Abstract
Previous simulations showed that the β-hairpin antimicrobial peptide (AMP) protegrin-1 can form stable octameric β-barrels and tetrameric arcs (half barrels) in both implicit and explicit membranes. Here, we extend this investigation to several AMPs of similar structure: tachyplesin, androctonin, polyphemusin, gomesin, and the retrocyclin θ-defensin. These peptides form short β-hairpins stabilized by 2–3 disulfide bonds. We also examine synthetic β-sheet peptides selected from a combinatorial library for their ability or inability to form pores in lipid membranes. When heptameric, octameric, and decameric β-barrels and tetrameric arcs of these peptides were embedded in preformed neutral or anionic lipid pores (i.e., pores in neutral or anionic membranes, respectively), a variety of behaviors and membrane binding energies were observed. Due to the cationic charge of the peptides, more favorable transfer energies and more stable binding were observed in anionic than neutral pores. The synthetic peptides bound very strongly and formed stable barrels and arcs in both neutral and anionic pores. The natural AMPs exhibited unfavorable or marginally favorable binding energy and kinetic stability in neutral pores, consistent with the lower hemolytic activity of some of them compared with protegrin-1. Binding to anionic pores was more favorable, but significant distortions of the barrel or arc structures were sometimes noted. These results are discussed in light of the available experimental data. The diversity of behaviors obtained makes it unlikely that the barrel and arc mechanisms are valid for the entire family of β-hairpin AMPs.
Keywords: molecular dynamics, molecular modeling, biological membranes, pore formation, protegrin, tachyplesin
Introduction
Antimicrobial peptides (AMPs) are small (12–50-amino-acid), usually cationic peptides that provide immunological defenses (Brogden 2005; Brown and Hancock 2006; Hancock 2001) against bacteria, fungi, parasites, the HIV virus, and even cancer cells (Bechinger 2004; Brown and Hancock 2006; Cole 2005; Hancock 2001; Matsuzaki et al. 1996; Papo and Shai 2005; Silphaduang 2001). They could serve as the basis of novel antibiotics (Bradshaw 2003). Their mechanism of action is not known precisely, but there is considerable evidence that they target cell membranes (Oren and Shai, 1998; Matsuzaki 1999).
The positive charge of AMPs offers an explanation for their selectivity for prokaryotic over eukaryotic cells (Matsuzaki 1999, 2009). Bacterial membranes tend to be anionic, i.e., rich in acidic phospholipids like phosphatidylglycerol (PG) and cardiolipin (Ratledge and Wilkinson 1988). In mammalian cells, acidic phospholipids are usually sequestered in the inner plasma membrane leaflet, whereas the outer leaflet is usually comprised of zwitterionic phosphatidylcholine and sphingomyelin molecules (Verkleij et al. 1973). Although the anionic composition of bacterial inner membranes varies widely, 30% anionic content is typical (Ratledge and Wilkinson 1988). For example, the composition of Escherichia coli inner membrane was found to be 70%–75% phosphatidylethanolamine (PE), 18%–22% PG, and 6%–8% cardiolipin (Morein et al. 1996), depending on temperature. Bilayers of PE and PG at a 7:3 ratio are commonly used to mimic bacterial cell membranes (Palermo et al. 2012).
AMPs may micellize and/or disintegrate the membrane (i.e., the carpet mechanism; Oren and Shai 1998) or aggregate to form pores that cause fatal ion leakage (Huang 2000, 2006). The pores may be cylindrical, lined by peptides (i.e., the barrel-stave model) or toroidal, stabilized by peptides and lined partially by lipids. There is evidence that alamethicin forms barrel-stave pores, whereas melittin and magainin form toroidal pores (Ludtke et al. 1996; Yang et al. 2001). Both cylindrical and toroidal pores were classically viewed as ordered structures, but molecular dynamics (MD) simulations have suggested that some AMPs may form disordered pores (Leontiadou et al. 2006; Sengupta et al. 2008; Thøgersen et al. 2008).
Most AMPs are helical when membrane-bound. However, some are disordered, and some form β-hairpins. The best-studied of the latter family are the protegrins, small β-hairpins stabilized by 2 disulfide bonds; the face with the disulfide bonds contains charged or polar residues, and the opposite face has a hydrophobic cluster flanked by charged residues. They are active against Gram-positive and Gram-negative bacteria and fungi in vitro (Kokryakov et al. 1993; Steinberg et al. 1997). A large number of variants (Chen et al. 2000; Ostberg and Kaznessis 2005) and synthetic analogs (Robinson et al. 2005) have been synthesized. Other β-hairpin AMPs include the retrocyclin θ-defensin (Lehrer et al. 2012), tachyplesin (Laederach et al. 2002), polyphemusin (Powers et al. 2004), gomesin (Mandard et al. 2002), and androctonin (Mandard et al. 2001).
Protegrin-1 (hereafter called protegrin) is the best-studied AMP of this family, both experimentally (Heller et al. 1998; Tang and Hong 2009; Lam et al. 2012) and computationally (Bolintineanu and Kaznessis 2011). It forms ion channels in membranes (Mangoni et al. 1996; Sokolov et al. 1999). MD simulations have been performed of protegrin monomers in micelles (e.g., Langham and Kaznessis 2006) and of monomers and dimers in bilayers with transmembrane and interfacial orientations (Jang et al. 2006; Jang et al. 2007; Kandasamy and Larson 2007; Khandelia and Kaznessis 2007; Rui et al. 2009). Tilting within the membrane (Rui and Im 2010) and association with and insertion into an anionic membrane (Vivcharuk and Kaznessis 2010; 2011) have also been calculated. The evidence that it oligomerizes into a closed β-barrel (Mani et al. 2006) inspired simulations of β-barrel models (Capone et al. 2010; Jang et al. 2008; Jang et al. 2010; Langham et al. 2008; Lazaridis et al. 2013).
Previous computational studies from this laboratory in implicit and explicit membranes examined the interaction of protegrin monomers with membranes and pores and the relative stability of different β-barrel topologies (Lazaridis et al. 2013). The NCNC parallel topology was most stable, because it allows immersion of the hydrophobic cluster of each peptide into the nonpolar membrane interior. Further, incomplete barrels (arcs) formed kinetically stable pores for 300 ns (Prieto et al. 2014). Extension of these simulations on the Anton supercomputer showed that the pore was still stable after 2 µs (Leveritt J et al., unpublished). Thus, the barrel or arc structures seem to constitute viable pore formation mechanisms for protegrin.
We presently ask whether the other β-hairpin AMPs could work by the same mechanism. While that would be an attractive proposal, experimental studies have suggested the carpet mechanism for gomesin (Domingues et al. 2010), non-pore-forming internalization for polyphemusin (Powers et al. 2005), and an orientation parallel to the membrane plane for tachyplesin (Doherty et al. 2006). It would be interesting to rationalize these differences through molecular modeling; we do this here using an implicit membrane approach (Lazaridis 2005b; Mihajlovic & Lazaridis 2010). We construct tetrameric arcs and heptameric, octameric, and decameric β-barrels of θ-defensin, tachyplesin, polyphemusin, gomesin, and androctonin, insert them into implicit toroidal pores of varying radii, and subject them to MD simulations. We observe their kinetic stability and estimate their thermodynamic stability by computing their transfer energies from the pores to bulk water. Our goal is to determine how structural differences are associated with differences in pore-forming ability.
In addition to the AMPs, we studied two 26-residue peptides selected from a combinatorial library based on known structures of membrane-spanning β-sheet peptides (Rausch et al. 2005). The first is a good pore former and contains the residues YGKRGF in the combinatorial sites; it has sterilizing antimicrobial activity and low activity against mammalian cell membranes (Rausch et al. 2007). The second peptide lacks pore-forming ability and contains AGGKGF in the combinatorial sites. Rausch et al. (2005) noted that the interfacial, hydrophobic sites in the library peptides support a membrane-spanning mechanism. However, fluorescence spectroscopy showed that the peptides were partially exposed to water on the bilayer surface rather than being in a membrane spanning-state, which suggested a carpet mechanism (Rausch et al. 2007). The authors also speculated that carpet model pores might be formed on a mechanistic pathway toward more structured β-barrel pores (Rausch et al. 2005).
Methods
Energy Functions
The simulations in this study employed Implicit Membrane Model 1 (IMM1; Lazaridis 2003), which is an extension of Effective Energy Function 1 (EEF1) for soluble proteins (Lazaridis and Karplus 1999). IMM1 extends EEF1 to heterogeneous membrane-water systems by making the solvation parameters dependent on vertical position. These are modeled as linear combinations of the values for water and cyclohexane; further, the dielectric’s dependence on vertical position accounts for strengthening of electrostatic interactions within the membrane. IMM1 has been extended to account for surface charge due to anionic lipids using Gouy-Chapman theory (Lazaridis 2005a), transmembrane voltage (Mottamal and Lazaridis 2006), membrane dipole potential (Zhan and Lazaridis 2012), and lateral pressure effects (Zhan and Lazaridis 2013). IMM1 can also accommodate pores (Lazaridis 2005b, Mihajlovic & Lazaridis 2010), whose shape can be adjusted by making the radius dependent on vertical position:
where Ro is the pore radius at the membrane center, R is the radius at a given z-level, T represents the hydrophobic thickness of the membrane, and k (curvature) determines pore shape. For example, Ro = 15 Å and k = 0 defines a cylindrical pore of radius 15 Å, whereas Ro = 15 Å and k = 20 Å defines a toroidal pore with radii of 15 and 35 Å at its center and rims, respectively.
Because Gouy-Chapman theory can no longer be used in pore geometries, the electrostatic potential in anionic pores (i.e., ones comprised of anionic lipids) is obtained by solution of the Poisson-Boltzmann (PB) equation (He et al. 2013); those potential values are applied as a static field in MD simulations. We solve the PB equation and use it similarly to the analytical Gouy-Chapman equations by adding an extra term to the effective energy function representing the interaction between solute charges and the electrostatic potential ϕPB(r̄):
This simple approximation gives acceptable results compared with the full nonlinear PB treatment (Ben-Tal et al. 1996). The bilayer’s dielectric properties are represented by a five-slab model (see Fig 2 from He et al. 2013), whereby ε(d) depends on distance (d) to the hydrophobic core’s surface:
where εmemb (the dielectric constant inside the membrane), εhead (that inside the interfacial region), and εwater (that in water) are 2, 10, and 80, respectively (He et al. 2013). Width D was set to 3.0 Å, localizing the boundary around the phosphate group. The ion accessibility factor is assigned values of
so that ions cannot penetrate below the phosphate groups; the ions were taken as monovalent with radius 2.0 Å. To reproduce the membrane dipole potential, two layers of charges were defined as Gaussian distribution functions:
where ρi, oi, and σi represent charge per unit area, offset of the charge layer from the membrane surface, and Gaussian width, respectively. Positive and negative charge layers, separated by 1.0 Å according to experimental data (He et al. 2013), represent the charge distribution in the membrane; i is + and − for the positive and negative charge layers, respectively. The negative and positive charge layers are localized on and below the plane of the lipid phosphate groups, respectively, to create a positive dipole potential in the membrane interior. ρ+ was set to +1q / A, where A is the area per lipid within the membrane (68 Å2 for 1,2-dioleylphosphatidylcholine [DOPC] and 1,2-dioleylphosphatidylglycerol [DOPG] bilayers), and ρ− was set to −(1 + ZI × anfr) q / A, where anfr is the fraction of anionic lipids in the membrane, and ZI is the charge of an anionic lipid molecule. The hydrocarbon core thickness was set at 26 Å (in accordance with the thickness of DOPC membranes; He et al. 2013), and anfr of 30% was used, in accordance with the typical composition of bacterial membranes (Ratledge and Wilkinson 1988).
Fig 2.
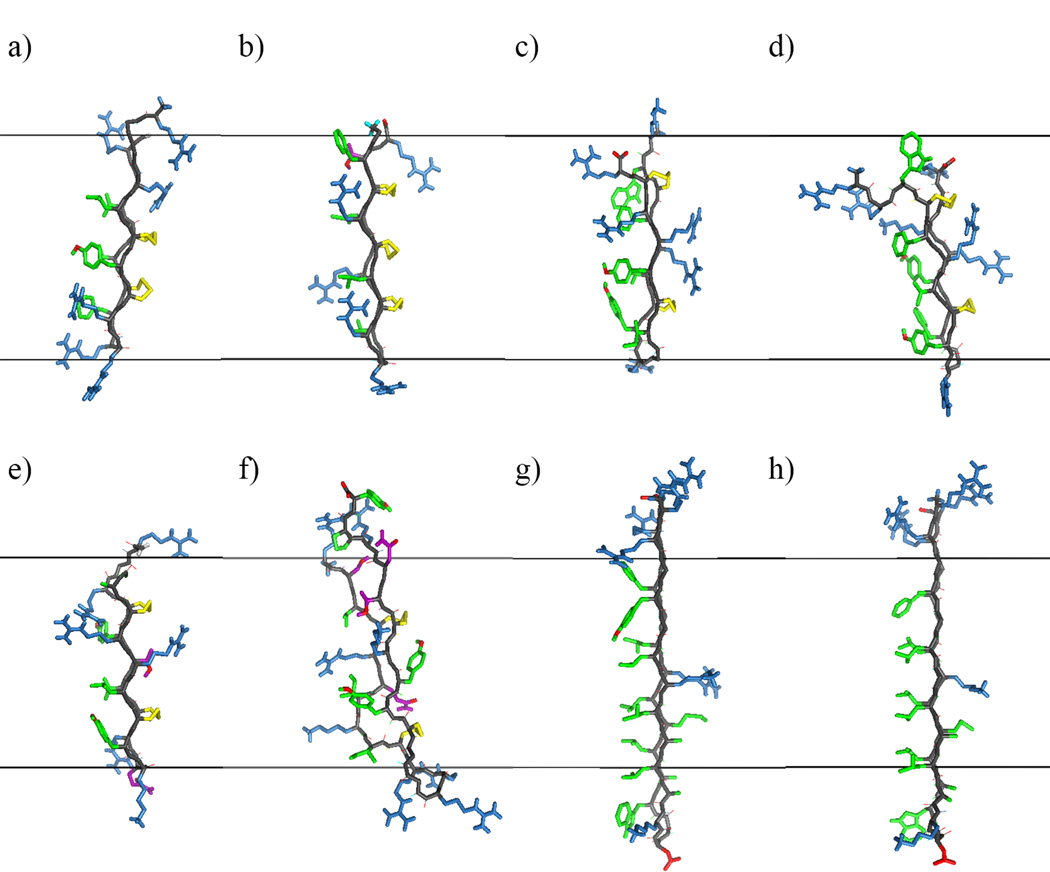
Visual structural comparison of the investigated peptides. Side views of one monomer from the initial β-barrel/arc conformations of each peptide after 200 ps of molecular dynamics (MD) simulation in water and 300 minimization steps but before insertion into a membrane: (a) protegrin-1; (b) θ-defensin; (c) tachyplesin; (d) polyphemusin; (e) gomesin; (f) androctonin; (g) YGKRGF; (h) AGGKGF. All peptides are oriented with the pore-facing side to the right and the turn region of the hairpin facing down. The black lines denote where the borders of a 26-Å-thick membrane region would be if the monomer were at center depth, the side chains and main chain are represented as sticks, and colors denote atom/side chain properties. Sky blue: basic side chain; red: acidic side chain or oxygen atom; purple: neutral, polar side chain; green: nonpolar side chain; cyan: glycine H; yellow: cysteine side chain/disulfide bond; gray: main chain. Figure created using MacPyMOL 1.7 (Schrödinger 2014).
The model accounts for the fact that head group density (and therefore charge density) may not be uniform on curved pore surfaces. It assumes that charge density on the pore rim is equal to that of a flat membrane (ρ0) and that charge density within the pore changes quadratically with |z′|, or vertical distance from the center of the membrane:
The homogeneity factor h (i.e., the ratio of charge density at the center of the pore to that in the intact membrane) was 0.6 in this study, according to all-atom simulation results (He et al. 2013).
The system was set up using mBuild, an in-house software package (He et al. 2013). The Poisson-Boltzmann equation was solved using the Advanced Poisson-Boltzmann Solver (Baker et al. 2001), taking average values in grid volumes after discretizing the space into a finite lattice box. The grid size was 161 × 161 × 161, with five focusing levels used to improve accuracy; that is, the calculations were run successively in cubic boxes of edge length 640 Å, 480 Å, 240 Å, 120 Å, and 80 Å (final resolution: 0.5 Å). For each run, the previous run’s potential was the boundary potential. Each volume was assigned values of dielectric constant, ion accessibility, and charge density by the distribution functions above; these simulated membranes were embedded into cubic boxes filled with 0.1 M aqueous salt ion solution. Multiple Debye-Hückel boundary conditions were used, and trilinear interpolation was used to obtain the ϕPB values and their derivatives. These are assumed to be steady state values, not changing throughout the simulation due to ion transport.
Initial Structures
The coordinate files for protegrin, θ-defensin, gomesin, polyphemusin, tachyplesin, and androctonin were downloaded from the Protein Data Bank (PDB; entries 1PG1 [Fahrner et al. 1996], 1HVZ [Trabi et al. 2001], 1KFP [Mandard et al. 2002], 1RKK [Powers et al. 2004], 1WO0 [Mizuguchi et al., unpublished], and 1CZ6 [Mandard et al. 1999], respectively). CHARMM version 39a2 (Brooks et al. 2009) was used to import the NMR model of each peptide monomer’s structure that seemed most conducive to β-barrel formation (i.e., the one in which the peptide’s structure was closest to a flat, ideal β-hairpin) and assign disulfide bonds between the appropriate residues. The peptides from the combinatorial library by Rausch et al. (2005) were initially built as β-strands. All charged residues were in their standard ionization states corresponding to pH ~7.
Then, the proper intramolecular hydrogen bonds for monomers of each peptide were imposed as distance constraints using a symmetrical potential well. For this, the values of kmin, rmin, kmax, and rmax within CHARMM’s Nuclear Overhauser Effect (NOE) facility were set to 1.0, 1.8, 5.0, and 2.3, respectively. Mean miscellaneous field potentials (MMFP) were applied to constrain the Cα carbons onto a plane, flattening the β-hairpins into conformations more conducive to barrel formation. After the above constraints were imposed, the energy of the peptides was minimized using the adopted basis Newton-Raphson algorithm (ABNER; used for all energy minimizations for 300 steps), followed by 200 ps of MD simulation using the Verlet integrator with a time step of 2 fs at 298.5 K (the same integrator, time step, and temperature were used in all MD simulations). Then, energy minimization yielded the final monomeric structures used to construct the β-barrels or arcs.
Simulations
The monomeric structures were arranged as tetrameric arcs or heptameric, octameric, or decameric β-barrels around the z-axis by translation of 11–14 Å in the x direction followed by rotation around the z-axis. For the β-barrels, the appropriate number of monomers was arranged in an evenly spaced cylinder around the z-axis in implicit water; for tetrameric arcs, four monomers were spaced evenly with z-axis rotations of 0°, 45°, 90°, and 135° (effectively forming half of an octamer barrel). We arranged the barrels and arcs in conformations analogous to those of protegrin: with the more hydrophobic side facing the membrane, the more hydrophilic side facing the pore, and the backbone hydrogen atoms both intermolecularly and intramolecularly H-bonded. Then, without any MMFP or NOE constraints, the β-barrel or arc underwent minimization in water with backbone constraints. Then, the appropriate intra- and intermolecular hydrogen bonds were imposed as NOE constraints, and minimization was performed without backbone constraints. Subsequently, 200 ps of MD simulation was run in water, followed by minimization.
After the β-barrel or arc underwent brief dynamics in water, the resulting structure was inserted into an implicit 26-Å-thick membrane and subjected to additional MD simulation. We set out to select the most realistic possible dimensions to represent the pores generated by the AMPs under investigation, so we conducted simulations with varying pore radii to find the conditions that provide optimal binding energies. We selected one value for which good protegrin octamer β-barrel binding results were previously obtained (Ro = 15 Å and k = 15 Å; Lazaridis et al. 2013). However, Rausch et al. (2007) suggested a pore radius of 10 Å, and initial simulations with YGKRGF indicated enhanced pore binding of the octamer β-barrel with Ro = 12 Å as compared with Ro = 15 Å. Therefore, for all peptides, we ran 2-ns simulations in pores of Ro = 10 Å, 12 Å, and 15 Å; we used both 30% anionic and zwitterionic membranes and four oligomeric states: tetramer arcs and heptamer, octamer, and decamer β-barrels. In all cases, k was 15 Å. For the arcs, shorter simulations were also run with k = 10 or 5 Å, but in no case was the stability in the pore better (and in some cases it was worse) than with k = 15 Å. (For our analysis, we selected the pore radius for each peptide and membrane charge condition that gave the best results: the most stable barrel/arc and most favorable binding energies.)
Once the β-barrel or arc had been inserted into the center of the pore, NOE and MMFP constraints were released for the production-length MD simulations. A simulation length of 2 ns was selected for all conformations. When we performed longer simulations on example peptides, we obtained results that converged onto those for the 2-ns simulations. In all conditions, the results seem to have stabilized by the end of a 2-ns MD simulation. The binding energy to the pore (ΔW) was estimated by averaging the energy of the barrel or arc at its position in the pore and subtracting the energy of the same conformation in water. In cases where the oligomer remained bound to the pore throughout the entire simulation, the value of ΔW was calculated by averaging values obtained every 1 ps throughout the last 1 ns of the simulation; in cases where the oligomer left the pore within 2 ns of MD simulation, the ΔW values were obtained by similar averaging during the period of 3–22 ps. Simulations using different random seeds and longer durations, run as checks, gave very similar results. At the end of the MD simulation, minimization provided the final post-MD image of the β-barrel or arc.
Results
Structural and Activity Comparison of the Peptides
In previous work, protegrin in NCNC parallel topology was observed to form stable β-barrels in both neutral and anionic membranes, with those in the latter being more stable (Lazaridis et al. 2013). In addition, four separate monomers in an implicit pore were observed to associate, forming a tetrameric arc of the same topology; the resulting structure formed stable pores in all-atom simulations (Prieto et al. 2014). In this study, we investigated the behavior of several similar peptides using simulations in implicit membrane pores. Before presenting the simulation results, we qualitatively compare the sequences (Fig 1) and structures of the peptides when configured as β-barrels in NCNC parallel topology (Figs 2–4). We also relate these comparisons to the peptides’ reported activity levels.
Fig 1.
Sequence alignment of the investigated peptides. Certain conserved residues are colored. The numbers represent residue positions in protegrin-1. The numbering reflects the sequence of protegrin-1. Figure created using STRAP (Gille 2012).
Fig 4.
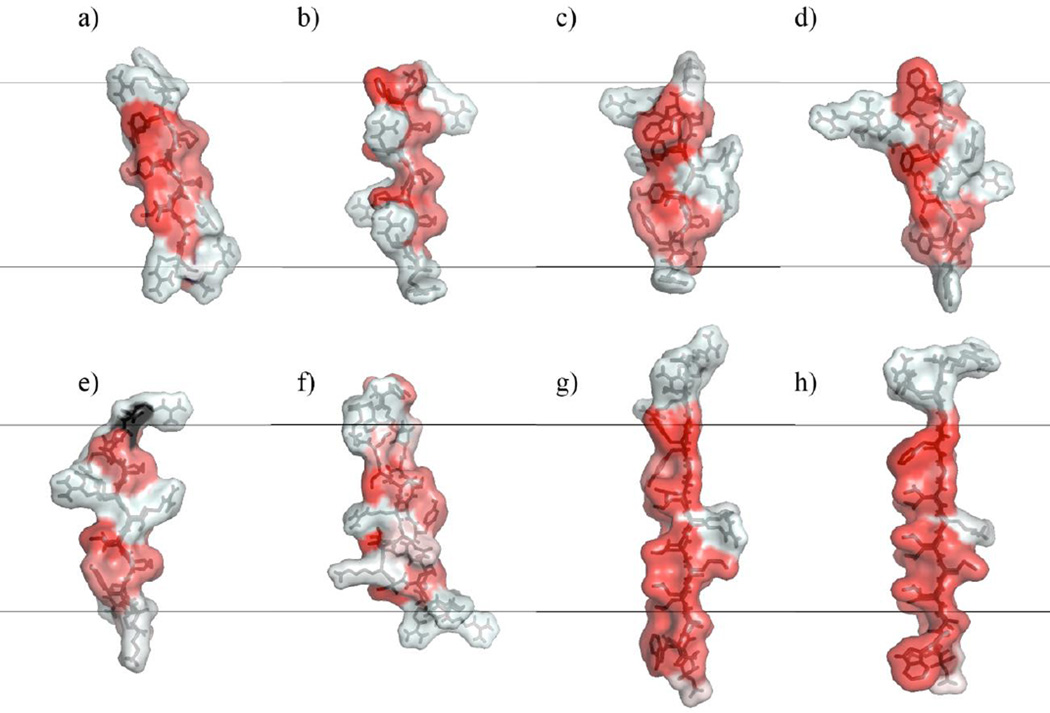
Comparison of the investigated peptides’ hydrophobicity levels at the molecular surface. Red: more hydrophobic; white: less hydrophobic. Side views of one monomer from the initial β-barrel/arc conformations of each peptide after 200 ps of molecular dynamics (MD) simulation in water and 300 minimization steps but before insertion into a membrane: (a) protegrin-1; (b) θ-defensin; (c) tachyplesin; (d) polyphemusin; (e) gomesin; (f) androctonin; (g) YGKRGF; (h) AGGKGF. All peptides are oriented with the pore-facing side to the right and the turn region of the hairpin facing down. The black lines denote where the borders of a 26-Å-thick membrane region would be if the monomer were at center depth, the chains are represented as black sticks, and the color surface overlay denotes hydrophobicity according to a normalized consensus hydrophobicity scale (Eisenberg et al. 1984). Figure created using MacPyMOL 1.7 (Schrödinger 2014).
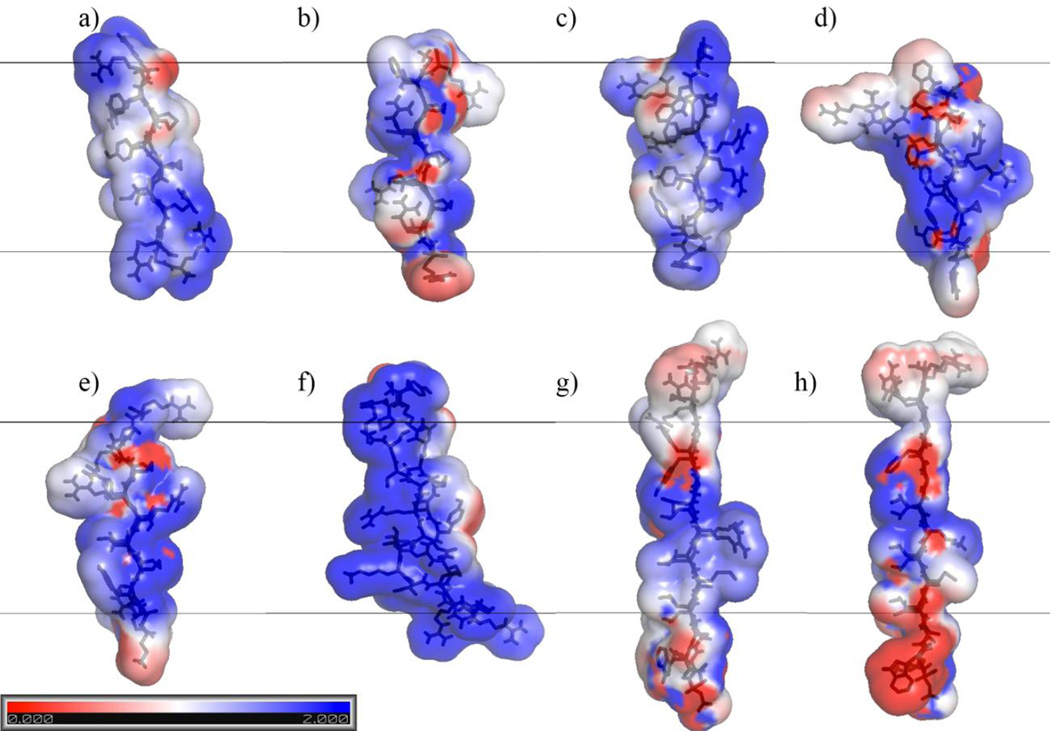
Experimental (Mani et al. 2006) and computational (Jang et al. 2008; Langham et al. 2008; Lazaridis et al. 2013) results indicate protegrin’s ability to form β-barrels in membrane pores. We thus infer that it has structural properties conducive to that arrangement. Protegrin’s β-hairpin conformation is constrained by two disulfide bonds, and the open ends of the hairpin both contain positively charged arginine residues (Fig 2a). The turn region of the hairpin is also highly concentrated in basic residues, containing three consecutive arginines; the resulting positive electrostatic potential in protegrin’s turn region can be seen upon solution of the PB equation at the solvent-accessible surface (Fig 3a). Further, protegrin has a relatively clear separation between polar and nonpolar regions. The β-sheet region between the turn and ends is relatively hydrophobic and nonpolar, with a clear division in hydrophobicity between the two sides of the molecule (Fig 4a): no charged or polar residues face the membrane, increasing its stability in membrane pores, while one positively charged arginine residue faces the pore in a 26-Å-thick membrane (all discussions of membrane–peptide structural proximity concern 26-Å-thick membranes; Fig 2a). Studies with truncated forms and disulfide variants of protegrin have shown the important role of the β-sheet and hairpin regions (especially the three consecutive arginine residues) for its activity against Neisseria gonorrheae (Roumestand et al. 1998). Protegrin’s minimal inhibitory concentrations (MICs) for Gram-positive and Gram-negative bacteria in one study were 0.3–0.8 µM and 0.7–2.8 µM, respectively (Tam et al. 2000). However, these values vary widely depending on experimental protocol (see Discussion). Protegrin also causes 50% hemolysis at 11.6 µM (Ishitsuka et al. 2006) and about 65% hemolysis at 46.4 µM (Tran et al. 2008).
Fig 3.
Comparison of the investigated peptides’ electrostatic potential at the solvent-accessible surface. Red: negative potential; white: middle; blue: positive potential. Potential denotes charge buildup at the surface. Side views of one monomer from the initial β-barrel/arc conformations of each peptide after 200 ps of molecular dynamics (MD) simulation in water and 300 minimization steps but before insertion into a membrane: (a) protegrin-1; (b) θ-defensin; (c) tachyplesin; (d) polyphemusin; (e) gomesin; (f) androctonin; (g) YGKRGF; (h) AGGKGF. All peptides are oriented with the pore-facing side to the right and the turn region of the hairpin facing down. The black lines denote where the borders of a 26-Å-thick membrane region would be if the monomer were at center depth, the chains are represented as black sticks, and the color surface overlay denotes electrostatic potential according to the scale shown. Figure created by solution of the Poisson-Boltzmann equation using the default parameters of the PyMOL APBS Tools plugin (Lerner and Carlson 2009) in MacPyMOL 1.7 (Schrödinger 2014).
θ-defensin has a cyclic backbone and contains three disulfide bonds (compared with two for protegrin and the other AMPs studied; Figs 1, 2b). In this study, we investigate the open chain analog of θ-defensin. The cyclic analog is required for antimicrobial activity in the presence of 150 mM sodium chloride and is three times as active as the acyclic analog (Tang et al. 1999). MIC values of 1.0 and 2.1 µM for Escherichia coli and Staphylococcus aureus, respectively, have been observed for θ-defensin (Tran et al. 2002). This peptide is relatively non-amphiphilic, accounting for its relatively low antimicrobial activity (Trabi et al. 2001; Selsted 2004), but it can also prevent HIV entry into cells, possibly as a competitive inhibitor of the gp120:gp41 membrane binding/fusion process (Penberthy et al. 2011). θ-defensin is much less hemolytic than protegrin; nevertheless, the cyclic analog caused 3% hemolysis at 5.5 µg/mL (Tran et al. 2008). When configured as a β-barrel, three of θ-defensin’s arginine side chains point towards the membrane rather than the top/bottom edges or pore interior (Fig 2b), and one additional polar group (Thr 17) is also oriented towards the membrane; this breaks the clear division in hydrophobicity between the membrane-facing and pore-facing sides seen in protegrin (Fig 4b). The electrostatic potential surface map (Fig 3b) also shows that θ-defensin has less positive charge concentration in the turn region than does protegrin. These features make θ-defensin less than ideal for the same type of pore-forming structure as protegrin in bacterial cells. Further, θ-defensin lacks the larger hydrophobic side chains that face the membrane in protegrin, which may negatively affect its affinity for the membrane.
Tachyplesin also exhibits less amphipathicity than protegrin (Fig 4c). One arginine residue (Arg 15) faces the membrane in embedded tachyplesin β-barrels. Further, in this arrangement, tachyplesin has two arginine residues (Arg 5 and Arg 14) facing the pore (compared with one for protegrin), which may cause crowding and electrostatic repulsion within the pore region and make β-barrel formation less favorable (Fig 2c); the electrostatic potential is also quite high in this region (Fig 3c). These structural differences may contribute to previous indications that tachyplesin orients parallel rather than normal to the membrane (Doherty et al. 2006, Boughton et al. 2011). Tachyplesin showed about 10% and 25% hemolysis at 100 and 150 µg/mL, respectively, but the hemolytic activity of a cysteine-deleted analog was abolished (Ramamoorthy et al. 2006). Nevertheless, the cysteine-deleted analog’s antibacterial activity was relatively intact (Ramamoorthy et al. 2006). MICs of 0.8–12.5, 3.1–12.5, and 1.6–3.1 µg/mL for Gram-negative bacteria, Gram-positive bacteria, and fungi, respectively, have been observed for tachyplesin (Miyata et al. 1989). Another study observed nearly linear dependence of hemolysis on tachyplesin’s concentration, finding 5% and 100% hemolysis at 20 and 100µM, respectively (Katsu et al. 1993).
Polyphemusin is extremely similar structurally to tachyplesin (Fig 1,2), but it contains an additional Arg residue at the N-terminus (Miyata et al. 1989). The disulfide bridge structure is identical between polyphemusin and tachyplesin, and each peptide has one charged residue facing the membrane in β-barrel conformation (Arg 15 and Lys 16 for tachyplesin and polyphemusin, respectively; Fig 2c–2d). MIC values of 3.1–12.5 µg/mL for Gram-negative bacteria and 6.3 µg/mL for Gram-positive bacteria and fungi have been observed for polyphemusin, indicating that its activity level is quite similar to tachyplesin’s (Miyata et al. 1989). Another study of polyphemusin obtained MICs of 0.125–0.5 µg/mL against Gram-negative and Gram-positive bacteria and 21.3 µg/mL against human red blood cells; a study using polyphemusin analogs indicated that the peptide binds to divalent cation binding sites on target cells’ lipopolysaccharide molecules, displacing divalent cations to penetrate/permeabilize the outer membrane (Zhang et al. 2000). Although polyphemusin and tachyplesin have quite similar structures, polyphemusin’s antimicrobial activity is dependent on disulfide bridges, as a linear analog with cysteine replaced by serine showed 4–16-fold less activity than polyphemusin; β-sheet structure is also required for polyphemusin to translocate past model membranes (Powers et al. 2004).
Gomesin has a relatively similar structure to protegrin (Figs 1–2) but contains a pyroglutamic acid residue at the N-terminus. In addition, two arginine residues face the membrane in β-barrel conformation (Fig 2e), which reduces the difference in hydrophobicity between the solvent-facing and membrane-facing sides of the molecule (Fig 4e). Gomesin has less positive electrostatic potential than protegrin in the turn region (Fig 3e). MICs of 0.4–6.25, 0.2–12.5, and 0.2–25 µg/mL for Gram-negative bacteria, Gram-positive bacteria, and fungi, respectively, have been measured for gomesin; direct comparison with androctonin revealed that gomesin was more active against most bacteria and fungi (Silva et al. 2000). Similarly to some other AMPs under investigation, gomesin’s disulfide bonds are important to its antimicrobial and hemolytic activity (Mandard et al. 2002; Fázio et al. 2006; Fázio et al. 2007): activity is markedly decreased upon reduction/alkylation of the disulfide bridges (Silva et al. 2000). Further, while gomesin is hemolytic at low concentrations (16% hemolysis at 1µM), hemolytic activity had little dependence on concentration (22% hemolysis at 100µM), making it significantly less hemolytic than protegrin in the high-concentration regime (Silva et al. 2000). One study showed that gomesin made giant unilamellar vesicles burst suddenly; stable pores were not observed, indicating that gomesin may work by the carpet mechanism (Domingues et al. 2010).
Androctonin shows some sequence homology to tachyplesin and polyphemusin (Ehret-Sabatier et al. 1996); however, it also has structural features that distinguish it from the other investigated peptides. Whereas gomesin, tachyplesin, and polyphemusin have three residues in each segment upstream and downstream of the disulfide bridges, androctonin has three residues in one segment and five in the other, with Cys 4 and Cys 10 bonded to Cys 20 and Cys 16, respectively (Fig 1; see Fig 5 from Silva et al. 2000). Probably because of this, androctonin’s NMR structure is a highly twisted antiparallel β-sheet with strands connected by a positively charged turn (Mandard et al. 1999). This feature might affect androctonin’s ability to form a β-barrel. In addition, androctonin has one arginine and two lysine residues facing the membrane in this arrangement, rather than large hydrophobic side chains (Fig 2f); this significantly reduces the hydrophobicity difference between the two sides of the molecule (Fig 4f), although in the conformation we used, the two sides of the molecule did vary in electrostatic potential (Fig 3f). MICs of 1.5–>30, 0.3–30, and 2–50 µg/mL for Gram-negative bacteria, Gram-positive bacteria, and fungi, respectively, have been measured for androctonin (Ehret-Sabatier et al. 1996); it was significantly less active than gomesin in most conditions that provided for direct comparison (Silva et al. 2000). Androctonin is not hemolytic even at 150 µM (Ehret-Sabatier et al. 1996). Silva et al. (2000) attributed the differences in hemolytic activity between androctonin on the one hand and gomesin and tachyplesin on the other to androctonin’s longer C-terminus and charge differences throughout the molecule. ATR-FTIR experiments seemed to indicate that androctonin is localized on the membrane surface and does not destabilize the bilayer structure (Hetru et al. 2000). Androctonin binds only to negatively charged lipid vesicles and seems to adopt a β-sheet structure while leaving the acyl chain order unaffected, suggesting a detergent-like mechanism (Hetru et al. 2000).
Fig 5.
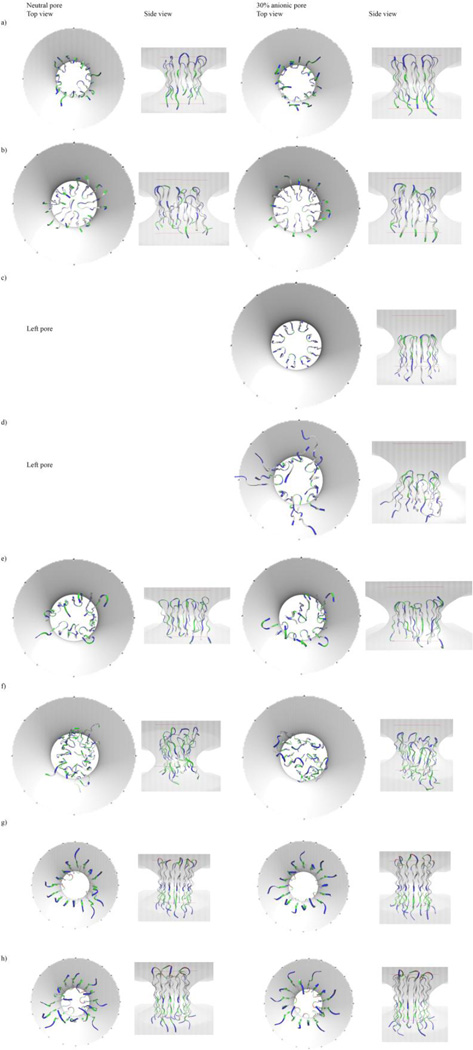
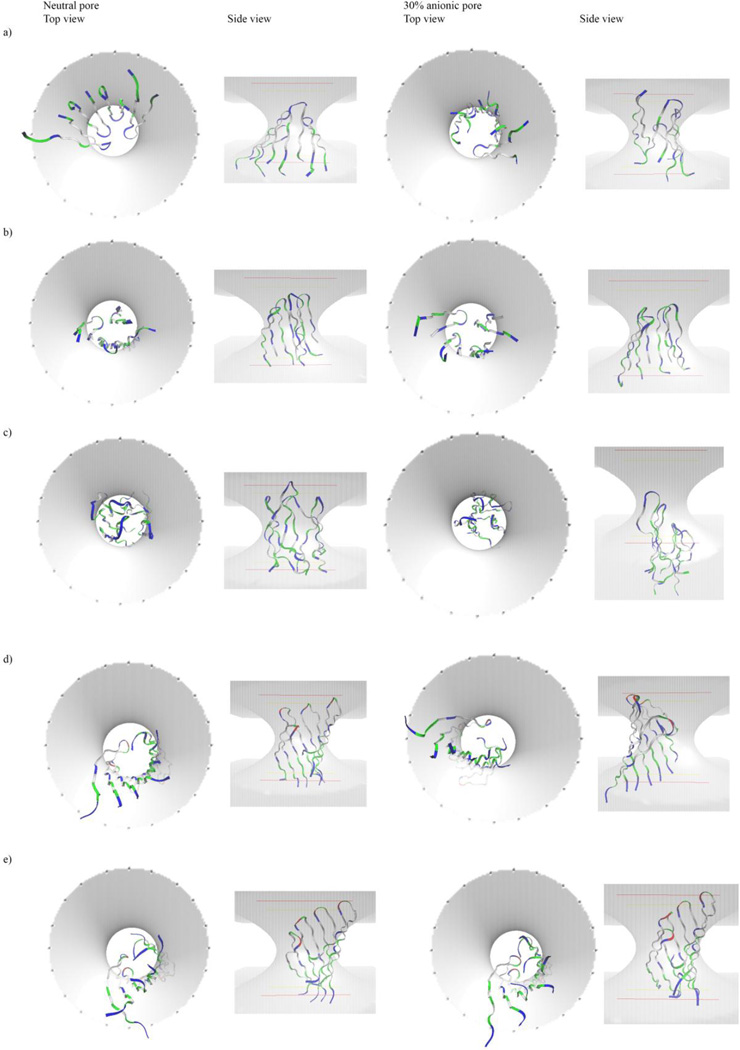
Top and side views of the minimized final conformations of octamer β-barrels of peptides in toroidal pores of optimal radius in zwitterionic and 30% anionic membranes, after molecular dynamics (MD) simulations. (a) protegrin-1; (b) θ-defensin; (c) tachyplesin; (d) polyphemusin; (e) gomesin; (f) androctonin; (g) YGKRGF; (h) AGGKGF. The barrels of polyphemusin and tachyplesin exited zwitterionic pores during the simulations, and those images are omitted. White: nonpolar; blue: basic; red: acidic; green: polar residues. These were the conformations of the peptides after the water-equilibrated monomers were arranged as octamer barrels, subject to 200 ps of MD in water and minimized, inserted into pre-formed toroidal pores in 26-Å-thick implicit membranes, and subject to 2 ns of MD followed by minimization. The shading and lines represent the boundaries of the toroidal pores in 26-Å-thick membranes. In the top views, the shading represents the inner and outer boundaries of the toroidal pore region; in the side views, the shaded volume represents the pore interior, and the lines represent the membrane edges. Figure created using VMD 1.9 (Humphrey et al. 1996).
YGKRGF and AGGKGF have some similarities with the protegrin family, such as the general locations of the positively charged and polar residues, but they also show several structural differences (Fig 1,2). These peptides are longer than protegrin (26 residues) and contain no disulfide linkages. However, the rational combinatorial library from which these peptides were extracted preserves certain key characteristics of the protegrin family. For example, aromatic residues are found at the lipid-exposed interfacial positions, and basic residues are found in the pore-lining region (Fig 2g–2h). The general locations of positive charges within the peptides have some similarity to those in protegrin: there is a pair of polar combinatorial sites in a position analogous to the pore-facing arginine residue in protegrin. Besides protegrin, these two peptides are the only ones studied that have no significant interruptions in the hydrophobicity of the molecule’s membrane-facing side (Fig 4g–4h). The turn region of YGKRGF and AGGKGF differs from the ones found in the AMPs we investigated in that they lack arginine and include a negative charge; this reduces the turn region’s overall electrostatic potential (Fig 3g–3h). While both YGKRGF and AGGKGF are active against Escherichia coli and show broad-spectrum antimicrobial activity, only YGKRGF causes measurable leakage of the contents of lipid vesicles, with the release of molecules as large as 3 kDa signifying a pore diameter of ~10 Å (Rausch et al. 2005). YGKRGF also had a low MIC against Staphylococcus aureus (2.1 µM; Rausch et al. 2005). In contrast, AGGKGF had the highest hemolytic activity, lysing human red blood cells at 15 µM. Relatively small structural differences differentiate YGKRGF, which forms functional pores, from AGGKGF, which does not. The presence of two charged side chains facing the pore (instead of one) and the replacement of an alanine with a tyrosine facing the membrane seem to increase the peptide’s pore-forming ability.
Simulation Results
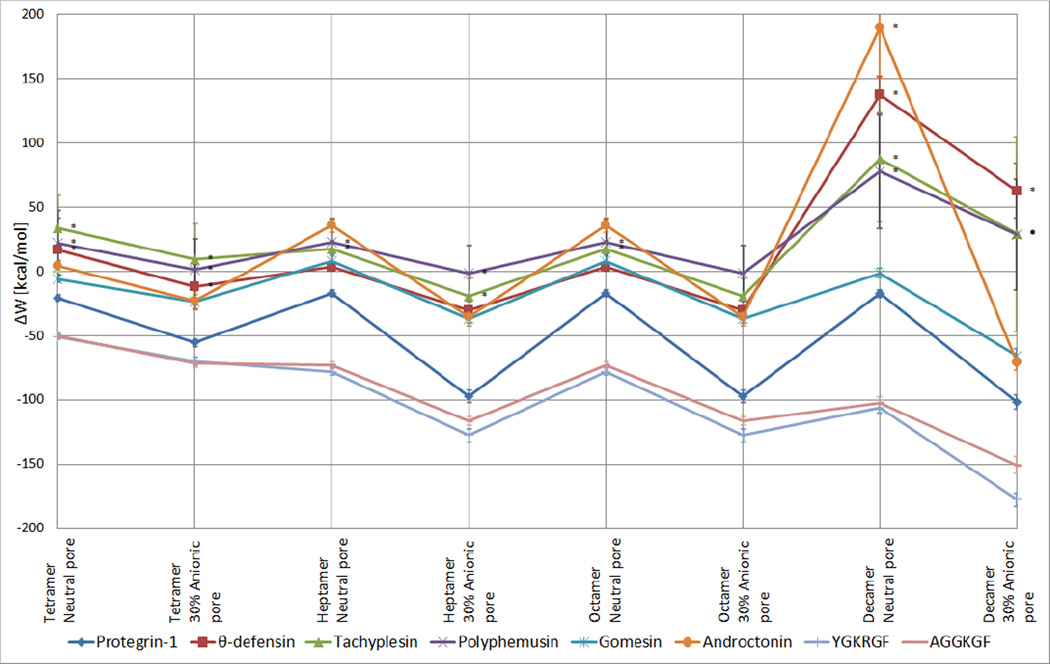
The tetramer arcs and heptamer, octamer, and decamer β-barrels of the peptides were subjected to 2-ns MD simulations in implicit toroidal pores with Ro = 10 Å, 12 Å, and 15 Å using both 30% anionic and zwitterionic membranes; we observed the peptides’ movement, structure, and the thermodynamic stability of their arrangement. For each peptide, the pore radius that gave the best energy in anionic membranes was chosen for further analysis (Table 1). The results for the pore radii not selected for further analysis and further notes on the peptides’ behavior during the simulations are found in Online Resource 1 (Tables S1–S8). Not all peptides remained within the pore for the duration of the simulation; notes on whether or not the peptides left the pore region or became distorted during the simulations are found in Table 1, and top-down and side views of the post-simulation conformations of the octamer barrels and tetramer arcs in pores of optimal radii are shown in Figs 5–6. The binding energies of these β-barrels and arcs to the pores were estimated by the transfer energy (ΔW) from water to pore (Tables 2–5, Fig 7). Negative values indicate favorable binding.
Table 1.
Behavior of selected tetramer arcs and heptamer, octamer, and decamer β-barrels during 2-ns molecular dynamics (MD) simulations
| Peptide | Pore radius (Å) |
Tetramer arc Pore charge |
Pore radius (Å) | Heptamer barrel Pore charge |
Pore radius (Å) | Octamer barrel Pore charge |
Pore radius (Å) | Decamer barrel Pore charge |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Neutral | 30% anionic | Neutral | 30% anionic | Neutral | 30% anionic | Neutral | 30% anionic | |||||||||||||
| Leaves pore |
Becomes distorted |
Leaves pore |
Becomes distorted |
Leaves pore |
Becomes distorted |
Leaves pore |
Becomes distorted |
Leaves pore |
Becomes distorted |
Leaves pore |
Becomes distorted |
Leaves pore |
Becomes distorted |
Leaves pore |
Becomes distorted |
|||||
| Protegrin-1 | 10 | 10 | 12 | 15 | x | |||||||||||||||
| θ-defensin | 12 | x | x | 15 | x | X | 15 | x | x | 15 | x | x | ||||||||
| Tachyplesin | 12 | x | x | 15 | x | x | x | X | 15 | x | x | 12 | x | x | ||||||
| Polyphemusin | 12 | x | x | x | x | 15 | x | x | x | X | 15 | x | x | x | 15 | x | x | x | x | |
| Gomesin | 10 | x | x | 12 | x | X | 15 | x | x | 15 | x | x | ||||||||
| Androctonin | 10 | x | x | 12 | x | X | 15 | x | x | 15 | x | x | x | |||||||
| YGKRGF | 10 | 10 | x | X | 10 | 12 | ||||||||||||||
| AGGKGF | 10 | 10 | 10 | 10 | x | x | ||||||||||||||
This table represents the characteristics of the final minimized conformations of peptide oligomers after MD simulations in implicit toroidal pores of the radii that gave the most favorable transfer energies from solvent to membrane. These were the positions and conformations of the peptides after the water-equilibrated monomers were arranged as octamer barrels, subject to 200 ps of MD in water, minimized, inserted into pre-formed toroidal pores in 26-Å-thick implicit membranes, and subject to 2 ns of MD followed by minimization. For each oligomeric state and pore charge condition, whether or not the peptide assembly left the pore region during the course of the simulation is represented by a checkbox in the “Leaves pore” column, and the presence of distortion is represented by a checkbox in the “Becomes distorted” column.
Fig 6.
Top and side views of the minimized final conformations of tetramer arcs of peptides in toroidal pores of optimal radius in zwitterionic and 30% anionic membranes, after molecular dynamics (MD) simulations. (a) protegrin-1; (b) gomesin; (c) androctonin; (d) YGKRGF; (e) AGGKGF. The corresponding arcs of polyphemusin, tachyplesin, and θ-defensin exited the membrane region during the simulation and are omitted. White: nonpolar; blue: basic; red: acidic; green: polar residues. These were the conformations of the peptides after the water-equilibrated monomers were arranged as octamer barrels, subject to 200 ps of MD in water and minimized, inserted into pre-formed toroidal pores in 26-Å-thick implicit membranes, and subject to 2 ns of MD followed by minimization. The shading and lines represent the boundaries of the toroidal pores in 26-Å-thick membranes. In the top views, the shading represents the inner and outer boundaries of the toroidal pore region; in the side views, the shaded volume represents the pore interior, and the lines represent the membrane edges. Figure created using VMD 1.9 (Humphrey et al. 1996).
Table 2.
Average membrane transfer energies (<ΔW>; kcal/mol) of tetramer arcs of peptides from water to toroidal pores (k = 15 Å) from 2-ns simulations.
| Tetramers: | Radius selected (Å) |
Neutral pore | 30% anionic pore |
|---|---|---|---|
| Protegrin-1 | 10 | −20.5 ± 2.1 | −55.6 ± 3.2 |
| θ-defensin | 12 | 16.7 ± 24.8 a | −11.9 ± 17.4 a |
| Tachyplesin | 12 | 34.0 ± 25.4 a | 9.7 ± 27.5 a |
| Polyphemusin | 12 | 22.4 ± 25.0 a | 1.2 ± 24.0 a |
| Gomesin | 10 | −5.7 ± 2.9 | −23.6 ± 2.8 |
| Androctonin | 10 | 4.2 ± 3.4 | −22.6 ± 7.2 |
| YGKRGF | 10 | −50.6 ± 2.2 | −70.2 ± 3.3 |
| AGGKGF | 10 | −50.0 ± 2.4 | −71.8 ± 3.2 |
Tetramer left the pore during the course of the simulation; values estimated using the time window of 3–22 ps. Other values estimated using the window 1001–2000 ps.
Table 5.
Average membrane transfer energies (<ΔW>; kcal/mol) of decamer barrels of peptides from water to toroidal pores (k = 15 Å) from 2-ns simulations.
| Decamers: | Radius selected (Å) |
Neutral pore | 30% anionic pore |
|---|---|---|---|
| Protegrin-1 | 15 | −17.2 ± 2.9 | −101.8 ± 5.3 |
| θ-defensin | 15 | 137.7 ± 14.2a | 62.8 ± 21.5a |
| Tachyplesin | 12 | 87.5 ± 48.6a | 29.3 ± 75.9a |
| Polyphemusin | 15 | 77.7 ± 43.9a | 28.8 ± 43.1a |
| Gomesin | 15 | −1.3 ± 3.8 | −65.7 ± 6.0 |
| Androctonin | 15 | 189.9 ± 39.1a | −70.4 ± 6.4 |
| YGKRGF | 12 | −106.4 ± 3.7 | −177.8 ± 5.0 |
| AGGKGF | 10 | −102.4 ± 4.6 | −150.7 ± 6.5 |
Decamer left the pore during the course of the simulation; values estimated using the time window of 3–22 ps. Other values estimated using the window 1001–2000 ps.
Fig 7.
Average membrane transfer energies (<ΔW>; kcal/mol) of tetramer arcs and heptamer, octamer, and decamer barrels of peptides from water to toroidal pores (k = 15 Å, pore radii as described in Tables 2–5) from 2-ns simulations. * The oligomer left the pore during the course of the simulation; values were estimated using the time window 3–22 ps. ΔW values in conditions in which the oligomer remained inside the pore were estimated using the window 1001–2000 ps. Error bars represent ± 1 SD.
Protegrin is the best studied among the peptides investigated. Throughout the 2-ns simulations, all studied protegrin oligomers remained bound to both neutral and anionic pores, and transfer energies to the pore-embedded state were favorable in all conditions (Tables 1–5). As expected, the transfer energies for all protegrin oligomers were significantly more favorable to anionic than neutral pores. (This trend also held for all other peptides in all conditions.) There was little distortion of the barrels/arcs throughout the simulations at the pore radii that gave optimum results (Figs 5a, 6a). The obtained ΔW values for protegrin matched well with the results from a previous study (Lazaridis et al. 2013).
The simulations of θ-defensin oligomers resulted in distortion of the peptide assemblies, and in some conditions, the barrels/arcs of θ-defensin exited the pore (Tables 1–5). Throughout the simulations, the θ-defensin octamer β-barrel remained bound to both neutral and anionic pores with 15 Å radii, but in the neutral case, some monomers moved to the center of the pore instead of remaining adjacent to the pore interface (Fig 5b), and there was an unfavorable transfer energy from water to pore; in an anionic pore, the octamer barrel distorted only slightly, and the transfer energy was favorable. The θ-defensin tetramer arcs and heptamer and decamer barrels displayed the same pattern of binding energies as the corresponding octamer barrels (i.e., unfavorable and favorable transfer energies in the neutral and anionic cases, respectively), but the other oligomeric states of θ-defensin did not show stable behavior during the simulations (Tables 1–5, Fig 7). The tetramer arcs migrated outside of both neutral and anionic pores within 70 ps of simulation onset. During the simulations of heptamer barrels, θ-defensin remained within the pore throughout the simulation, but the integrity of the barrel was not maintained: there was barrel shear in a neutral pore, and the ring collapsed in an anionic pore. The decamer barrels of θ-defensin exited the pore rapidly at the onset of the simulation (Table 1).
Although the oligomers of tachyplesin remained relatively coherent throughout the simulations in most conditions, some conditions resulted in exit from the membrane, even when the transfer energy was favorable (Tables 1–5, Fig 7). For heptamer and octamer β-barrels of tachyplesin, transfer energies were favorable in anionic and unfavorable in neutral pores. The tetramer arcs and decamer β-barrels had unfavorable transfer energies to both neutral and anionic pores. The octamer barrel in an anionic pore was the only condition in which tachyplesin remained at the pore interface throughout the simulation (Fig 5c); the octamer barrel exited a neutral pore within 50 ps of simulation onset. Tachyplesin tetramer arcs also exited the pore rapidly, but whereas the tetramer drifted away from a neutral membrane, it remained as a coherent arc with its hydrophobic side adjacent to the anionic membrane’s surface. Similarly, the heptamer barrel of tachyplesin left a neutral pore rapidly (Table 1), whereas it migrated to the membrane surface of an anionic pore and remained there while the barrel opened. Decamer barrels of tachyplesin exited the pore region rapidly in all conditions (Table 1).
Polyphemusin oligomers generally displayed unstable behavior in pores, with distortions observed in all conditions (Table 1). Polyphemusin’s pattern of favorable vs. unfavorable binding energies was exactly the same as that for tachyplesin: the octamer and heptamer β-barrels had favorable and unfavorable binding energies for anionic and neutral pores, respectively, and tetramer arcs and decamer barrels had unfavorable transfer energies in all pores (Tables 2–5, Fig 7). Polyphemusin oligomers left the pore in all conditions except for the octamer in an anionic pore, where the barrel moved toward the surface of the membrane with only the membrane-facing end of the barrel remaining coherent; the solvent-facing end became distorted, and the ends of the monomers spread apart (Fig 5d). The polyphemusin barrels did not degrade completely; although there was some distortion, the assemblies remained somewhat coherent after migrating into the solvent. The tetramer arc in an anionic pore, however, became highly distorted and remained with the hydrophobic side adjacent to the membrane surface.
The simulations of gomesin oligomers revealed favorable transfer energies in all conditions except the heptamer barrel in a neutral pore (Tables 2–5, Fig 7). The gomesin octamer β-barrel remained bound to both neutral and anionic pore interfaces throughout the simulations, but some monomers migrated to the center of the pore and away from the pore interface (Fig 5e), indicating partial collapse of the octamer barrel. The tetramer arcs of gomesin remained bound to both neutral and anionic pores throughout the simulation; however, the arcs did not remain coherent, reconfiguring as groups of three and one monomers that remained orthogonal to the membrane surface (Fig 6b). The heptamer and decamer barrels also remained bound to the pore throughout the 2-ns MD simulations, but barrel collapse and distortion occurred in the heptameric and decameric conditions, respectively (Table 1).
For all oligomeric states, androctonin showed unfavorable and favorable transfer energies for neutral and anionic pores, respectively (Tables 2–5, Fig 7). Although the conformation of androctonin was altered from its native one to facilitate barrel formation (which may have led to the high standard deviation in the ΔW values), the androctonin oligomers maintained at least some coherence throughout the 2 simulations (Fig 5f, Fig 6c), and they generally remained bound to the pore throughout the simulation: the only condition in which androctonin left the pore was the decamer from a neutral membrane. However, distortion of the androctonin oligomers occurred in all conditions: there was moderate distortion of the octamer barrels, and in the heptamer and decamer barrel conditions in which androctonin did not leave the pore, the barrels collapsed, indicating a lack of ring stability (Table 1). The tetramer arcs of androctonin reconfigured as barrel-like structures (Fig 6c).
YGKRGF and AGGKGF showed remarkably stable behavior in all simulated conditions. They remained bound to both neutral and anionic pores throughout the 2-ns simulations for all oligomeric states (Table 1). In several conditions, there was little distortion of the arc/barrel conformation: the octamer barrels and tetramer arcs remained intact, with the monomers of the AGGKGF tetramer tilting in an anionic pore (Figs 5g–h, 6d–e). Although the heptamer barrels of YGKRGF and AGGKGF both remained within the pore, the heptamer barrel of YGKRGF opened even with Ro = 10 Å. The decamer barrel of AGGKGF became distorted in an anionic pore, but the decamer of YGKRGF remained stable and retained its shape throughout simulations in both neutral and anionic pores. These peptides’ transfer energies were negative to both neutral and anionic pores in all conditions (Tables 2–5, Fig 7); in fact, they had the most favorable ΔW values of any peptides studied.
Discussion
The results indicate that β-barrels and arcs of θ-defensin, tachyplesin, gomesin, polyphemusin, and androctonin are not as stable as those of protegrin. In many cases, we observed exit from the pore, significant distortions of the barrels, and/or unfavorable binding energies. None of them had transfer energies as favorable as those of protegrin under any conditions. However, two combinatorial library peptides surpassed protegrin in this measure. We discuss the results in light of available experimental data.
One reason for the stability of protegrin and the synthetic peptides as β-barrels is that they fit a pattern of amphipathicity wherein charged/polar residues are predominantly found at the ends or turn of the hairpin and nonpolar residues in the middle of one face of the hairpin. These peptides have no polar/charged residues facing the membrane in barrel configuration, whereas ach of the other studied AMPs have at least one. The desolvation cost of a polar side chain sequestered among nonpolar membrane lipids likely contributes significantly to the instability of these peptides in pores. Further, θ-defensin and androctonin lack sufficient large hydrophobic residues facing the membrane for efficient membrane binding in β-barrel conformation. Another factor could be the amount of charge in the pore: protegrin has one Arg residue facing the pore, whereas tachyplesin and polyphemusin have two in a more central location, which might create electrostatic repulsion. There is experimental evidence that tachyplesin (Doherty et al. 2006, Boughton et al. 2011) and androctonin (Hetru et al. 2000) orient parallel to the membrane surface instead of adopting a transmembrane orientation, and a carpet mechanism was suggested for gomesin at high peptide concentrations (Domingues et al. 2010). The inability of polyphemusin to cause dye leakage from vesicles led other authors to propose an internalization mechanism that does not involve pore formation (Powers et al. 2005).
All peptides considered here show less favorable binding in neutral than 30% anionic pores. This is not surprising, considering that they are all positively charged. A practical goal in antibiotic development is to target microbes’ anionic membranes while leaving mammalian cells’ zwitterionic membranes intact. Thus, if the ΔW values in zwitterionic and anionic membranes reflect antimicrobial and hemolytic activity, respectively, and if the β-barrel mechanism is valid, one would expect a correlation between these values and the experimental biological activities. Among the natural AMPs studied, protegrin had the most favorable ΔW values in both anionic and neutral pores. Thus, the other natural AMPs are expected to be less hemolytic than protegrin. This seems mostly consistent with available experimental data: θ-defensin is not hemolytic (Tran et al. 2008), and gomesin is less hemolytic than protegrin, notably in the high-concentration regime (Silva et al. 2000). Androctonin’s relatively low hemolytic activity levels are in accordance with its relatively hydrophilic region facing the membrane and moderately unfavorable ΔW in neutral pores. On the other hand, tachyplesin’s hemolytic activity is comparable to protegrin’s (Ramamoorthy et al. 2006), and polyphemusin is more hemolytic than protegrin (Zhang et al. 2000), so alternative explanations are needed to rationalize these findings, such as toroidal pore stabilization without peptide assembly into a well-defined structure.
It is also instructive to review the relationship between ΔW in anionic pores and activity against bacteria (He and Lazaridis 2013). Because there are many species of bacteria against which the activity levels of several of these peptides have been tested, we select Staphylococcus aureus (SA) and Escherichia coli (EC) as examples of Gram-positive and Gram-negative bacteria, respectively. Protegrin has exhibited a range of observed MIC values: one study observed MICs of 30 µM against both EC and SA using a broth dilution technique (Mangoni et al. 1996); however, another study using the same technique observed an MIC of 4.6 µM for EC (Wessely-Szponder et al. 2010), and another study observed MICs of 2.9 and 5.8 µM for EC and SA, respectively (Lai et al. 2006). On the low end, a study using a radial diffusion assay measured MICs of 0.62–0.68 and 0.82–0.87 µM for EC and SA, respectively (Tam et al. 2000). For θ-defensin, researchers have observed MIC values of 1.0 and 2.1 µM for EC and SA, respectively (Tran et al. 2002). For tachyplesin, the results for SA have depended on whether the peptide was natural or synthetic and the strain of bacteria used; MIC values have ranged 1.4–5.5 µM, and values for EC ranged 0.7–1.4 µM regardless of whether the peptide was natural or synthetic (Miyata et al. 1989). For polyphemusin, the MIC values were 2.6 µM for both EC and SA. In the study in which gomesin was originally isolated, its MIC against EC ranged 0.4–1.6 µM, depending on strain, and that for SA was 1.6–3.15 µM (Silva et al. 2000). The same study measured MIC values for androctonin of 3–15 µM and 15–30 µM, respectively, for EC and SA (Silva et al. 2000). YGKRGF and AGGKGF have minimal sterilizing concentrations (MSC) of 2.1 µM (Rausch et al. 2005) and 1.0 µM (Rausch et al. 2007), respectively, against SA, and both peptides have MSC values of 0.5 µM against EC (Rausch et al. 2007). If we adopt the low-end values for protegrin (Tran et al. 2002), then a correlation is observed between the ΔW values of the octamer barrels in anionic pores and activity against both types of bacteria, with the same peptides (protegrin, YGKRGF, and AGGKGF) having the most negative ΔW values and the lowest MIC values. The other peptides, with higher ΔW values of the octamer barrel in anionic pores, also have higher MIC values.
The binding energy values for YGKRGF and AGGKGF may be misleading, because those quantities do not contain any conformational, translational, or rotational entropy contributions. These peptides’ lack of disulfide bonds makes constraining them into hairpin conformations entropically costly; thus, their pore binding energies are likely significantly less favorable than those we computed. Nevertheless, the fact that the negative control (AGGKGF) exhibits less favorable binding energies than the positive control is gratifying. The observed stability of these peptides’ pore structures contrasts with the experimental observation of graded and incomplete dye leakage from vesicles (Rausch et al. 2005, 2007), which suggested a carpet or “sinking raft” mechanism (although that could occur as a mechanistic step toward pore formation; Rausch et al. 2005). A high energetic barrier (enthalpic for dissociation and entropic for association) may separate the barrel/arc states from the rest of the configurational space; this would need to be considered in a complete computational characterization. It would also be interesting to synthesize disulfide-bonded versions of the combinatorial library peptides and test their activity.
Mechanisms other than membrane pore formation are possible for many of the peptides investigated here. For example, polyphemusin might bind to divalent cation binding sites on target cells’ lipopolysaccharide molecules to penetrate and permeabilize the membrane (Zhang et al. 2000). Tachyplesin activates annexin labeling and caspase-3 activation (Paredes-Gamero et al. 2012). Not only gomesin (Soletti et al. 2010) but also protegrin itself (Paredes-Gamero et al. 2012) causes calcium influx through L-type channels and generates reactive oxygen species. Gomesin’s mechanism may depend on concentration, as lower concentrations promote apoptosis, whereas higher concentrations result in cell membrane disruption (Paredes-Gamero et al. 2012). Thus, higher concentrations of peptide may make pore formation more likely. There were certain conditions in which non-protegrin AMPs remained within the pore without becoming distorted, such as the tachyplesin octamer in an anionic membrane; it may be worth investigating whether pore formation occurs at high concentrations of tachyplesin, with monomers orienting parallel to the membrane as a first step.
The initial structures in this study had zero tilt angle for the monomers, which implies zero β-sheet twist in the barrel (Murzin et al., 1994). Unconstrained simulations did not produce significant systematic change in tilt, except local distortions. However, many of the tetramer arcs spontaneously rearranged into configurations with substantial tilt angles. Thus, an energetic barrier might prevent barrel reconfiguration. It would be useful to consider starting structures with tilted β-strands in subsequent studies. Membrane thickness was also not varied in this study: all simulations used a thickness of 26 Å. In addition, we used a generalization regarding the lipid composition of mammalian and bacterial cells, assuming zwitterionic and 30% anionic lipids in those respective cases. However, those values are not exact for all cells, so it would be useful to investigate how changes in membrane charge affect the results. Further, we did not systematically investigate pore curvature, mainly studying toroidal pores with k = 15 Å, whereas in the previous study of protegrin, we considered cylindrical pores in addition to toroidal pores with k = 10, 15, and 20 Å. We did vary the pore radius, but it may be useful to simulate even smaller radii than 10 Å for arcs. Indeed, tetramer arcs of many of these peptides seemed to curl naturally into incomplete cylindrical conformations with tilted monomers, some of which had significantly smaller radii than the corresponding octamer barrels. Additional oligomeric states could also be considered. These additional model parameter variations could supplement the results obtained in this study.
Supplementary Material
Table 3.
Average membrane transfer energies (<ΔW>; kcal/mol) of heptamer barrels of peptides from water to toroidal pores (k = 15 Å) from 2-ns simulations.
| Heptamers: | Radius selected (Å) |
Neutral pore | 30% anionic pore |
|---|---|---|---|
| Protegrin-1 | 10 | −17.1 ± 2.5 | −97.3 ± 4.7 |
| θ-defensin | 15 | 3.5 ± 3.4 | −30.4 ± 3.6 |
| Tachyplesin | 15 | 17.9 ± 6.1a | −19.1 ± 13.8a |
| Polyphemusin | 15 | 22.5 ± 18.5a | −1.5 ± 21.4a |
| Gomesin | 12 | 7.7 ± 5.6 | −36.5 ± 4.0 |
| Androctonin | 12 | 36.1 ± 5.5 | −35.6 ± 7.0 |
| YGKRGF | 10 | −77.8 ± 3.1 | −127.7 ± 4.9 |
| AGGKGF | 10 | −73.1 ± 2.8 | −116.4 ± 3.5 |
Heptamer left the pore during the course of the simulation; values estimated using the time window of 3–22 ps. Other values estimated using the window 1001–2000 ps.
Table 4.
Average membrane transfer energies (<ΔW>; kcal/mol) of octamer barrels of peptides from water to toroidal pores (k = 15 Å) from 2-ns simulations.
| Octamers: | Radius selected (Å) |
Neutral pore | 30% Anionic pore |
|---|---|---|---|
| Protegrin-1 | 12 | −23.3 ± 2.6 | −103.9 ± 4.9 |
| θ-defensin | 15 | 21.3 ± 3.7 | −11.7 ± 7.5 |
| Tachyplesin | 15 | 51.0 ± 35.0 a | −44.1 ± 4.7 |
| Polyphemusin | 15 | 35.8 ± 29.1a | −36.3 ± 3.3 |
| Gomesin | 15 | −3.3 ± 4.3 | −40.5 ± 4.6 |
| Androctonin | 15 | 34.5 ± 6.1 | −24.7 ± 12.6 |
| YGKRGF | 10 | −93.8 ± 3.4 | −147.7 ± 5.3 |
| AGGKGF | 10 | −80.9 ± 3.3 | −134.4 ± 4.1 |
Octamer left the pore during the course of the simulation; values estimated using the time window of 3–22 ps. Other values estimated using the window 1001–2000 ps.
Acknowledgments
Funding for this research was provided by the National Science Foundation (MCB 1244207). Infrastructure support was provided in part by RCMI grant 2G12RR03060-26A1/8G12MD007603-27 from the National Institutes of Health. No human or animal studies were carried out by the authors for this article.
Footnotes
Authors RBL and TL declare that they have no conflict of interest.
Online Resource 1 Average total effective energies (<W>; kcal/mol) and membrane transfer energies (<ΔW>; kcal/mol) of octamer barrels and tetramer arcs of peptides from water to toroidal pores of Ro = 10 Å, 12 Å, and 15 Å (k = 15 Å) from 2-ns simulations. Description of the behavior of all octamer β-barrels and tetramer arcs during these simulations.
References
- Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc Natl Acad Sci USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechinger B. Structure and function of membrane-lytic peptides. Crit Rev Plant Sci. 2004;23:271–292. [Google Scholar]
- Ben-Tal N, Honig B, Peitzsch RM, Denisov G, McLaughlin S. Binding of small basic peptides to membranes containing acidic lipids: Theoretical models and experimental results. Biophys J. 1996;71:561–575. doi: 10.1016/S0006-3495(96)79280-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolintineanu DS, Kaznessis YN. Computational studies of protegrin antimicrobial peptides: A review. Peptides. 2011;32:188–201. doi: 10.1016/j.peptides.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boughton AP, Nguyen K, Andricioaei I, Chen Z. Interfacial orientation and secondary structure change in tachyplesin I: Molecular dynamics and sum frequency generation spectroscopy studies. Langmuir. 2011;27:14343–14351. doi: 10.1021/la203192c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradshaw JP. Cationic antimicrobial peptides. Issues for potential clinical use. Biodrugs. 2003;17:233–240. doi: 10.2165/00063030-200317040-00002. [DOI] [PubMed] [Google Scholar]
- Brogden K. Antimicrobial peptides: Pore formers or metabolic inhibitors in bacteria? Nat Rev Microbiol. 2005;3:238–250. doi: 10.1038/nrmicro1098. [DOI] [PubMed] [Google Scholar]
- Brooks BR, Brooks CL, III, MacKerrel AD, Jr, Nilsson L, Petrella RJ, et al. CHARMM: The biomolecular simulation program. J Comput Chem. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown KL, Hancock REW. Cationic host defense (antimicrobial) peptides. Curr Opin Immunol. 2006;18:24–30. doi: 10.1016/j.coi.2005.11.004. [DOI] [PubMed] [Google Scholar]
- Capone RM, Mustata M, Jang H, Arce FT, Nussinov R, Lal R. Antimicrobial protegrin-1 forms ion channels: Molecular dynamic simulation, atomic force microscopy, and electrical conductance studies. Biophys J. 2010;98:2644–2652. doi: 10.1016/j.bpj.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Falla TJ, Liu H, Hurst MA, Fujii CA, et al. Development of protegrins for the treatment and prevention of oral mucositis: Structure-activity relationships of synthetic protegrin analogues. Biopolymers. 2000;55:88–98. doi: 10.1002/1097-0282(2000)55:1<88::AID-BIP80>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Cole AM. Antimicrobial peptide microbicides targeting HIV. Protein Pept Lett. 2005;12:41–47. doi: 10.2174/0929866053406101. [DOI] [PubMed] [Google Scholar]
- Doherty T, Waring AJ, Hong M. Membrane-bound conformation and topology of the antimicrobial peptide tachyplesin I by solid-state NMR. Biochemistry. 2006:13323–13330. doi: 10.1021/bi061424u. [DOI] [PubMed] [Google Scholar]
- Domingues TM, Riske KA, Miranda A. Revealing the lytic mechanism of the antimicrobial peptide gomesin by observing giant unilamellar vesicles. Langmuir. 2010;26:11077–11084. doi: 10.1021/la100662a. [DOI] [PubMed] [Google Scholar]
- Ehret-Sabatier L, Loew D, Goyffon M, Fehlbaum P, Hoffmann JA, et al. Characterization of novel cysteine-rich antimicrobial peptides from scorpion blood. J Biol Chem. 1996;271:29537–29544. doi: 10.1074/jbc.271.47.29537. [DOI] [PubMed] [Google Scholar]
- Eisenberg D, Schwarz E, Komarony M, Wall R. Amino acid scale: Normalized consensus hydrophobicity scale. J Mol Biol. 1984;179:125–142. doi: 10.1016/0022-2836(84)90309-7. [DOI] [PubMed] [Google Scholar]
- Fahrner RL, Dieckmann T, Harwig SSL, Lehrer RI, Eisenberg D, Feigon J. Solution structure of protegrin-1, a broad-spectrum antimicrobial peptide from porcine leukocytes. J Chem Biol. 1996;3:543–550. doi: 10.1016/s1074-5521(96)90145-3. [DOI] [PubMed] [Google Scholar]
- Fázio MA, Jouvensal L, Vovelle F, Bulet P, Miranda TM, et al. Biological and structural characterization of new linear gomesin analogues with improved therapeutic indices. Pept Sci. 2007;88:386–400. doi: 10.1002/bip.20660. [DOI] [PubMed] [Google Scholar]
- Fázio MA, Oliveira VX, Bulet P, Miranda MTM, Daffre S, Miranda A. Structure–activity relationship studies of gomesin: Importance of the disulfide bridges for conformation, bioactivities, and serum stability. Pept Sci. 2006;84:205–218. doi: 10.1002/bip.20396. [DOI] [PubMed] [Google Scholar]
- Gille C. STRAP: Interactive structure based sequences alignment program. [Accessed 7 November, 2014];Institut für Biochemie, Charité Berlin. 2012 http://www.bioinformatics.org/strap/index2.html. [Google Scholar]
- Hancock REW. Cationic peptides: Effectors in innate immunity and novel antimicrobials. Lancet Infect Dis. 2001;1:156–164. doi: 10.1016/S1473-3099(01)00092-5. [DOI] [PubMed] [Google Scholar]
- He Y, Prieto L, Lazaridis T. Modeling peptide binding to anionic membrane pores. J Comput Chem. 2013;34:1463–1475. doi: 10.1002/jcc.23282. [DOI] [PubMed] [Google Scholar]
- He Y, Lazaridis T. Activity Determinants of Helical Antimicrobial Peptides: A Large-scale Computational Study. PLOS One. 2013;8(6):e66440. doi: 10.1371/journal.pone.0066440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller WT, Waring AJ, Lehrer RI, Huang HW. Multiple states of β-sheet peptide protegrin in lipid bilayers. Biochemistry. 1998;37:17331–17338. doi: 10.1021/bi981314q. [DOI] [PubMed] [Google Scholar]
- Hetru C, Letellier L, Oren Z, Hoffmann JA, Shai Y. Androctonin, a hydrophilic disulphide-bridged non-haemolytic anti-microbial peptide: A plausible mode of action. Biochem J. 2000;345:653–664. [PMC free article] [PubMed] [Google Scholar]
- Huang HW. Action of antimicrobial peptides: Two-state model. Biochemistry. 2000;39:8347–8352. doi: 10.1021/bi000946l. [DOI] [PubMed] [Google Scholar]
- Huang HW. Molecular mechanism of antimicrobial peptides: The origin of cooperativity. Biochim Biophys Acta. 2006;1758:1292–1302. doi: 10.1016/j.bbamem.2006.02.001. [DOI] [PubMed] [Google Scholar]
- Humphrey W, Dalke A, Schulten K. VMD – Visual Molecular Dynamics. J Molec Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Ishitsuka Y, Pham DS, Waring AJ, Lehrer RI, Lee KYC. Insertion selectivity of antimicrobial peptide protegrin-1 into lipid monolayers: Effect of head group electrostatics and tail group packing. Biochim Biophys Acta. 2006;1758:1450–1460. doi: 10.1016/j.bbamem.2006.08.001. [DOI] [PubMed] [Google Scholar]
- Jang H, Ma B, Lal R, Nussinov R. Models of toxic beta-sheet channels of protegrin-1 suggest a common subunit organization motif shared with toxic Alzheimer beta-amyloid ion channels. Biophys J. 2008;95:4631–4642. doi: 10.1529/biophysj.108.134551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang H, Ma BY, Nussinov R. Conformational study of the protegrin-I (PG-I) dimer interaction with lipid bilayers and its effect. BMC Struct Biol. 2007;7:20. doi: 10.1186/1472-6807-7-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang H, Ma B, Woolf TB, Nussinov R. Interaction of protegrin-1 with lipid bilayers: Membrane thinning effect. Biophys J. 2006;91:2848–2859. doi: 10.1529/biophysj.106.084046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang H, Teran Arce F, Ramachandran S, Capone R, Lal R, Nussinov R. Structural convergence among diverse, toxic β-sheet ion channels. J Phys Chem B. 2010;114:9445–9451. doi: 10.1021/jp104073k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandasamy SK, Larson RG. Binding modes of protegrin-1, a β-strand antimicrobial peptide, in lipid bilayers. Mol. Simul. 2007;33:799–807. [Google Scholar]
- Katsu T, Nakao S, Iwanaga S. Mode of action of an antimicrobial peptide, tachyplesin I, on biomembranes. Bio Pharm Bull. 1993;16:178–181. doi: 10.1248/bpb.16.178. [DOI] [PubMed] [Google Scholar]
- Khandelia H, Kaznessis YN. Structure of the antimicrobial β-hairpin peptide protegrin-1 in a DLPC lipid bilayer investigated by molecular dynamics simulation. Biochim Biophys Acta. 2007;1768:509–520. doi: 10.1016/j.bbamem.2006.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokryakov VN, Harwig SSL, Panyutich EA, Shevchenko AA, Aleshina GM, et al. Protegrins: Leukocyte antimicrobial peptides that combine features of corticostatic defensins and tachyplesins. FEBS Lett. 1993;327:231–236. doi: 10.1016/0014-5793(93)80175-t. [DOI] [PubMed] [Google Scholar]
- Laederach A, Andreotti AH, Fulton DB. Solution and micelle-bound structures of tachyplesin I and its active aromatic linear derivatives. Biochemistry. 2002;41:12359–12368. doi: 10.1021/bi026185z. [DOI] [PubMed] [Google Scholar]
- Lai JR, Epand RF, Weisblum B, Epand RM, Gellman SH. Roles of salt and conformation in the biological and physicochemical behavior of protegrin-1 and designed analogues: Correlation of antimicrobial, hemolytic, and lipid bilayer-perturbing activities. Biochemistry. 2006;45:15718–15730. doi: 10.1021/bi0617759. [DOI] [PubMed] [Google Scholar]
- Lam KLH, Wang H, Siaw TA, Chapman MR, Waring AJ, Kindt JT, Lee KYC. Mechanism of structural transformations induced by antimicrobial peptides in lipid membranes. Biochimica Et Biophysica Acta-Biomembranes. 2012;1818:194–204. doi: 10.1016/j.bbamem.2011.11.002. [DOI] [PubMed] [Google Scholar]
- Langham AA, Ahmad AS, Kaznessis YN. On the nature of antimicrobial activity: A model for protegrin-1 pores. J Am Chem Soc. 2008;130:4338–4346. doi: 10.1021/ja0780380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langham AA, Kaznessis YN. Effects of mutations on the C-terminus of protegrin-1: A molecular dynamics simulation study. Mol Simul. 2006;32:193–201. [Google Scholar]
- Lazaridis T. Effective energy function for proteins in lipid membranes. Proteins. 2003;52:176–192. doi: 10.1002/prot.10410. [DOI] [PubMed] [Google Scholar]
- Lazaridis T. Implicit solvent simulations of peptide interactions with anionic lipid membranes. Proteins. 2005a;58:518–527. doi: 10.1002/prot.20358. [DOI] [PubMed] [Google Scholar]
- Lazaridis T. Structural determinants of transmembrane beta-barrels. Journal of Chemical Theory and Computation. 2005b;1:716–722. doi: 10.1021/ct050055x. [DOI] [PubMed] [Google Scholar]
- Lazaridis T, He Y, Prieto L. Membrane interactions and pore formation by the antimicrobial peptide protegrin. Biophys J. 2013;104:633–642. doi: 10.1016/j.bpj.2012.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazaridis T, Karplus M. Effective energy function for proteins in solution. Proteins. 1999;35:133–152. doi: 10.1002/(sici)1097-0134(19990501)35:2<133::aid-prot1>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- Lehrer RI, Cole AM, Selsted ME. Theta-defensins: Cyclic peptides with endless potential. J Biol Chem. 2012;287:27014–27019. doi: 10.1074/jbc.R112.346098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leontiadou H, Mark AE, Marrink SJ. Antimicrobial peptides in action. J Am Chem Soc. 2006;128:12156–12161. doi: 10.1021/ja062927q. [DOI] [PubMed] [Google Scholar]
- Lerner MG, Carlson HA. PyMOL APBS tools. [Accessed 7 November, 2014];2009 http://www.pymolwiki.org/index.php/Apbs. [Google Scholar]
- Ludtke SJ, He K, Heller WT, Harroun TA, Yang L, Huang HW. Membrane pores induced by magainin. Biochemistry. 1996;35:13723–13728. doi: 10.1021/bi9620621. [DOI] [PubMed] [Google Scholar]
- Mandard N, Bulet P, Caille A, Daffre S, Vovelle F. The solution structure of gomesin, an antimicrobial cysteine-rich peptide from the spider. Eur J Biochem. 2002;269:1190–1198. doi: 10.1046/j.0014-2956.2002.02760.x. [DOI] [PubMed] [Google Scholar]
- Mandard N, Labbe H, Da Silva P, Hetru C, Landon C, Vovelle F. Role of disulfide bridges in the hairpin fold of androctonin. Structure-activity relationships. Comptes Rendus De L Academie Des Sciences Serie Ii Fascicule C-Chimie. 2001;4:735–738. [Google Scholar]
- Mandard N, Sy D, Maufrais C, Monmatin JM, Bulet P, et al. Androctonin, a novel antimicrobial peptide from scorpion Androctonus australis: Solution structure and molecular dynamics simulations in the presence of a lipid monolayer. J Biomol Struct Dyn. 1999;17:367–380. doi: 10.1080/07391102.1999.10508368. [DOI] [PubMed] [Google Scholar]
- Mangoni ME, Aumelas A, Charnet P, Roumestand P, Chiche L, et al. Change in membrane permeability induced by protegrin 1: Implication of disulphide bridges for pore formation. FEBS Lett. 1996;383:93–98. doi: 10.1016/0014-5793(96)00236-0. [DOI] [PubMed] [Google Scholar]
- Mani R, Cady SD, Tang M, Waring AJ, Lehrer RI, Hong M. Membrane-dependent oligomeric structure and pore formation of beta-hairpin antimicrobial peptide in lipid bilayers from solid-state NMR. P Natl Acad Sci USA. 2006;103:16242–16247. doi: 10.1073/pnas.0605079103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki K. Why and how are peptide–lipid interactions utilized for self-defense? Magainins and tachyplesins as archetypes. Biochim Biophys Acta. 1999;1462:1–10. doi: 10.1016/s0005-2736(99)00197-2. [DOI] [PubMed] [Google Scholar]
- Matsuzaki K. Control of cell selectivity of antimicrobial peptides. Biochim. Biophys Acta. 2009;1788:1687–1692. doi: 10.1016/j.bbamem.2008.09.013. [DOI] [PubMed] [Google Scholar]
- Matsuzaki K, Murase O, Fujii N, Miyajima K. An antimicrobial peptide, magainin 2, induced rapid flip-flop of phospholipids coupled with pore formation and peptide translocation. Biochemistry. 1996;35:11361–11368. doi: 10.1021/bi960016v. [DOI] [PubMed] [Google Scholar]
- Mihajlovic M, Lazaridis T. Antimicrobial peptides bind more strongly to membrane pores. Biochim Biophys Acta. 2010;1798:1494–1502. doi: 10.1016/j.bbamem.2010.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata T, Tokunaga F, Yoneya T, Yoshikawa K, Iwanaga S, et al. Antimicrobial peptides, isolated from horseshoe crab hemocytes, tachyplesin II, and polyphemusins I and II: Chemical structures and biological activity. J Biochem. 1989;106:663–668. doi: 10.1093/oxfordjournals.jbchem.a122913. [DOI] [PubMed] [Google Scholar]
- Mizuguchi M, Kamata S, Kawabata S, Fujitani N, Kawano K. Solution structure of tachyplesin I in H2O. PDB 1WO0. (unpublished) [Google Scholar]
- Morein S, Andersson A-S, Rilfors L, Lindblom G. Wild-type Escherichia coli cells regulate the membrane lipid composition in a “window” between gel and non-lamellar structures. J Biol Chem. 1996;271:6801–6809. doi: 10.1074/jbc.271.12.6801. [DOI] [PubMed] [Google Scholar]
- Mottamal M, Lazaridis T. Voltage-dependent energetics of alamethicin monomers in the membrane. Biophys Chem. 2006;122:50–57. doi: 10.1016/j.bpc.2006.02.005. [DOI] [PubMed] [Google Scholar]
- Murzin AG, Lesk AM, Chothia C. Principles determining the structure of beta-sheet barrels in proteins. I. A theoretical analysis. J Mol Biol. 1994;236:1369–1381. doi: 10.1016/0022-2836(94)90064-7. [DOI] [PubMed] [Google Scholar]
- Oren Z, Shai Y. Mode of action of linear amphipathic alpha-helical antimicrobial peptides. Biopolymers. 1998;47:451–463. doi: 10.1002/(SICI)1097-0282(1998)47:6<451::AID-BIP4>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Ostberg N, Kaznessis Y. Protegrin structure-activity relationships: Using homology models of synthetic sequences to determine structural characteristics important for activity. Peptides. 2005;26:197–206. doi: 10.1016/j.peptides.2004.09.020. [DOI] [PubMed] [Google Scholar]
- Palermo EF, Vemparala S, Kuroda K. Cationic spacer arm design strategy for control of antimicrobial activity and conformation of amphiphilic methacrylate random copolymers. Biomacromolecules. 2012;13:1632–1641. doi: 10.1021/bm300342u. [DOI] [PubMed] [Google Scholar]
- Papo N, Shai Y. Host defense peptides as new weapons in cancer treatment. Cell Mol Life Sci. 2005;62:784–790. doi: 10.1007/s00018-005-4560-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paredes-Gamero EJ, Martins MNC, Cappabianco FAM, Ide JS, Miranda A. Characterization of dual effects induced by antimicrobial peptides: Regulated cell death or membrane disruption. Biochim Biophys Acta. 2012;1820:1062–1072. doi: 10.1016/j.bbagen.2012.02.015. [DOI] [PubMed] [Google Scholar]
- Penberthy WT, Chari S, Cole AL, Cole AM. Retrocyclins and their activity against HIV-1. Cell Mol Life Sci. 2011;68:2231–2242. doi: 10.1007/s00018-011-0715-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers JPS, Rozek A, Hancock REW. Structure-activity relationships for the beta-hairpin cationic antimicrobial peptide polyphemusin I. Biochim Biophys Acta. 2004;1698:239–250. doi: 10.1016/j.bbapap.2003.12.009. [DOI] [PubMed] [Google Scholar]
- Powers JPS, Tan A, Ramamoorthy A, Hancock REW. Solution structure and interaction of the antimicrobial polyphemusins with lipid membranes. Biochemistry. 2005;44:15504–15513. doi: 10.1021/bi051302m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto L, He Y, Lazaridis T. Protein arcs may form stable pores in lipid membranes. Biophys J. 2014;106:154–161. doi: 10.1016/j.bpj.2013.11.4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramamoorthy A, Thennarasu S, Tan A, Gottipati K, Sreekumar S, Heyl DL, et al. Deletion of all cysteines in tachyplesin I abolishes hemolytic activity and maintains antimicrobial activity and lipopolysaccharide selective binding. Biochemistry. 2006;45:6529–6540. doi: 10.1021/bi052629q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratledge C, Wilkinson SG. Microbial Lipids. Vol. 1. London: Academic Press; 1988. [Google Scholar]
- Rausch JM, Marks JR, Rathinakumar R, Wimley WC. β-sheet pore-forming peptides selected from a rational combinatorial library: Mechanism of pore formation in lipid vesicles and activity in biological membranes. Biochemistry. 2007;46:12124–12139. doi: 10.1021/bi700978h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausch JM, Marks JR, Wimley WC. Rational combinatorial design of pore-forming β-sheet peptides. P Natl Acad Sci USA. 2005;102:10511–10515. doi: 10.1073/pnas.0502013102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JA, Shankaramma SC, Jetter P, Kienzl U, Schwendener RA, et al. Properties and structure-activity studies of cyclic b-hairpin peptidomimetics based on the cationic antimicrobial peptide protegrin I. Bioorg Med Chem. 2005;13:2055–2064. doi: 10.1016/j.bmc.2005.01.009. [DOI] [PubMed] [Google Scholar]
- Roumestand C, Louis V, Aumelas A, Grassy G, Calas B, Chavanieu A. Oligomerization of protegrin-1 in the presence of DPC micelles. A proton high-resolution NMR study. FEBS Lett. 1998;421:263–267. doi: 10.1016/s0014-5793(97)01579-2. [DOI] [PubMed] [Google Scholar]
- Rui H, Lee J, Im W. Comparative molecular dynamics simulation studies of protegrin-1 monomer and dimer in two different lipid bilayers. Biophys J. 2009;97:787–795. doi: 10.1016/j.bpj.2009.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rui HA, Im W. Protegrin-1 Orientation and Physicochemical Properties in Membrane Bilayers Studied by Potential of Mean Force Calculations. Journal of Computational Chemistry. 2010;31:2859–2867. doi: 10.1002/jcc.21580. [DOI] [PubMed] [Google Scholar]
- Schrödinger LLC. The PyMOL Molecular Graphics System, Version 1.7. Schrödinger LLC; 2014. [Google Scholar]
- Selsted ME. θ-defensins: Cyclic antimicrobial peptides produced by binary ligation of truncated α-defensins. Curr Protein Pept Sci. 2004;5:365–371. doi: 10.2174/1389203043379459. [DOI] [PubMed] [Google Scholar]
- Sengupta D, Leontiadou H, Mark AE, Marrink SJ. Toroidal pores formed by antimicrobial peptides show significant disorder. Biochim Biophys Acta. 2008;1778:2308–2317. doi: 10.1016/j.bbamem.2008.06.007. [DOI] [PubMed] [Google Scholar]
- Silphaduang U, Noga EJ. Peptide antibiotics in mast cells of fish. Nature. 2001;414:268–269. doi: 10.1038/35104690. [DOI] [PubMed] [Google Scholar]
- Silva PI, Daffre S, Bulet P. Isolation and characterization of gomesin, an 18-residue cysteine-rich defense peptide from the spider Acanthoscurria gomesiana hemocytes with sequence similarities to horseshoe crab antimicrobial peptides of the tachyplesin family. J Biol Chem. 2000;275:33464–33470. doi: 10.1074/jbc.M001491200. [DOI] [PubMed] [Google Scholar]
- Sokolov Y, Mirzabekov T, Martin DW, Lehrer RI, Kagan BL. Membrane channel formation by antimicrobial protegrins. Biochim Biophys Acta. 1999;1420:23–29. doi: 10.1016/s0005-2736(99)00086-3. [DOI] [PubMed] [Google Scholar]
- Soletti RC, del Barrio L, Daffre S, Miranda A, Borges HL, et al. Peptide gomesin triggers cell death through L-type channel calcium influx, MAPK/ERK, PKC and PI3K signaling and generation of reactive oxygen species. Chemico-Biological Interactions. 2010;186:135–143. doi: 10.1016/j.cbi.2010.04.012. [DOI] [PubMed] [Google Scholar]
- Steinberg DA, Hurst MA, Fujii CA, Kung AH, Ho JF, et al. Protegrin-1: A broad-spectrum, rapidly microbicidal peptide with in vivo activity. Antimicrob Agents Chemother. 1997;41:1738–1742. doi: 10.1128/aac.41.8.1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam JP, Wu CW, Yang JL. Membranolytic selectivity of cystine-stabilized cyclic protegrins. Eur J Biochem. 2000;267:3289–3300. doi: 10.1046/j.1432-1327.2000.01359.x. [DOI] [PubMed] [Google Scholar]
- Tang Y-Q, Yuan J, Ösapay G, Ösapay K, Tran D, et al. A cyclic antimicrobial peptide produced in primate leukocytes by the ligation of two truncated α-defensins. Science. 1999;286:498–502. doi: 10.1126/science.286.5439.498. [DOI] [PubMed] [Google Scholar]
- Tang M, Hong M. Structure and mechanism of beta-hairpin antimicrobial peptides in lipid bilayers from solid-state NMR spectroscopy. Mol Biosyst. 2009;5:317–322. doi: 10.1039/b820398a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thøgersen L, Schiøtt B, Vosegaard T, Nielsen NC, Tajkhorshid E. Peptide aggregation and pore formation in a lipid bilayer: A combined coarse-grained and all atom molecular dynamics study. Biophys J. 2008;95:4337–4347. doi: 10.1529/biophysj.108.133330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trabi M, Schirra HJ, Craik DJ. Three-dimensional structure of RTD-1, a cyclic antimicrobial defensin from rhesus macaque leukocytes. Biochemistry. 2001;40:4211–4221. doi: 10.1021/bi002028t. [DOI] [PubMed] [Google Scholar]
- Tran D, Tran P, Roberts K, Ösapay G, Schaal J, et al. Microbicidal proterties and cytocidal selectivity of rhesus macaque theta defensins. Antimicrob Agents Chemother. 2008;52:944–953. doi: 10.1128/AAC.01090-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkleij AJ, Zwaal RFA, Roelofsen B, Comfurius P, Kastelijn D, Deenen LLMV. The asymmetric distribution of phospholipids in the human red cell membrane. A combined study using phospholipases and freeze-etch electron microscopy. Biochim Biophys Acta. 1973;323:178–193. doi: 10.1016/0005-2736(73)90143-0. [DOI] [PubMed] [Google Scholar]
- Vivcharuk V, Kaznessis Y. Free energy profile of the interaction between a monomer or a dimer of protegrin-1 in a specific binding orientation and a model lipid bilayer. J Phys Chem B. 2010;114:2790–2797. doi: 10.1021/jp909640g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivcharuk V, Kaznessis Y. Thermodynamic analysis of protegrin-1 insertion and permeation through a lipid bilayer. J Phys Chem B. 2011;115:14704–14712. doi: 10.1021/jp205153y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessely-Szponder J, Majer-Dziedzic B, Smolira A. Analysis of antimicrobial peptides from porcine neutrophils. J Microbiol Meth. 2010;83:8–12. doi: 10.1016/j.mimet.2010.07.010. [DOI] [PubMed] [Google Scholar]
- Yang L, Harroun TA, Weiss TM, Ding L, Huang HW. Barrel-stave model or toroidal model? A case study on melittin pores. Biophys J. 2001;81:1475–1485. doi: 10.1016/S0006-3495(01)75802-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan H, Lazaridis T. Influence of the membrane dipole potential on peptide binding to lipid bilayers. Biophys Chem. 2012;161:1–7. doi: 10.1016/j.bpc.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan H, Lazaridis T. Inclusion of lateral pressure/curvature stress effects in implicit membrane models. Biophys J. 2013;104:643–654. doi: 10.1016/j.bpj.2012.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Scott MG, Yan H, Mayer LD, Hancock REW. Interaction of polyphemusin I and structural analogs with bacterial membranes, lipopolysaccharide, and lipid monolayers. Biochemistry. 2000;39:14504–14514. doi: 10.1021/bi0011173. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.