Abstract
The recently developed CARD-FISH protocol was refined for the detection of marine Archaea by replacing the lysozyme permeabilization treatment with proteinase K. This modification resulted in about twofold-higher detection rates for Archaea in deep waters. Using this method in combination with microautoradiography, we found that Archaea are more abundant than Bacteria (42% versus 32% of 4′,6′-diamidino-2-phenylindole counts) in the deep waters of the North Atlantic and that a larger fraction of Archaea than of Bacteria takes up l-aspartic acid (19% versus 10%).
Over the past decade, our knowledge of the phylogenetic composition of marine prokaryotic communities, including those inhabiting the deep ocean, has increased considerably due to the application of molecular tools such as fingerprinting techniques, cloning, and sequencing (7, 9, 10, 12, 20, 21). Fluorescence in situ hybridization (FISH) can directly assess the abundance of specific prokaryotes, but it frequently yields a very low recovery of Bacteria and Archaea compared to the total number of 4′,6′-diamidino-2-phenylindole (DAPI)-stainable cells (3). Only recently, the use of polynucleotide probes allowed the assessment of the abundance of Bacteria and Archaea in the meso- and bathypelagic waters of the Pacific (18) and in Antarctic marine waters (5). These authors found that the relative abundance of Crenarchaea increased significantly with depth, comprising up to 39% of total picoplankton cells down to 500 m, whereas the abundance of Euryarchaea was very low (<10%) throughout the water column. By contrast, very little is known about the metabolic function of specific prokaryotic groups in natural conditions (6, 23). The combination of fluorescence in situ techniques and microautoradiography has been used to determine the specific uptake of a given substrate in natural assemblages (6, 8, 14, 15, 19, 22, 23) but, to our knowledge, never in the deeper layers of the ocean (below 200 m).
The recently developed catalyzed reporter deposition FISH (CARD-FISH) method allows the use of oligonucleotide probes labeled with horseradish peroxidase (HRP), resulting in a sensitivity comparable to that of polynucleotide probes (25). The original CARD-FISH protocol included a permeabilization step for prokaryotic cells that uses lysozyme. However, archaeal cell walls do not contain murein (16, 17), rendering archaeal cells insensitive to lysozyme.
In this paper we describe a modified CARD-FISH protocol for more efficient detection of marine Archaea, replacing the lysozyme permeabilization treatment with a proteinase K treatment. We further combined the CARD-FISH protocol with microautoradiography (MICRO-CARD-FISH) to determine the number of bacterial and archaeal cells taking up the amino acid aspartic acid (Asp) in the deep waters of the North Atlantic down to a 4,000-m depth.
Samples were collected from 5 depths (100 to 4,000 m) at 10 stations in the North Atlantic during the TRANSAT-2 cruise (May and June 2003). We selected two stations (41.15°N, 62.43°W and 62.53°N, 30.35°W) in order to compare the efficiency of the lysozyme versus the proteinase K treatment. Water samples of 20 to 40 ml were spiked with l-[3H]Asp (specific activity, 37 Ci/mmol; final concentration, 10 nM; Amersham) and incubated in the dark at in situ temperature for 8 to 10 h. We experimentally estimated that samples reached saturation levels of radioactivity after 6 to 8 h. The concentration of l-[3H]Asp inoculated was ∼2 to 10 times higher than the in situ concentration (24). The uptake rates of the bulk prokaryotic community, obtained by adding l-[3H]Asp at a 1 nM final concentration, and the percentage of cells taking up l-[3H]Asp at a 10 nM final concentration, determined using microautoradiography, correlated well (r = 0.86; P < 0.001; n = 64). Controls consisted of 20- to 40-ml samples of cells killed with paraformaldehyde (final concentration, 2%) 20 min prior to addition of l-[3H]Asp. Incubations were terminated by adding paraformaldehyde (final concentration, 2%), and the samples were stored at 4°C in the dark for 12 to 18 h. Fixation times of more than 3 h minimize the amount of tritium-labeled compounds leaking from the cells (22). Thereafter, samples were filtered through 0.2-μm-pore-size polycarbonate filters (Millipore) supported by cellulose acetate filters (0.45-μm pore size; Millipore), washed twice with Milli-Q water, dried, and stored at −20°C until further processed.
CARD-FISH.
Filters were dipped in low-gelling-point agarose (0.1% [wt/vol] in Milli-Q water), dried upside down on a glass petri dish at 37°C, and subsequently dehydrated in 95% (vol/vol) ethanol. For cell wall permeabilization, filters were incubated either with lysozyme (catalogue number 7651, 10 mg/ml; Sigma) or with proteinase K (1,844 U/mg, 10.9 mg/ml, catalogue number 82456, 0.2 μl/ml; Fluka) solution (0.05 EDTA, 0.1 Tris-HCl [pH 8]) at 37°C for 1 h. Subsequently, the filters were washed three times with Milli-Q water and incubated in 0.01 M HCl at room temperature for 20 min. Incubation in HCl inhibits potentially present intracellular peroxidases and proved to also effectively inhibit residual proteinase K. Previous attempts to use proteinase K treatment to detect Archaea failed (26), probably because of the incomplete inactivation of the proteinase K after the permeabilization step. Complete inactivation of proteinase K is of crucial importance to avoid degradation of the HRP linked to the oligonucleotide probe. After incubation in HCl, filters were washed twice with Milli-Q water, dehydrated with 95% ethanol, and dried at room temperature.
Filters were cut in sections for hybridization with the oligonucleotide probes Eub338 (targeting Bacteria), Non338 (1), Cren537 (5′-TGACCACTTGAGGTGCTG-3′; targeting Crenarchaea), and Eury806 (5′-CACAGCGTTTACACCTAG-3′; targeting Euryarchaea). The probes target the same cells as the polynucleotide probes used in previous studies (11, 18). Three hundred microliters of hybridization buffer (0.9 M NaCl, 20 mM Tris-HCl [pH 7.5], 10% [wt/vol] dextran sulfate, 0.02% [wt/vol] sodium dodecyl sulfate, 1% blocking reagent [Boehringer Mannheim, Mannheim, Germany], and 55% [vol/vol] formamide [for Eub338 and Non338] or 20% [vol/vol] formamide [for Cren537 and Eury806]) was dispensed into a 0.5-ml reaction vial. The HRP probe was added at a final DNA concentration of 0.28 ng/μl (0.05 μM). The hybridization was performed at 35°C for 8 to 12 h. Thereafter, the sections were transferred into 50 ml of prewarmed washing buffer (5 mM EDTA [pH 8], 20 mM Tris-HCl [pH 7.4 to 7.6], 0.01% [wt/vol] sodium dodecyl sulfate) at 37°C for 10 to 15 min. Two different washing buffers were prepared, containing 13 mM NaCl (for Eub338 and Non338) and 145 mM NaCl (for Cren537 and Eury806) (24). Sections were then placed in phosphate-buffered saline (PBS) (145 mM NaCl, 1.4 mM NaH2PO4, 8 mM Na2HPO4 [pH 7.6]) amended with 0.05% Triton X-100 (PBS-T) at room temperature for 15 min. After removal of excess buffer, the filter sections were immediately transferred to a 1.5-ml reaction vial containing 493 μl of amplification buffer (10% [wt/vol] dextran sulfate, 2 M NaCl, 0.1% [wt/vol] blocking reagent, and 0.0015% H2O2 in PBS) and 5 μl of tyramide-Alexa488 (1 mg/ml) and incubated at 37°C for 30 to 40 min. H2O2 was freshly prepared daily. Alexa dyes are more photostable than Cy dyes (2). After amplification, filters were washed in PBS-T (room temperature, 15 min), Milli-Q water, and 95% ethanol. Finally, filter sections were air dried and stored at −20°C until further processed.
Microautoradiography.
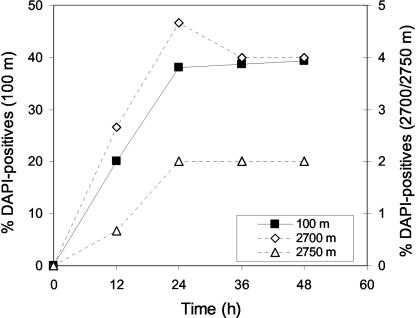
In the darkroom, kept at 15°C, the photographic emulsion (type NTB-2; Kodak) was melted in a water bath set at 43°C for 1 h. Afterwards, the emulsion was mixed with ultrapure water (Sigma) in a 1:3 (vol/vol) ratio, divided into small aliquots (10.5 ml each), and stored at 4°C. Each aliquot was melted only up to three times to avoid high background levels of silver grains. The previously hybridized filter sections were transferred onto slides coated with photographic emulsion. Subsequently, the slides were dried on ice-cold aluminum plates for 5 min. Coating of the slides was done in complete darkness. The transfer of the filter sections into the emulsion was done under a 15-W lamp using a safelight red filter (Hama-8194). The slides were then placed in a light-tight box with silica gel as a drying agent and kept at 4°C for exposure. The slides were developed and fixed using the specifications of Kodak. Before the slides were completely dry, filter sections were peeled off and the cells were counterstained with a DAPI mix (5.5 parts Citifluor [Citifluor, Ltd.], 1 part Vectashield [Vector Laboratories, Inc.], 0.5 parts PBS with DAPI at a final concentration of 1 μg/ml). We evaluated the influence of exposure time on the percentage of cells taking up l-[3H]Asp with water from different depths. Slides were developed at 12-h intervals, and the percentage of total DAPI-stained cells with associated silver grains was enumerated (Fig. 1). The percentage of cells with attached silver grains increased nearly linearly from 0 to 24 h. Thereafter, the number of active cells detected by microautoradiography did not further increase. Consequently, we routinely used an exposure time of 36 to 48 h.
FIG. 1.
Dependence of the percentage of cells taking up l-Asp (cells with more than three associated silver grains related to the abundance of DAPI-stained cells) on the duration of the exposure for autoradiographic development. Prokaryotic assemblages collected from a 100-, 2,700-, or 2,750-m depth were analyzed.
The slides were examined under a Zeiss Axioplan 2 microscope equipped with a 100-W Hg lamp and appropriate filter sets for DAPI and Alexa488. The presence of silver grains surrounding cells was checked by using the transmission mode of the microscope (4). In the killed controls, less than 0.5% of the total DAPI-stained cells were associated with two or more silver grains. More than 800 DAPI-stained cells were counted per sample. The paired t test was used for mean comparison after confirming the normal distribution of the data set.
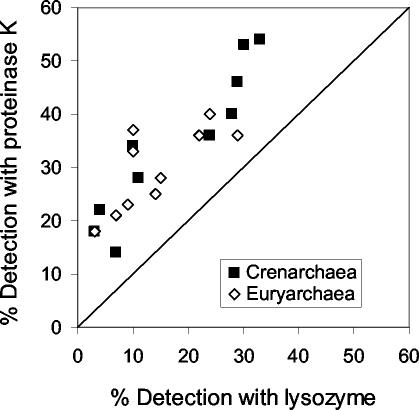
The percentages of DAPI-stainable cells detected with the specific archaeal probes were significantly higher (t test; P < 0.0001; n = 20) with proteinase K than with the lysozyme treatment (Fig. 2). The mean percentage of DAPI-stainable cells identified as Euryarchaea was 14% ± 2% (± standard error) with lysozyme and 30% ± 2% with proteinase K. Similarly, the Crenarchaea detection rate was 18% ± 2% with lysozyme and 35% ± 5% with proteinase K. Negative control counts (hybridization with the Non338-HRP probe) were on average 1% ± 0.2% with proteinase K and 0.5% ± 0.3% with lysozyme.
FIG. 2.
Comparison of the detection rates (percentage of probe-hybridized cells normalized to total DAPI-stained cells) by CARD-FISH for Crenarchaea (Cren537) and Euryarchaea (Eury806) when using lysozyme and proteinase K treatment for cell wall permeabilization.
Using polynucleotide probes to enumerate Euryarchaea in coastal and open oceanic waters yielded only <10% of DAPI-stained cells (5, 11, 18), which is similar to the euryarchaeotal abundance we detected with the lysozyme treatment but substantially lower than that obtained with the proteinase K permeabilization.
Proteinase K treatment of Bacteria using the Eub338 probe did not yield higher bacterial numbers than the lysozyme treatment (data not shown) but led to a certain disruption of bacterial cells at proteinase K concentrations used for Archaea. No disruption of archaeal cells was observed, however, even at higher concentrations than used routinely. The disruption of bacterial cells did not affect the fluorescence of the DAPI-stained cells. Thus, to enumerate Bacteria, lysozyme permeabilization was used. As previously reported (26), we did not detect a significant cell loss in agarose-embedded samples after an incubation period of 1 h either with lysozyme or with proteinase K. There was no significant difference detectable between total DAPI counts in samples treated with lysozyme or proteinase K (t test; P = 0.455; n = 43).
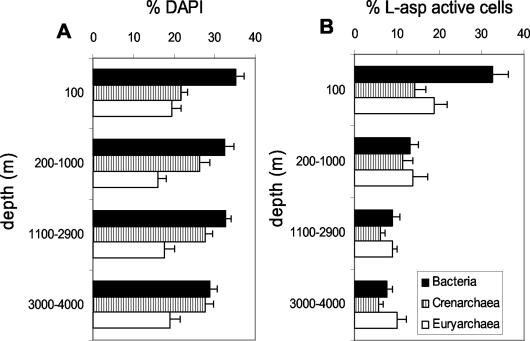
We applied this procedure to samples collected in the North Atlantic (Fig. 3A). The percentage of Bacteria decreased with depth, whereas Crenarchaea abundance increased with depth. This pattern of abundance was similar to that seen with polynucleotide probes (18). The total DAPI-stained cells detected with the three oligonucleotides averaged 71% (±2%) and is similar to that obtained with polynucleotide probes (5, 18).
FIG. 3.
Percentages of Bacteria, Crenarchaea, and Euryarchaea normalized to total DAPI counts (A) and percentages of Bacteria, Crenarchaea, and Euryarchaea taking up l-Asp normalized to the total number of Bacteria, Euryarchaea, and Crenarchaea (B), respectively, in different depth layers of the North Atlantic. Bars represent the mean (± standard error) of data from 10 different stations.
Although microautoradiography has been previously combined with FISH techniques (6, 8, 14, 15, 19, 23), to our knowledge, this is the first attempt to combine microautoradiography with CARD-FISH. Prior to combining the two techniques, we checked for possible effects of the different permeabilization treatments on the development of silver grains associated with DAPI-stained cells taking up l-Asp. No significant differences were found in the percentage of cells taking up Asp between untreated and lysozyme-treated samples (t test; P = 0.73; n = 7). Treatment with proteinase K could a priori be more problematic given the lower specificity of proteinase K than of lysozyme; however, no significant differences were found between the numbers of active cells in samples treated with lysozyme and those treated with proteinase K (t test; P = 0.78; n = 17).
We used MICRO-CARD-FISH for the detection of specific l-Asp uptake by Bacteria and Archaea. A previous work combining microautoradiography and FISH showed that marine Archaea from a 200-m depth took up l-amino acids (23). At the base of the euphotic zone, at a 100-m depth, the percentages of Bacteria and Archaea taking up l-Asp were similar (∼33%). In deeper waters, however, a higher proportion of Archaea (16 to 24%) took up l-Asp than Bacteria (∼10%) (Fig. 3B).
The percentage of total DAPI-stained cells taking up l-Asp ranged from 2 to 40%, which is in contrast to the rather high percentage of detection of DAPI-stained cells with CARD-FISH (Eub338 plus Cren537 plus Eury806; range, 40 to 102%). Such disagreement was also reported by others (6, 13, 14, 15, 23) and has given rise to a discussion of the limitation of microautoradiography for detection of active cells. On the one hand, prokaryotes with uptake rates below a certain threshold might not be detectable with microautoradiography. On the other hand, uptake rates for different compounds can greatly vary among different prokaryotic groups and environments (6). Despite the potential limitations inherent in microautoradiography, our results suggest that Archaea could play a major role in the oceanic biogeochemical cycles and indicate the great potential of the described MICRO-CARD-FISH approach for studying specific substrate uptake by prokaryotes in the deep ocean.
Acknowledgments
We thank the captain and crew of the R/V Pelagia for their help during work at sea.
This research was supported through a European Community Marie Curie Fellowship to E.T., by grants from the NWO-ALW (project NWO-ALW 811.33.004) to G.J.H., and by the Commission of the EU (BASICS project). The help and suggestions of J. M. Arrieta during the development of the modified protocol are gratefully acknowledged.
REFERENCES
- 1.Amann, R. I., W. Ludwig, and K. H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berlier, J. E., A. Rothe, B. Buller, J. Bradford, D. R. Gray, B. J. Filanoski, W. G. Telford, S. Yue, J. X. Liu, C. Y. Cheung, W. Chang, J. D. Hirsch, J. M. Beechem, and R. P. Haugland. 2003. Quantitative comparison of long-wavelength Alexa fluor dyes to Cy dyes: fluorescence of the dyes and their bioconjugates. J. Histochem. Cytochem. 51:1699-1712. [DOI] [PubMed] [Google Scholar]
- 3.Bouvier, T., and P. A. del Giorgio. 2003. Factors influencing the detection of bacterial cells using fluorescence in situ hybridization (FISH): a quantitative review of published reports. FEMS Microbiol. Ecol. 44:3-15. [DOI] [PubMed] [Google Scholar]
- 4.Carman, K. R. 1993. Microautoradiographic detection of microbial activity, p. 397. In P. F. Kemp, B. F. Sherr, E. B. Sherr, and J. F. Cole (ed.), Handbook of methods in aquatic microbial ecology. Lewis Publishers, Boca Raton, Fla.
- 5.Church, M. J., E. F. DeLong, H. W. Ducklow, M. B. Karner, C. M. Preston, and D. M. Karl. 2003. Abundance and distribution of planktonic Archaea and Bacteria in the waters west of the Antarctic Peninsula. Limnol. Oceanogr. 48:1893-1902. [Google Scholar]
- 6.Cottrell, M. T., and D. L. Kirchman. 2000. Natural assemblages of marine proteobacteria and members of the Cytophaga-Flavobacter cluster consuming low- and high-molecular-weight dissolved organic matter. Appl. Environ. Microbiol. 66:1692-1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cottrell, M. T., and D. L. Kirchman. 2000. Community composition of marine bacterioplankton determined by 16S rRNA gene clone libraries and fluorescence in situ hybridization. Appl. Environ. Microbiol. 66:5116-5122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cottrell, M. T., and D. L. Kirchman. 2003. Contribution of major bacterial groups to bacterial biomass production (thymidine and leucine incorporation) in the Delaware estuary. Limnol. Oceanogr. 48:168-178. [Google Scholar]
- 9.Crump, B. C., and J. A. Baross. 2000. Archaeaplankton in the Columbia River, its estuary and the adjacent coastal ocean, USA. FEMS Microbiol. Ecol. 31:231-239. [DOI] [PubMed] [Google Scholar]
- 10.DeLong, E. F., K. Y. Wu, B. B. Prézelin, and R. V. M. Jovine. 1994. High abundance of Archaea in Antarctic marine picoplankton. Nature 371:695-697. [DOI] [PubMed] [Google Scholar]
- 11.DeLong, E. F., L. T. Taylor, T. L. Marsh, and C. M. Preston. 1999. Visualization and enumeration of marine planktonic archaea and bacteria by using polyribonucleotide probes and fluorescent in situ hybridization. Appl. Environ. Microbiol. 65:5554-5563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fuhrman, J. A., and C. C. Ouverney. 1998. Marine microbial diversity studied via 16S rRNA sequences: cloning results from coastal waters and counting of native Archaea with fluorescent single cell probes. Aquat. Ecol. 32:3-15. [Google Scholar]
- 13.Gray, N. D., R. Howarth, R. W. Pickup, J. G. Jones, and I. M. Head. 1999. Substrate uptake by uncultured bacteria form the genus Achromatium determined by microautoradiography. Appl. Environ. Microbiol. 65:5100-5106. [DOI] [PMC free article] [PubMed]
- 14.Gray, N. D., R. Howarth, R. W. Pickup, J. G. Jones, and I. M. Head. 2000. Use of combined microautoradiography and fluorescence in situ hybridization to determine carbon metabolism in mixed natural communities of uncultured bacteria from the genus Achromatium. Appl. Environ. Microbiol. 66:4518-4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ito, T., J. L. Nielsen, S. Okabe, Y. Watanabe, and P. H. Nielsen. 2002. Phylogenetic identification and substrate uptake patterns of sulfate-reducing bacteria inhabiting an oxic-anoxic sewer biofilm determined by combining microautoradiography and fluorescent in situ hybridization. Appl. Environ. Microbiol. 68:356-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kandler, O. 1994. Cell wall biochemistry in Archaea and its physical implications. J. Biol. Phys. 20:165-169. [Google Scholar]
- 17.Kandler, O., and H. Konig. 1998. Cell walls polymers in Archaea (Archaeabacteria). Cell Mol. Life Sci. 54:305-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karner, M. B., E. F. DeLong, and D. M. Karl. 2001. Archaeal dominance in the mesopelagic zone of the Pacific Ocean. Nature 409:507-510. [DOI] [PubMed] [Google Scholar]
- 19.Lee, N., P. H. Nielsen, K. H. Andreasen, S. Juretschko, J. L. Nielsen, K.-H. Schleifer, and M. Wagner. 1999. Combination of fluorescent in situ hybridization and microautoradiography—a new tool for structure-function analyses in microbial ecology. Appl. Environ. Microbiol. 65:1289-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Massana, R., A. E. Murray, C. M. Preston, and E. F. DeLong. 1997. Vertical distribution and phylogenetic characterization of marine planktonic Archaea in the Santa Barbara Channel. Appl. Environ. Microbiol. 63:50-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Massana, R., L. T. Taylor, A. E. Murray, K. Y. Wu, W. H. Jeffrey, and E. F. DeLong. 1998. Vertical distribution and temporal variation of marine planktonic archaea in the Gerlache Strait, Antarctica, during early spring. Limnol. Oceanogr. 43:607-617. [Google Scholar]
- 22.Nielsen, J. L., D. Christensen, M. Kloppenborg, and P. H. Nielsen. 2003. Quantification of cell-specific substrate uptake by probe-defined bacteria under in situ conditions by microautoradiography and fluorescence in situ hybridization. Environ. Microbiol. 5:202-211. [DOI] [PubMed] [Google Scholar]
- 23.Ouverney, C. C., and J. A. Fuhrman. 2000. Marine planktonic archaea take up amino acids. Appl. Environ. Microbiol. 66:4829-4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pérez, M. T., C. Pausz, and G. J. Herndl. 2003. Major shift in bacterioplankton utilization of enantiomeric amino acids between surface waters and the ocean's interior. Limnol. Oceanogr. 48:755-763. [Google Scholar]
- 25.Pernthaler, A., C. M. Preston, J. Pernthaler, E. F. DeLong, and R. Amann. 2002. Comparison of fluorescently labeled oligonucleotide and polynucleotide probes for the detection of pelagic marine bacteria and archaea. Appl. Environ. Microbiol. 68:661-667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pernthaler, A., J. Pernthaler, and R. Amann. 2002. Fluorescence in situ hybridization and catalyzed reporter deposition for the identification of marine bacteria. Appl. Environ. Microbiol. 68:3094-3101. [DOI] [PMC free article] [PubMed] [Google Scholar]