
- 23.McLellan, W., W. R. Hartley, and M. Brower. 1988. Health advisory for octahydro-1,3,5,7-tetranitro-1,3,5,7-tetrazocine. Technical report PB90-273533. Office of Drinking Water, U.S. Environmental Protection Agency, Washington, D.C.
- 24.Myler, C. A., and W. Sisk. 1991. Bioremediation of explosive-contaminated soils (scientific questions/engineering realities), p. 137-146. In G. S. Sayler, R. Fox, and J. W. Blackburn (ed.), Environmental bio/technology for waste treatment. Plenum Press, New York, N.Y.
- 25.Nielsen, A. T., A. P. Chafin, S. L. Christian, D. W. Moore, M. P. Nadler, R. A. Nissan, and D. J. Vanderah. 1998. Synthesis of polyazapolycyclic caged polynitramines. Tetrahedron 54:11793-11812. [Google Scholar]
- 26.O'Donnell, V. B., G. C. Smith, and O. T. Jones. 1994. Involvement of phenyl radicals in iodonium inhibition of flavoenzymes. Mol. Pharmacol. 46:778-785. [PubMed] [Google Scholar]
- 27.Patil, D. G., and T. B. Brill. 1991. Thermal decomposition of energetic materials. 53. Kinetics and mechanisms of thermolysis of hexanitrohexaazaisowurtzitane. Combust. Flame 87:145-151. [Google Scholar]
- 28.Seth-Smith, H. M. B., S. J. Rosser, A. Basran, E. R. Travis, E. R. Dabbs, S. Nicklin, and N. C. Bruce. 2002. Cloning, sequencing and characterization of the hexahydro-1,3,5-trinitro-1,3,5-triazine degradation gene cluster from Rhodococcus rhodochrous. Appl. Environ. Microbiol. 68:4764-4771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suzuki, K., T. Gomi, T. Kaidoh, and E. Itagaki. 1991. Hydroxylation of o-halogenophenol and o-nitrophenol by salicylate hydroxylase. J. Biochem. (Tokyo) 109:348-353. [PubMed] [Google Scholar]
- 30.Talmage, S. S., D. M. Opresko, C. J. Maxwel, C. J. E. Welsh, F. M. Cretella, P. H. Reno, and F. B. Daniel. 1999. Nitroaromatic munition compounds: environment effects and screening values. Rev. Environ. Contam. Toxicol. 161:1-156. [DOI] [PubMed] [Google Scholar]
- 31.Trott, S., S. F. Nishino, J. Hawari, and J. C. Spain. 2003. Biodegradation of the nitramine explosive CL-20. Appl. Environ. Microbiol. 69:1871-1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang, L. H., S. C. Tu, and R. C. Lusk. 1984. Apoenzyme of Pseudomonas cepacia salicylate hydroxylase. Preparation, fluorescence property, and nature of flavin binding. J. Biol. Chem. 259:1136-1142. [PubMed] [Google Scholar]
- 33.White-Stevens, R. H., and H. Kamin. 1972. Studies of a flavoprotein, salicylate hydroxylase. I. Preparation, properties, and the uncoupling of oxygen reduction from hydroxylation. J. Biol. Chem. 247:2358-2370. [PubMed] [Google Scholar]
- 34.Woody, R. C., G. L. Kearns, M. A. Brewster, C. P. Turley, G. B. Sharp, and R. S. Lake. 1986. The neurotoxicity of cyclotrimethylenetrinitramine (RDX) in a child: a clinical and pharmacokinetic evaluation. J. Toxicol. Clin. Toxicol. 24:305-319. [DOI] [PubMed] [Google Scholar]
- 35.Xinqi, Z., and S. Nicheng. 1996. Crystal and molecular structures of ɛ-HNIW. Chin. Sci. Bull. 41:574-576. [Google Scholar]
- 36.Yinon, J. 1990. Toxicity and metabolism of explosives, p. 145-170. CRC Press, Inc., Boca Raton, Fla.