Abstract
Sine oculis-related homeobox 3 (SIX3) and SIX6, 2 closely related homeodomain transcription factors, are involved in development of the mammalian neuroendocrine system and mutations of Six6 adversely affect fertility in mice. We show that both small interfering RNA knockdown in gonadotrope cell lines and knockout of Six6 in both embryonic and adult male mice (Six6 knockout) support roles for SIX3 and SIX6 in transcriptional regulation in gonadotrope gene expression and that SIX3 and SIX6 can functionally compensate for each other. Six3 and Six6 expression patterns in gonadotrope cell lines reflect the timing of the expression of pituitary markers they regulate. Six3 is expressed in an immature gonadotrope cell line and represses transcription of the early lineage-specific pituitary genes, GnRH receptor (GnRHR) and the common α-subunit (Cga), whereas Six6 is expressed in a mature gonadotrope cell line and represses the specific β-subunits of LH and FSH (LHb and FSHb) that are expressed later in development. We show that SIX6 repression requires interaction with transducin-like enhancer of split corepressor proteins and competition for DNA-binding sites with the transcriptional activator pituitary homeobox 1. Our studies also suggest that estradiol and circadian rhythm regulate pituitary expression of Six6 and Six3 in adult females but not in males. In summary, SIX3 and SIX6 play distinct but compensatory roles in regulating transcription of gonadotrope-specific genes as gonadotrope cells differentiate.
Infertility can be caused at the neuroendocrine level by defects in pituitary gonadotropes, GnRH neurons, or circadian pacemaker neurons (1, 2). An early step in commitment of the developing pituitary is expression of the common α-subunit (Cga, αGSU) in presumptive gonadotropes and thyrotropes on embryonic day (e) 11.5 in the mouse (3). LH, FSH, and TSH are heterodimeric glycoprotein hormones containing the common α-subunit of the glycoprotein hormones (CGA), in combination with a β-subunit specific to LH, FSH, or TSH. The individual β-subunits are transcribed from separate genes that are initially expressed in the mouse at e16.5 for LHb and e17.5 for FSHb (3).
Molecular investigation of the regulation of gonadotrope gene expression is greatly facilitated by the use of validated, cultured cell lines that represent differentiated cell types (4–7). The αT1-1 cell line represents a precursor to the gonadotrope-thyrotrope lineages (5) and expresses only one glycoprotein hormone subunit gene, Cga (8). The immature gonadotrope αT3-1 cell line expresses both Cga and GnRH receptor (GnRHR), and the mature gonadotrope LβT2 cell line expresses the 4 gonadotrope-specific genes Cga, GnRHR, LHb, and FSHb (5, 9, 10), whereas the TαT1 cell line represents a thyrotrope and expresses Cga and TSHb (5).
Mammalian sine oculis-related homeobox (SIX) 6 and SIX3 are a closely related subfamily of the SIX proteins that are vertebrate homologues of Drosophila Optix (11) with 2 highly conserved domains: a homeodomain (HD) for DNA-binding and a “Six” domain for protein-protein interaction. Although other SIX proteins, such as SIX1, SIX2, SIX4, and SIX5, all show broad expression during embryogenesis, SIX3 and SIX6 are restricted to the developing eye, brain, and pituitary (11–14). When compared with Six6, the expression pattern of Six3 is highly similar, but with a generally broader transcript distribution in both the brain and visual system during development (11). During formation of Rathke's pouch, Six3 and Six6 expression becomes detectable at e11.5, with Six3 appearing to be expressed at a higher level early in development, including at e13.5 (Allen Brain Atlas [www.brain-map.org] and Refs. 15, 16). Expression of both SIX proteins emerges in the pituitary precursors around the lumen and is found in a subset of pituitary precursors by e15.5. As development progresses, both Six3 and Six6 are expressed in some, but not all, of the differentiating anterior lobe cells.
Six3 knockout (KO) mice die at birth, lacking most head structures anterior to the midbrain, although the rest of the body appears normal (17). In contrast, Six6-KO mice have hypoplastic pituitaries (15) and are infertile but appear otherwise healthy (1), although some might be blind (15). In previous studies, we found that female Six6-KO mice fail to progress through the estrous cycle, show any signs of successful ovulation, or produce litters, although their serum LH and FSH levels are normal. Male Six6-KO mice are subfertile, produce substantially fewer litters than wild-type (WT) mice, and have lower levels of FSH (1). Analysis of GnRH (GnRH) expression in adult mice reveals a dramatic decrease (90%) in total GnRH mRNA and GnRH neuron numbers in the hypothalamus (1). Finally, Six6-KO mice lack normal morphology and gene expression in the suprachiasmatic nucleus, the main pacemaker for circadian rhythms, which is also required for normal fertility (2).
Using well-characterized animal and cell culture models, we have investigated the molecular mechanisms of Six3 and Six6 action in pituitary during development and adulthood. Here, we show that both Six3 and Six6 are specifically expressed in a differentiated pituitary gonadotrope cell line and regulate transcription of gonadotrope-specific genes. SIX3 and SIX6 play distinct roles in pituitary lineage specification during development and compensation by increased Six3 expression within the gonadotrope may contribute to the normal/undisrupted gonadotropin hormone expression seen in Six6-KO mice.
Materials and Methods
Materials
Mouse Six6 and Six3 expression vectors (1), mouse FSHb (18) and GnRHR (19), human Cga (20), and rat LHb (21) luciferase reporter constructs, the HD-binding element (HDBE) multimer (22) and ATTA multimer (1) have been previously described. The Six6 eh1 mutant and the Ptx1 promoter element mutations were previously described or constructed by QuikChange Site-Directed Mutagenesis kit from Agilent Technologies (20). Oligonucleotides were obtained from IDT. DNA-modifying enzymes were obtained from New England Biolabs. Small interfering RNAs (siRNA) and transfection reagents were obtained from Dharmacon.
Animals
All animal procedures were performed in accordance with the University of California, San Diego Institutional Animal Care and Use Committee regulations. All mice were group housed on a 12-hour light, 12-hour dark cycle with ad libitum chow (11% of calories fat, 17% of calories protein) and water.
Analysis of developing and adult pituitaries
Adult Six6 heterozygous mice were set up in timed matings. On e18.5, the pregnant females were euthanized by carbon dioxide inhalation and the embryos were extracted. Pituitaries were collected and placed individually in tubes on dry ice. The embryos were genotyped for Six6 (1) and sex determining region of Chr Y (Sry) (SRY forward 5′AGAGATCAGCAAGCAGCTGG3′, SRY reverse 5′TCTTGCCTGTATGTGATGGC3′) from tail biopsies. Single pituitaries of e18.5 male WT and Six6-KO mice were dissected and used individually. The pituitaries of 4- to 6-month-old adult male mice were collected in a similar manner and each sample represents a single pituitary and n was always more than or equal to 3.
Steroid feedback paradigms were as previously described (see reference 30 below). Briefly, C57BL/6J mice of 2–3 months of age were anesthetized by isoflurane inhalation, then females were ovariectomized (OVX) and males were castrated (CX). At the time of surgery, animals were sc implanted with capsules containing either vehicle (blank) or corresponding steroids. One week after surgery, animals were killed in the morning (am, before 11:30) or evening (pm, 30 minutes after lights out) by CO2 inhalation followed by exsanguination.
RNA was isolated from adult pituitaries using a QIAshredder and RNeasy mini kit (QIAGEN Sciences), or TRIzol (Invitrogen) as directed by the manufacturers. For embryonic individual pituitaries, total RNA was isolated by RNaqueos-Micro kit (Ambion), according to manufacturer's protocol. A total of 100 ng of total RNA was reverse transcribed using an iScript cDNA Synthesis kit (Bio-Rad). Samples without reverse transcriptase served as a negative control. For quantitative polymerase chain reaction (qPCR), cDNA was diluted 1:10 in water. qPCR was performed using SYBR Green supermix and an iQ5 real-time PCR machine (Bio-Rad). GAPDH, PPIA, and/or H2AFZ was used as internal controls (as indicated in the legends) for analysis of Six3, Six6, LHb, FSHb, GnRHR, and Cga. Intron-spanning primer sequences have previously been described (1, 23).
RiboTag/αGSU-iCre mice
RiboTag mice were bred to the αGSU-iCre recombinase-expressing mouse and genotyped for the presence of the iCre recombinase gene and the RiboTag allele (24, 25). Both genes were heterozygous. As described previously in detail (23), homogenates were prepared as follows: 4–5 pituitary samples per group were rapidly removed from 3- to 4-month-old Cre+ male mice and weighed before Dounce homogenization (2%–3% wt/vol) in polysome buffer (23). Pituitaries from Cre− mice were also collected and used as negative controls. For immunoprecipitations, 100-μL protein G magnetic beads (Dynabeads; Invitrogen) were coupled directly to 2-μL mouse monoclonal anti-human influenza hemagglutinin (HA) antibody (HA.11; Covance) for 4 hours in homogenization buffer. Supernatants were then added directly to the antibody-coupled protein G magnetic beads and rotated overnight at 4°C. The next day, samples were placed in a magnet on ice and supernatants were recovered. RNA was isolated using an RNeasy Mini plus kit (QIAGEN) and reverse transcribed with iScript kit (Bio-Rad) to generate cDNA. Samples without reverse transcriptase served as a negative control. Resulting cDNA was subjected to qPCR using specific primers (1, 23).
Cell culture, transient transfections, and luciferase assays
Cell lines were maintained in a humidified atmosphere of 5% CO2 in air at 37°C. αT1-1, αT3-1, LβT2, TαT1, NIH 3T3, CV1, and COS cells were maintained in high-glucose DMEM containing 10% fetal bovine serum and 100-U/mL penicillin/streptomycin as previously described (26). Transient transfection overexpression assays were performed according to the FuGENE 6 protocol (Promega). TK-β-galactosidase expression vector (100 ng) was used as the internal control for transfection efficiency. Forty-eight hours after transfection, cells were harvested in lysis buffer (100mM potassium phosphate [pH 7.8] and 0.2% Triton X-100). Luciferase assays were performed as previously described (27), and β-galactosidase assays were performed using the Galacto-Light Plus assay system as directed by the manufacturer (Applied Biosystems). All transfections were repeated at least 4 times, and values are presented as the mean ± SEM.
RNA isolation and RT-PCR from cultured cell lines
Total RNA was harvested and isolated from αT1-1, αT3-1, LβT2, TαT1, and NIH 3T3 cells using TRIzol reagent (Sigma-Aldrich). RNA was quantified and treated to remove DNA with Turbo DNA-free from Ambion according to manufacturer's protocol. Purified RNA was then reverse transcribed with iScript (Bio-Rad), or mock reverse transcribed as a negative control, to generate cDNA. Resulting cDNA was subject to 35 cycles of qPCR using specific primers previously described (1), and the coding sequence of GAPDH was used as control.
Six6 and Six3 siRNA knockdown
αT3-1 and LβT2 cells were transfected for 48 or 72 hours with 100nM ON-TARGET SMARTpool scrambled control, ON-TARGET plus SMARTpool Six6 or Six3 siRNA, or ON-TARGET SMARTpool cyclophilin B purchased from Dharmacon. DharmaFECT 1 transfection reagent was used according to manufacturer's protocol.
Total RNA was harvested with RNeasy plus mini kit (QIAGEN) according to manufacturer's protocol. Two nanograms of purified RNA was reverse transcribed with iScript kit (Bio-Rad) to generate cDNA, and samples without reverse transcriptase served as negative controls. Resulting cDNA was subjected to qPCR using intron-spanning primers for Six6, Six3, LHb, FSHb, GnRHR, Cga, Cyclophilin B, and GAPDH (1). Values represent the average of 4 independent experiments, and all samples are normalized to GAPDH levels. Cyclophlin B siRNA did not affect expression of any mRNA other than Cyclophilin B (data not shown). All values are normalized to scrambled siRNA control and are expressed as mean ± SEM.
Nuclear extracts and EMSA
COS cells were transfected with cytomegalovirus (CMV)-flag or Six6-flag expression vectors for 48 hours. Nuclear protein was extracted and prepared from COS cells as previously described (28). Protein concentration was determined using the Bio-Rad Protein Assay (Bio-Rad). Oligonucleotide probes are shown in the figures. Oligonucleotide probes were annealed, end labeled using T4 Polynucleotide Kinase (New England Biolabs) and [γ32P]ATP (7000 Ci/mmol; MP Biomedicals), and then purified using G25 Probe Quant columns (Amersham). Binding reactions were carried out using 2 μg of nuclear protein and 4 fmol of labeled oligonucleotide in a 10-μL reaction containing 1mM Dithiothreitol, 0.0125-μg/μL Poly dIdC, and binding buffer (50mM HEPES [pH 7.8], 250 mM KCl, 5 mM EDTA, and 30% glycerol). For competition assays, 1000-fold excess of double-stranded unlabeled oligonucleotides were annealed and used in binding reactions. After the addition of probe and nuclear protein, binding reactions were incubated 5 minutes before electrophoresis on a 5% nondenaturing polyacrylamide gel in 0.25× Tris/Borate/EDTA buffer. For supershift EMSA assays, 2 μL of mouse anti-FLAG (F3165; Sigma-Aldrich) or normal mouse IgG (sc-2025; Santa Cruz Biotechnology, Inc) were added to reactions before the addition of nuclear protein and incubated for 10 minutes. Gels were run at 250 V for approximately 2 hours, dried under vacuum, and exposed to autoradiographic film overnight.
Statistical analysis
Quantitative RT-PCR and transient transfection experiments were performed in triplicate and each experiment was repeated at least 3 times. Data were analyzed by Student's t test, one-way ANOVA, followed by Tukey honestly significant difference or two-way ANOVA, followed by a post hoc analysis as indicated in figure legends. Statistical analysis was performed using GraphPad Prism 5. For all analyses, the result was considered significant if P ≤ .05.
Results
SIX6 regulation of mRNA expression in developing and adult male pituitary
To determine the transcriptional roles of SIX6 in gonadotrope development in vivo, we examined the expression of Cga, GnRHR, LHb, FSHb, and Six3, in both developing (e18.5) and adult Six6-KO male mouse pituitary. Expression of Six6 was absent at both developmental stages from Six6-KO pituitary as expected (data not shown).
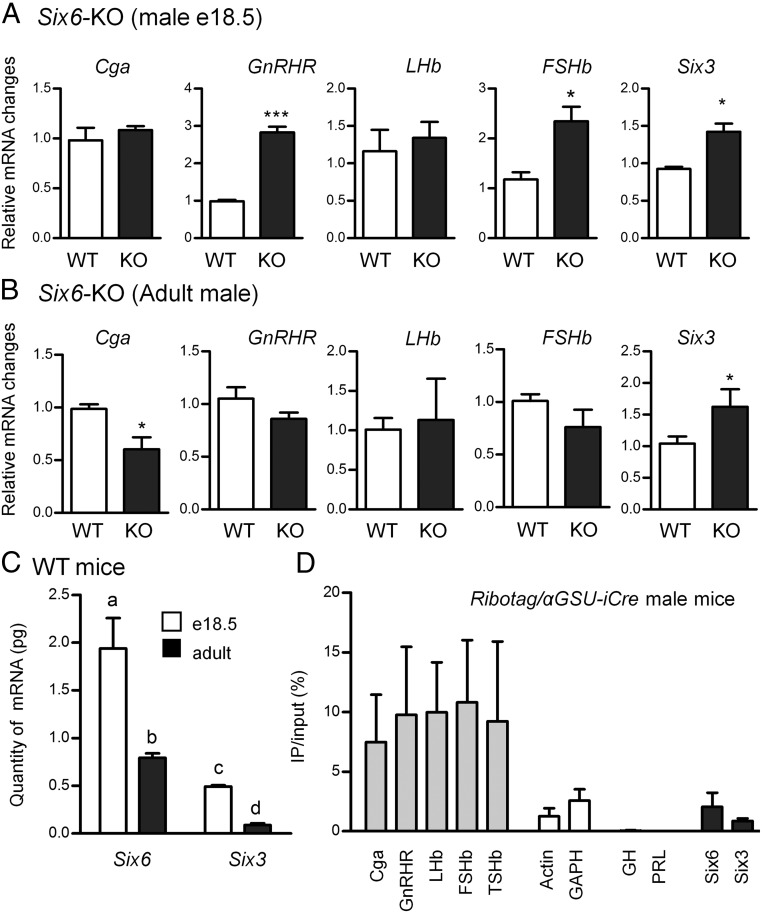
In the developing male pituitary (e18.5), mRNA expression of GnRHR, FSHb, and Six3 was significantly increased in Six6-KO embryonic pituitaries at e18.5 compared with WT (Figure 1A). However, Six6-KO pituitaries did not have a statistically significant difference in LHb or Cga expression from WT. We next analyzed the expression of pituitary mRNAs from 4- to 6-month-old male mice. Increased Six3 expression was also found in adult male mice (Figure 1B), consistent with the results observed in the developing pituitary (Figure 1A). The only difference between adult WT and KO pituitaries was the decreased expression of Cga mRNA in the adult male Six6-KO pituitary. No changes in GnRHR and FSHb expression were identified.
Figure 1.
The role of SIX6 in pituitary development. A, Individual embryonic e18.5 developing pituitaries from WT and Six6-KO (KO) male mice were analyzed (n ≥ 3). All target genes were analyzed using the 2−ΔΔCT method by normalizing the GOI to the housekeeping gene GAPDH. B, Four- to 6-month-old male pituitaries from WT and Six6-KO mice. Data were analyzed by the 2−ΔΔCT method by normalizing the GOI to the average of housekeeping genes PPIA and H2AFZ. A and B, Data are expressed as fold change compared with WT. *, P < .05; ***, P < .001 different from WT (Student's t test). C, Six3 and Six6 mRNA quantitation. Quantitation of Six6 and Six3 mRNA from both embryonic e18.5 and adult mice was normalized to the mRNA of the housekeeping gene GAPDH using comparison with a standard curve of plasmid DNA. Different letters represent significant differences by two-way ANOVA followed by Bonferroni post hoc (GraphPad Prism 5). D, Enrichment of mRNAs isolated from RiboTag/αGSU-iCre pituitaries. qPCR analysis of mRNAs from αGSU-iCre+ pituitary cells immunoprecipitated with HA antibody. The immunoprecipitated RNA samples were compared with the input sample in each case. All cell-specific marker genes and control genes in the target cells were analyzed using the 2−ΔΔCT method.
Expression of Six3 and Six6 in pituitary
To further address the roles of SIX3 and SIX6 on gonadotropes during pituitary development, we determined the levels of mRNA expression in vivo. Using quantitative PCR relative to a standard curve of plasmid DNA, we found that the levels of Six6 are at least 4-fold higher than those of Six3 in both adult and e18.5 pituitary (Figure 1C). This is in contrast to earlier in development when Six3 is predominant (Allen Brain Atlas). Furthermore, both Six3 and Six6 expression are decreased in adult pituitaries compared with e18.5 pituitary (Figure 1C).
The anterior pituitary has 5 different endocrine cell types. The gonadotropes emerge late in development at approximately e16.5 and comprise approximately 5%–10% of pituitary cells in adulthood. Until now, no efficient method has been established to isolate gonadotropes from the pituitary. The RiboTag technique has been established to tag ribosome-associated mRNAs of specific cell types, then mRNAs can be isolated and purified for further analyses (24). Because all gonadotropes express Cga, we crossed RiboTag mice with αGSU-iCre mice to isolate ribosome-associated transcripts in Cga+ pituitary cell types, which include both gonadotropes and thyrotropes. Cga, GnRHR, FSHb, and LHb were used as specific markers for gonadotropes and TSHb for thyrotropes. GAPDH and Actin were used as controls as they should be pulled down in all cell types. GH and prolactin (PRL) were used as negative controls as they are produced by somatotropes and lactotropes in the pituitary, which do not express Cga. We have previously described in detail the specificity of this approach and validated the antibody used in this experiment (23). Both Six3 and Six6 mRNA were enriched in αGSU-iCre+ cells, indicating their specific expression in gonadotropes and thyrotropes in adult male mice (Figure 1D). Based on this study, we cannot rule out the possibility that Six3 and Six6 are also expressed in other pituitary cell types, but these data show that they are enriched in a sample containing a mixture of gonadotropes and thyrotropes.
Six3 and Six6 are expressed in immortalized mouse gonadotrope cell lines
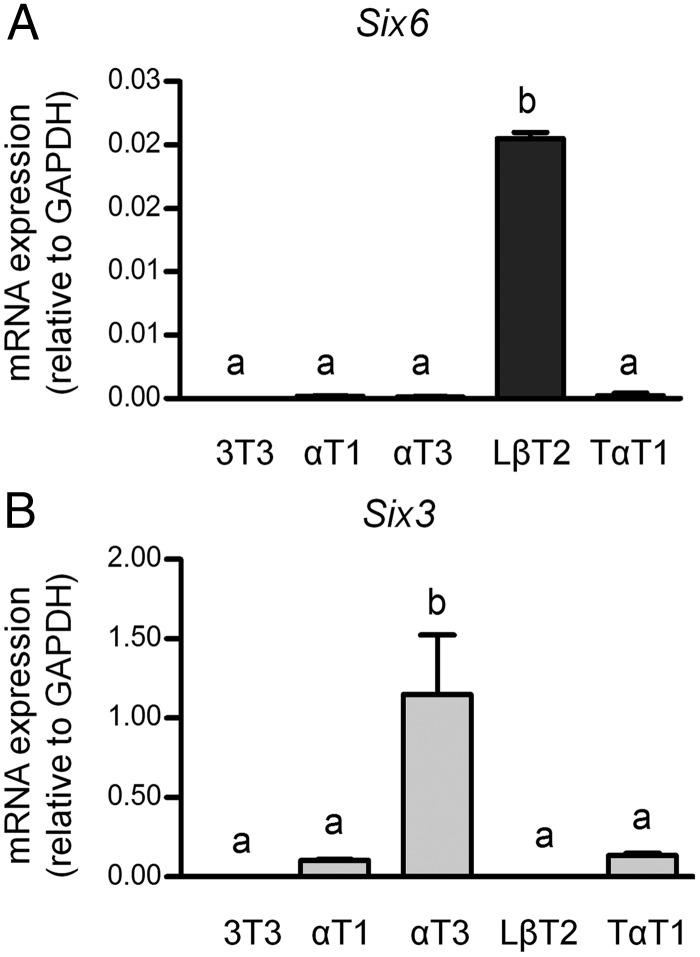
Based on their expression pattern and structural similarity, we hypothesized that both SIX3 and SIX6 are required for pituitary development. We therefore tested immortalized gonadotrope cell lines in vitro as potential model systems. Using validated Six3- and Six6-specific primers (1), qPCR analysis was performed on 3 mouse gonadotrope-derived cell lines that represent different developmental stages, including progenitor αT1-1, immature αT3-1, and mature LβT2 cells. A mature thyrotrope cell line TαT1 and fibroblast NIH 3T3 cells were used as controls. The data show that Six6 is highly expressed in gonadotropes LβT2, whereas Six3 is highly expressed in αT3-1 (Figure 2). For gonadotropes, Six6 was approximately 200-fold higher in mature LβT2 cells compared with immature and progenitor cells (Figure 2A). Conversely, Six3 was highly expressed in immature αT3-1 cells, then dramatically reduced by approximately 500-fold in mature LβT2 cells (Figure 2B), which strongly suggests that SIX3 functions in immature early gonadotropes rather than mature gonadotropes. Mature thyrotropes, TαT1 cells, only expressed very low levels of Six3 and Six6 mRNAs, at a similar level to the progenitor αT1-1 cells (Figure 2, A and B). The expression of Six3 and Six6 in gonadotrope cell lines but not in the thyrotrope cell line (TαT1) suggests that gonadotropes likely express Six3 and Six6 in vivo. Our data also suggest that the expression of HD proteins SIX3 and SIX6 is stage-specific during gonadotrope development with SIX3 expressed earlier than SIX6, as appears to be the case in vivo (Figure 1C and Allen Brain Atlas). In addition, this shows that gonadotrope-derived cell lines can be used to study the role of SIX3 and SIX6 in vitro. The expression levels of Six3 and Six6 in these cell lines indicate that they may play overlapping, but distinct, roles at different stages of pituitary development.
Figure 2.
Six6 and Six3 mRNA expression in the gonadotrope-derived cell lines. Six6 (A) and Six3 (B) mRNA expressions in the gonadotrope-derived cell lines are shown relative to the level of GAPDH. Total RNAs were harvested from gonadotrope-derived cell lines: progenitor αT1-1, immature αT3-1, and mature LβT2. Fibroblast NIH 3T3 cells and thyrotrope TαT1 cells served as controls. Values are the mean ± SEM of at least 4 independent experiments. Different letters represent significant differences by one-way ANOVA (GraphPad Prism 5).
siRNA-mediated knockdown of Six3 and Six6 in αT3-1 and LβT2 cells
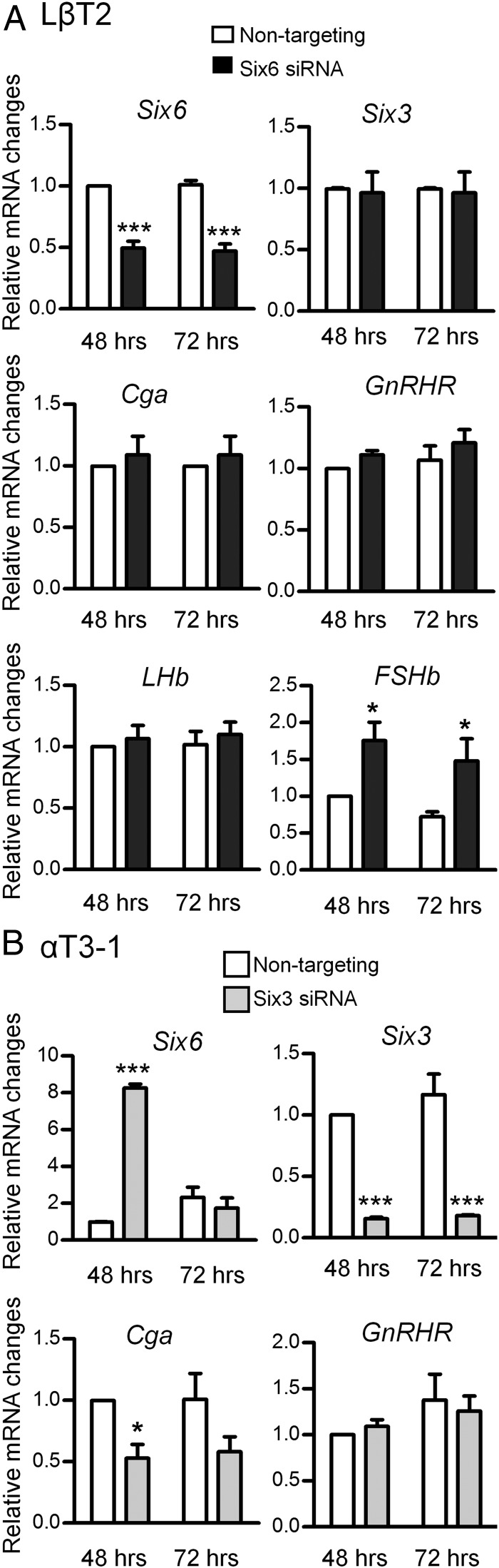
To investigate the actions of endogenous SIX3 and SIX6, siRNA-mediated knockdown was used to attenuate their function in either immature αT3-1 cells that express Six3 (Figure 2B) or mature LβT2 cells that express Six6 (Figure 2A). Nontargeting siRNA and Cyclophilin B siRNA were used as negative and positive controls respectively. No changes were observed with the Cyclophilin B siRNA other than to Cyclophilin B mRNA, which was reduced by approximately 75%–80% (data not shown). Transfection of Six6 siRNA into LβT2 cells reduces Six6 mRNA approximately 50%–60% due to the low transfection efficiency of this cell line and has no effect on Six3 mRNA, after either 48 or 72 hours of knockdown (Figure 3A). Of the 4 target genes, only FSHb mRNA expression showed a modest, although significant, increase (Figure 3A). Thus, SIX6 acts to suppress expression of FSHb in the LβT2 gonadotrope lineage cells.
Figure 3.
Knockdown of endogenous Six6 and Six3 mRNAs in gonadotrope cell lines affects gonadotrope-specific mRNA expression. A, LβT2 cells were transfected with SMARTpools of nontargeting or Six6 siRNA for either 48 or 72 hours. B, αT3-1 cells were transfected with SMARTpools of nontargeting or Six3 siRNA for either 48 or 72 hours. All values represent the SQ ± means from 4 independent experiments with all means adjusted to corresponding GAPDH values within experiment, using relative standard curve for analysis. Statistical analysis by one-way ANOVA (GraphPad Prism 5). *, P < .05; ***, P < .001 as compared with nontargeting siRNA.
Transfection of Six3 siRNA into αT3-1 cells results in an approximately 85% reduction of Six3 mRNA. Interestingly, a dramatic reduction of Six3 mRNA induces Six6 mRNA expression to approximately 8-fold higher than in the nontargeting control, which indicates that SIX6 may compensate for the dramatic loss of SIX3 expression and this may account for the lack of effect on mRNA expression of GnRHR. There was also a significant decrease in endogenous Cga mRNA expression. Both changes of Six6 and Cga are temporary, and lost after 72 hours of siRNA knockdown (Figure 3B).
Overexpression of Six3 and Six6 regulates transcription of gonadotrope-specific genes
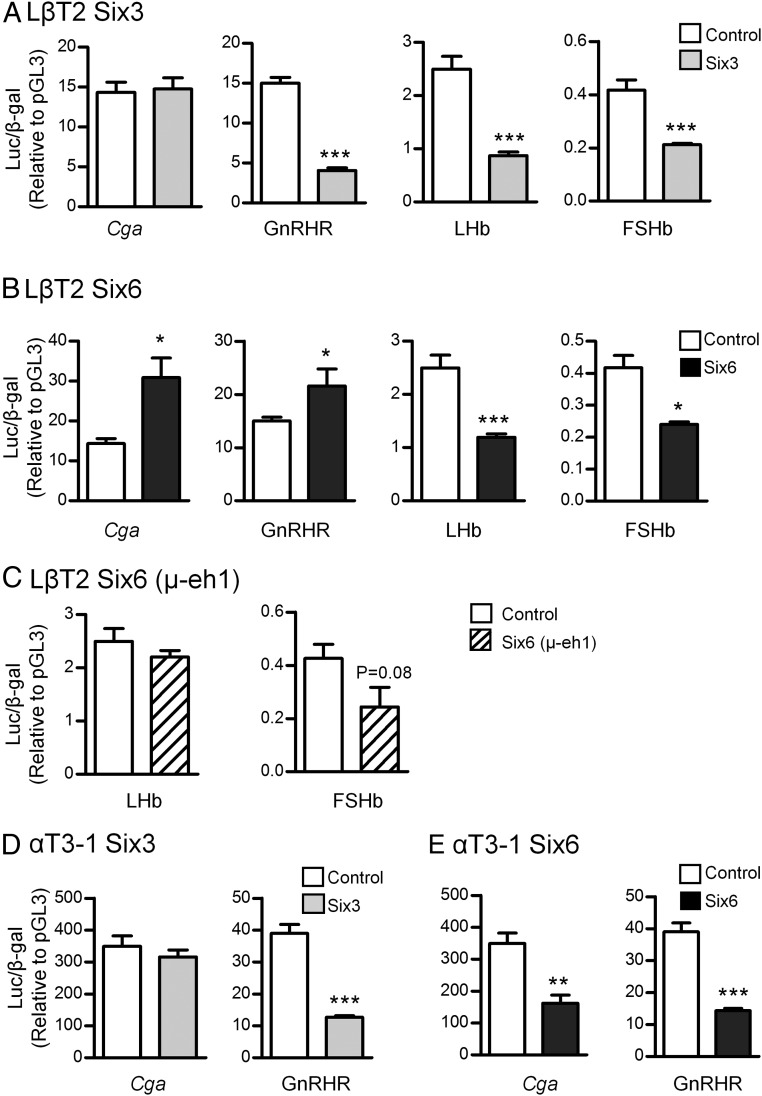
SIX family members are HD proteins that function as transcription factors. We have shown that SIX6 activates and SIX3 represses the GnRH enhancer and promoter in GT1–7 hypothalamic cells and that SIX6 relieves repression by SIX3 (1). Based on the studies from Six6-KO mice and siRNA knockdown, we next determined transcriptional regulation by SIX3 and SIX6 of 4 gonadotrope-specific genes, Cga, GnRHR, LHb, and FSHb in the mature LβT2 (Figure 4, A–C) and immature αT3-1 (Figure 4, D and E) cell lines. Our results show that both SIX3 and SIX6 repress LHb and FSHb transcription in LβT2 cells, which is consistent with the increased FSHb expression by Six6 knockdown (Figure 3A). For the Cga and GnRHR promoters, although SIX6 activated transcription of the Cga and GnRHR promoters, SIX3 repressed GnRHR and had no effect on Cga transcription (Figure 4, A and B). SIX proteins recruit transducin-like enhancer of split (TLE)/Grg corepressors through the eh1 domain for transcriptional repression (29, 30). Mutation of the eh1 domain of Six6 (μ-eh1) disrupted transcriptional regulation of LHb and FSHb (Figure 4C). Therefore, the eh1 domain is required for SIX6-mediated repression of gonadotrope target genes LHb and FSHb.
Figure 4.
Transcriptional regulation of gonadotrope-specific genes by Six3 and Six6 in gonadotrope-derived cell lines. The WT luciferase reporters driven by the promoters from the human Cga, mouse GnRHR, mouse FSHb, or rat LHb were cotransfected with either Six6 (A) or Six3 (B) expression vectors or equal mass of empty expression vector (control) into LβT2 cells. C, The transcriptional regulation of promoters mediated by μ-eh1 in LβT2 cells. The transcriptional regulation by SIX6 (D) and SIX3 (E) in the αT3-1 cell line. The fold change is shown relative to pGL3 empty vector control. Values are the mean ± SEM of at least 3 independent experiments. Statistical analysis by one-way ANOVA (GraphPad Prism 5). *, P < .05; ***, P < .001 represents significant difference from empty vector.
Because Six3 is highly expressed in immature αT3-1 cells, we also tested the role of SIX3 and SIX6 in this developmentally earlier gonadotrope cell line. SIX3 plays the same roles during gonadotrope maturation: it had no effect on the Cga promoter and repressed GnRHR (Figure 4, A and D), which is consistent with specific expression of Cga in the αT3-1 cell line. In contrast, overexpressed Six6 plays distinct roles in regulation of Cga and GnRHR at different developmental stages: although SIX6 activated Cga and GnRHR promoters in LβT2 cells, it repressed both promoters in αT3-1 cells (Figure 4E). Therefore, SIX3 and SIX6 exhibit different effects in the transcriptional regulation of gonadotrope-specific genes in different developmental and promoter contexts.
SIX3 and SIX6 regulate transcription by interference with PITX1 transcriptional activation
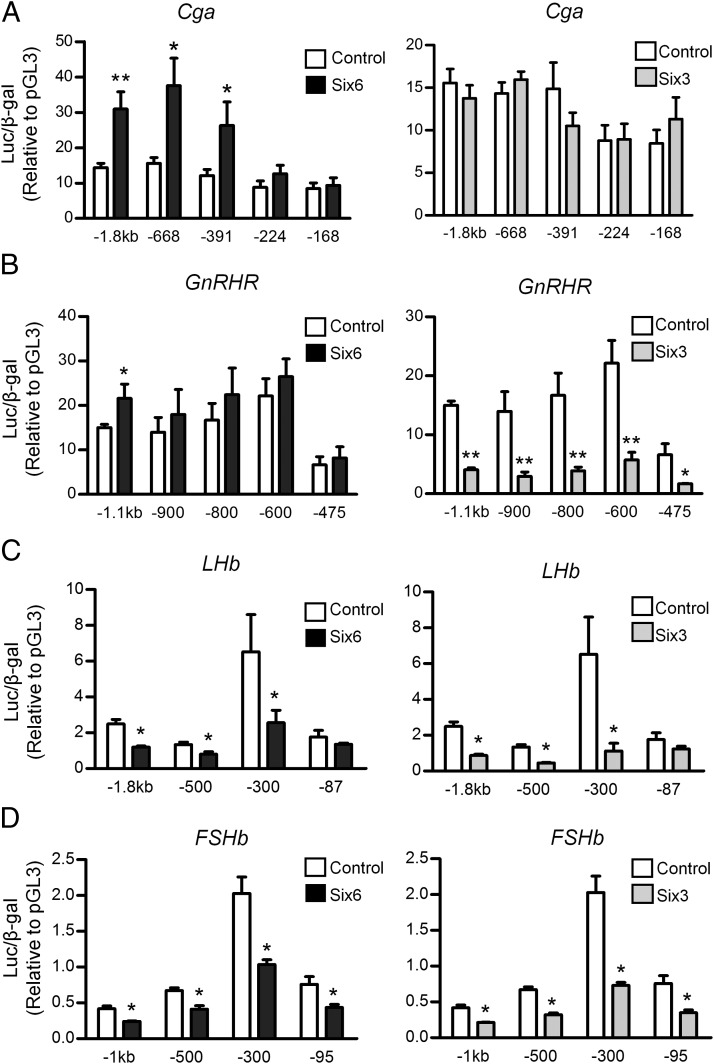
To map the Six regulatory elements in the promoters of the 4 gonadotrope-specific genes, promoter truncation analysis was used to define the regions of importance (20, 28). We found that in LβT2 cells, Cga activation by SIX6 is mediated by elements downstream of −391 bp, with a trend towards induction on the −224 bp as well, although SIX3 had no effect even on the full-length promoter (Figure 5A). Activation of the GnRHR reporter by SIX6 reached significance only for the full-length promoter, whereas repression by SIX3 mapped to the proximal promoter inside of −475 bp (Figure 5B). Repression of the LHb promoter mapped in between −300 and −87 bp (Figure 5C) and the FSHb promoter to inside of −95 bp (Figure 5D).
Figure 5.
Mapping Six3- and Six6-responsive regions in 4 gonadotrope-specific promoters. A, The full-length −1.8-kb human Cga promoter and its truncations. B, The −1.1-kb GnRHR promoter and its truncations. C, The −1.8-kb rat LHb promoter and its truncations. D, The −1-kb mouse FSHb promoter and its truncations. Luciferase reporter genes were cotransfected with either Six6 or Six3 expression vector or an equal mass of empty expression vector (control) into LβT2 cells. The fold change is relative to pGL3 empty vector control. Values are the mean ± SEM of at least 3 independent experiments. Statistical analysis by one-way ANOVA (GraphPad Prism 5). *, P < .05; ***, P < .001 represents significant difference from empty vector.
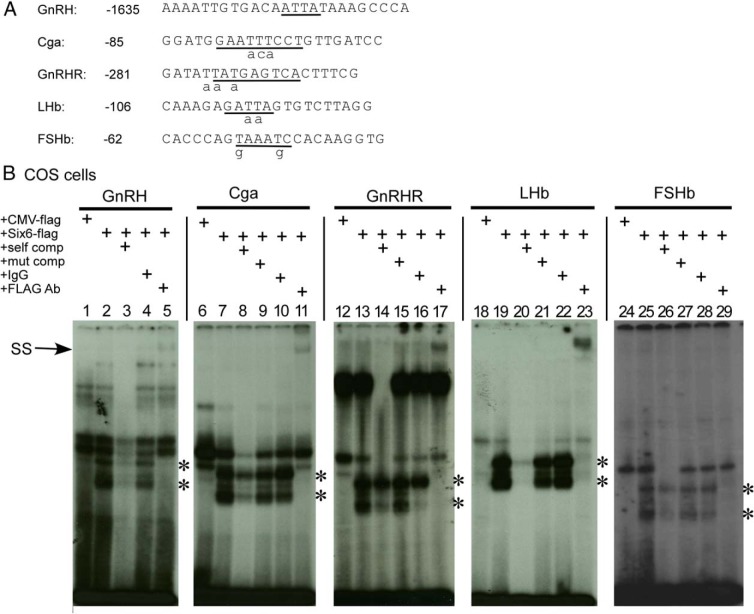
Within the most proximal responsive regions of these 4 promoters, HDBEs known to bind pituitary homeobox 1 (PITX1)/2 and/or ISL1 transcription factor (ISL1) tissue-specific HD activators are found (31–35), and these binding elements are also consistent with the Six6 consensus (Figure 6A) (31). To investigate whether SIX6 can directly bind to the known Pitx sites on the promoters of the 4 gonadotrope-specific genes, we transfected heterologous COS cells, which are fibroblasts from monkey kidney that do not express Six3 or Six6 (data not shown), with a Flag-tagged Six6 expression vector. The nuclear extract was then incubated with excess oligonucleotide probes representing WT human Cga (Figure 6B, lanes 6–11), mouse GnRHR (Figure 6B, lanes 12–17), LHb (Figure 6B, lanes 18–23), and FSHb (Figure 6B, lanes 24–29) containing the known PITX-binding sites in these genes. The GnRH promoter Six-binding element (−1635 bp) was used as positive control (Figure 6A, 6B, lanes 1–5) (1). A CMV-flag empty vector was also transfected into COS-1 cells as negative control (Figure 6B, lanes 1, 6, 12, 18, and 24). The WT competitor for each Pitx-binding element (Figure 6, A and B, lanes 3, 8, 14, 20, and 26) dramatically blocked binding to the oligo. However, mutation of the Pitx site (mutations are indicated by lowercase letters in Figure 6A) in each of the 4 probes almost totally blocked this competition (Figure 6B, lanes 9, 15, 21, and 27). Inclusion of an antibody directed against the Flag tag (FLAG Ab) resulted in decreases in the level of the bands due to SIX6 (indicated by *) on all of the probes and the formation of a new supershift complex of markedly reduced mobility on all probes except for FSHb, demonstrating the highly specific binding of SIX6 to the individual probes (Figure 6B, lanes 5, 11, 17, 23, and 29).
Figure 6.
The binding of FLAG-tagged SIX6 protein to Pitx elements from the LHb, FSHb, GnRHR, and Cga promoters. A, The oligonucleotides containing the Pitx sites from the proximal GnRH, Cga, GnRHR, LHb, and FSHb promoters and their corresponding competition mutants are listed. The sequences of Pitx-binding elements on positive control (GnRH) and 4 gonadotrope-specific promoters are underlined and the competition mutants are marked as lowercase letters underneath the sequence. B, Competition EMSA and supershift assays. COS-1 cells were transfected with either CMV-flag empty and CMV-Six6-flag expression plasmids and their nuclear extracts were isolated for EMSA. The GnRH enhancer 1 SIX6-binding element (−1635 bp) was used as positive control (1). Specific protein/DNA complexes were identified by competition with 250-fold excess unlabeled WT (self comp) or mutant Pitx-site oligonucleotides (mut comp). Confirmation of the presence of SIX6 in specific complexes was shown by inclusion of an antibody against FLAG or mouse IgG as a control. Asterisks represent the DNA/protein complexes, and the arrow marks the SIX6 supershifted complex (SS) after binding with FLAG antibody.
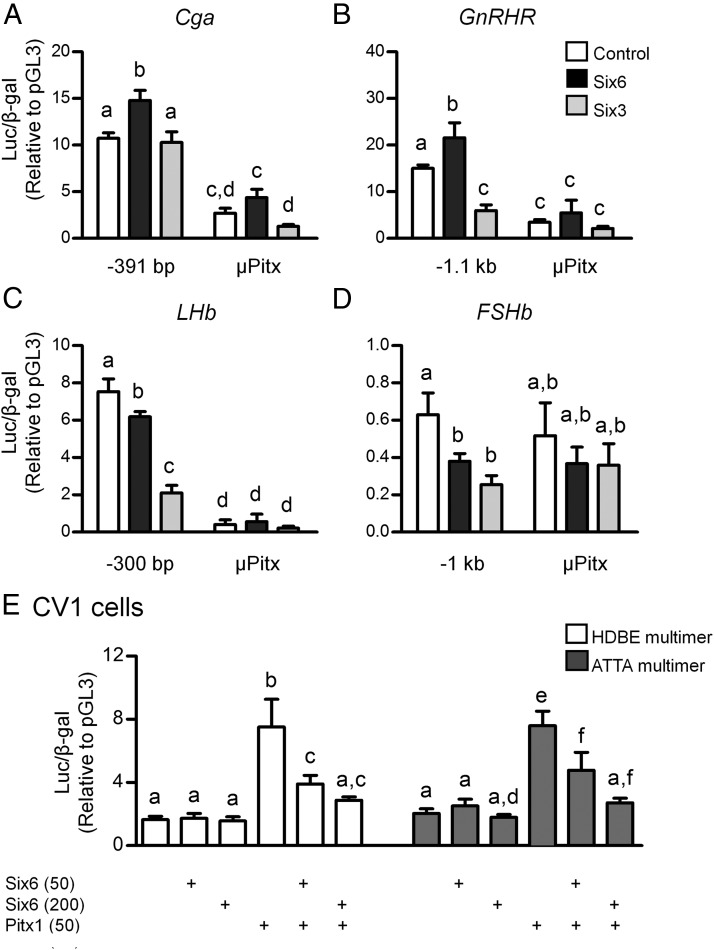
To test the hypothesis that SIX proteins repress via interference or displacement of PITX1 or other HD activators, CV-1 cells were used due to their lack of endogenous Pitx1, Six3 and Six6 expression (data not shown). Mutation of the proximal Pitx-binding elements (Figure 6A) within all 4 promoters disrupted transcriptional regulation by both SIX3 and SIX6 for all 4 promoters, but also abolished most basal transcriptional activity for all studied promoters, except FSHb, which may be due to its low basal transcription (Figure 7, A–D). Therefore, a HDBE multimer was used to test the competitive binding between SIX6 with HD activators. 4xHDBE-Luc contains 4 repeats of a consensus bicoid-related HDBE (ACTAATCCCT). The ATTA multimer contains 5 copies of the known Six-binding sequence from GnRH enhancer 1 (ctcATTAaat) (1). SIX6 significantly activated this luciferase plasmid in the GnRH hypothalamic cell line, GT1–7 cells (1). However, in CV-1 cells, SIX6 alone did not regulate either the ATTA or HDBE multimer (Figure 7E). PITX1 robustly stimulated transcription of both multimers and this activation was repressed by coexpression of SIX6 (Figure 7E). Our data from both the HDBE and ATTA multimers indicates that SIX6 may regulate transcription by interference with PITX1-mediated transcriptional activation in a dose-dependent manner.
Figure 7.
The transcriptional regulation by SIX proteins of Cga, GnRHR, LHb, and FSHb promoters requires Pitx1 HDBEs. Reporters, including the WT or Pitx cis-mutation of the Cga (−391 bp) (A), GnRHR (−1.1 kb) (B), LHb (−300 bp) (C), and FSHb (−1 kb) (D) promoters, were cotransfected with either Six6 or Six3 expression vectors or an equal mass of empty expression vector (control) into LβT2 cells. E, The HDBE multimer and GnRH ATTA multimer were cotransfected with 2 different amounts of the Six6 expression vector (50 or 200 ng), and/or with the Pitx1 expression vector (50 ng) or the appropriate empty vectors into CV-1 cells as indicated below the graph. The fold change is relative to pGL3 empty vector control. Values are the mean ± SEM of at least 3 independent experiments. Different letters represents significant differences by two-way ANOVA (GraphPad Prism 5).
Steroid regulation of Six6 expression in female but not in male mice
Because we had previously shown that SIX3 and SIX6 were regulated in a circadian manner in the hypothalamus (2) and circadian regulation might affect the pituitary response in the estrous cycle, we tested whether the Six mRNAs were regulated by circadian rhythms or steroid hormones in the adult mouse pituitary. It is unknown whether expression of Six3 and Six6 are influenced by a combination of either circadian factors or high steroid levels that are present during the LH surge in the pituitary. Young C57Bl/6 males were CX and implanted with blank (oil), dihydrotestosterone (DHT), or testosterone (T) capsules. Pituitaries were collected and qPCR was performed for Six3 and Six6. No significant differences were observed with either T or DHT replacement in CX mice (Figure 8A). Young C57Bl/6 females were OVX and implanted with blank or estradiol (E2) capsules. Pituitaries were collected in the morning (am, representing low LH levels due to negative feedback) or evening (pm, representing the LH surge due to positive feedback). Six6 transcript levels were higher overall in the am than pm in female pituitary, and E2 was able to significantly reduce Six6 transcript levels in both the am and pm groups (Figure 8B). Six3 transcript levels were higher overall in the am than pm in female pituitary as well, but no significant effect was observed with E2 (Figure 8B). These results indicate that both Six3 and Six6 have a circadian expression in the pituitary, but only Six6 is responsive to estrogen.
Figure 8.
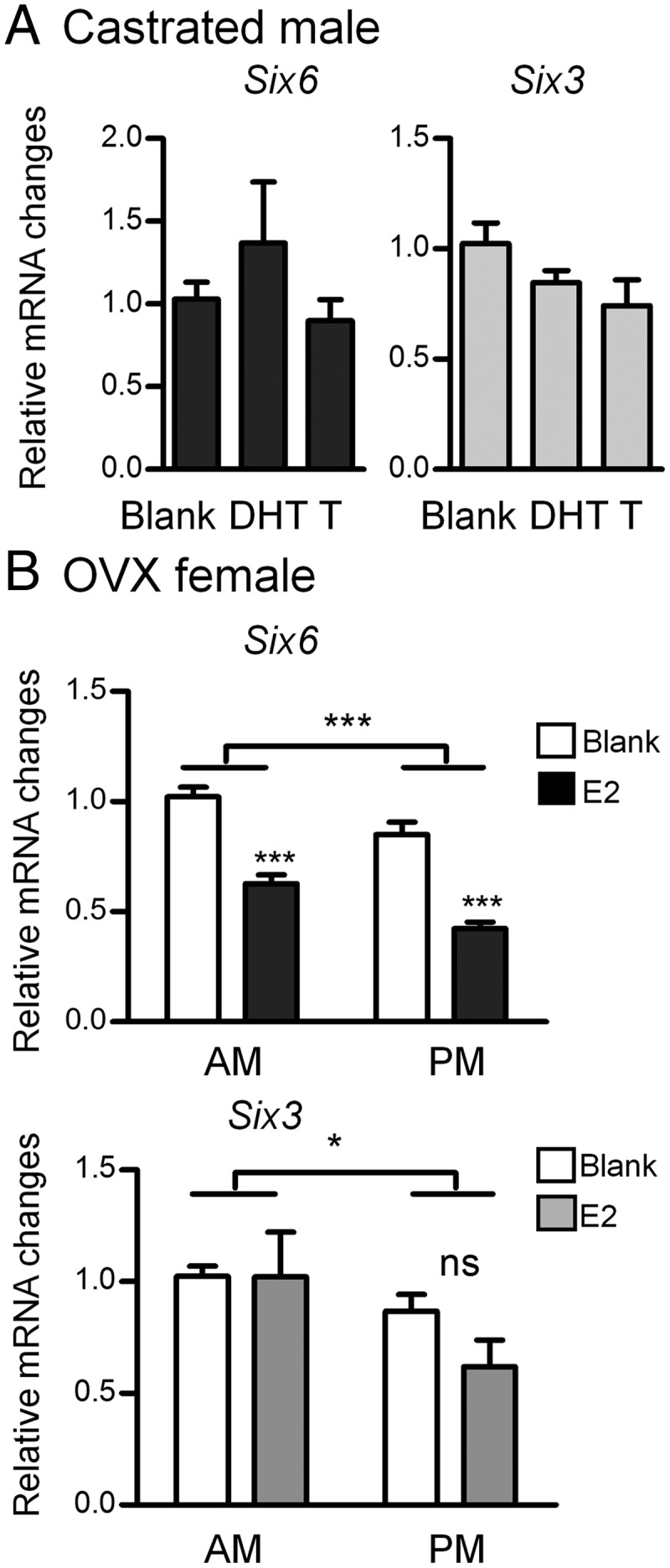
Regulation of Six6 and Six3 expression by steroids in the pituitary. A, Pituitaries were collected from 2- to 3-month-old CX males that had been implanted with vehicle (blank), DHT, or T (n = 4–6). B, Pituitaries were collected from 2- to 3-month-old OVX females that had been implanted with vehicle (blank) or E2, for both am (11) and pm (6:30) groups (n = 7–16). Six6 and Six3 genes were normalized using 2−ΔΔCt method and values represent the SQ ± means adjusted to corresponding average of housekeeping genes PPIA and H2AFZ. Statistical analysis by two-way ANOVA followed by Bonferroni post hoc (GraphPad Prism 5). *, P < .05; ***, P < .001 represents significant difference from corresponding am control or as indicated by bar.
Discussion
Elucidation of the molecular and cellular mechanisms underlying pituitary development and specification of the 5 individual hormone-secreting cell types is critical to our understanding of reproduction and infertility. Proper expression of Cga, GnRHR, LHb, and FSHb subunits is critical for mammalian reproductive function and the mature gonadotropes respond to hypothalamic GnRH input via GnRHR to modulate expression and release of LH and FSH. A particularly important set of transcription factors for gonadotrope development is the homeobox proteins, which can be either stimulatory, such as LHX3, PIT1, ISL1, and PITX1, or inhibitory, such as Msx homeobox 1 (MSX1) (28, 36–39).
In this study, we dissect the functions of the closely related HD proteins SIX3 and SIX6 in the development and function of the pituitary gonadotrope in vivo and in vitro with a focus on direct regulation of gene expression during gonadotrope development. SIX3 and SIX6 belong to the same subfamily of SIX proteins and have highly conserved Six elements and HDs. Their expression patterns are overlapping in the hypothalamus, pituitary, and eye (11–13, 40). Here, we have shown that Six3 is preferentially expressed in immature gonadotrope cells (αT3-1), whereas Six6 is specifically expressed in differentiated pituitary gonadotrope cells (LβT2) (Figure 2). Our RiboTag data confirmed this restricted expression in gonadotropes and thyrotropes in vivo in adult male mice (Figure 1C). Because the gonadotrope/thyrotrope progenitor cell line, αT1-1, and the mature thyrotrope TαT1 cell line have undetectable Six3 and Six6 mRNA compared with the gonadotrope cell lines (Figure 2), the pulldown of Six6 and Six3 transcripts in aGSU-iCre in pituitary most likely reflects Six6 and Six3 mRNA levels expressed in gonadotropes rather than thyrotropes. It should be noted that our studies, both in vitro and in vivo, do not allow us to rule out the possibility that both Six3 and Six6 are expressed in other anterior pituitary cells, including somatotropes, corticotropes, and lactotropes.
We also find that SIX3 and SIX6 can compensate for each other, both in vitro and in vivo. The siRNA knockdown experiments show that Six6 mRNA is dramatically induced (∼8-fold) 48 hours after Six3 knockdown in immature αT3-1 cells. However, knockdown of Six6 at several different concentrations (data not shown) in mature LβT2 cells did not induce Six3 mRNA expression. These differential effects might be explained by the approximately 200-fold higher Six6 expression as compared with Six3 in LβT2 cells (Figure 2A), and, thus, knockdown of only 50% of endogenous Six6 would not necessarily lead to a compensatory expression of Six3 mRNA. Indeeḑ a compensatory increase of Six3 was observed in Six6-KO pituitary, where Six3 mRNA expression is 50% higher in both developing (e18.5) and adult male pituitaries (Figure 1) in the complete absence of Six6. Despite the fertility defects in Six6-KO mice, we found that adult Six6-KO mice have normal LH expression in their hypoplastic pituitaries and that LH secretion responds to GnRH (1). Only male Six6-KO mice had a significant reduction in serum FSH (1), suggesting that pituitary function in Six6-KO mice is relatively normal, but that the hypothalamic signal to the pituitary is diminished. Based on our observations in this paper, we suspect that the accompanying increase of Six3 may be compensating for the loss of Six6 in Six6-KO pituitary and might allow the relatively normal gonadotropin transcription in the adult.
We then addressed the overlapping but distinct roles of SIX3 and SIX6 in gonadotrope development. We hypothesized that the sequential expression of Six3 then Six6 would prevent premature differentiation of the gonadotrope by transcriptional activation/repression of specific genes early in development, including Cga, GnRHR, LHb, and FSHb. Based on their expression levels and the results of siRNA knockdown in the αT3-1 vs LβT2 cell lines, our data suggest that SIX proteins are required for regulation of the gonadotrope-specific genes. Consistent with the distinct expression patterns of Six3 and Six6 during different stages of gonadotrope development in vitro (Figure 2), our data indicate that SIX3 functions during an early developmental stage by repressing both Cga and GnRHR transcription. Then, SIX6 replaces SIX3 during differentiation eliminating the repression of Cga and GnRHR and instead represses LHb and FSHb (Figures 1 and 4).
TLE/Groucho homologues control many embryonic and postembryonic processes such as differentiation, cell specification, embryonic patterning, and apoptosis (41–44). The TLE family of corepressors down-regulate transcription by inhibiting the basal transcriptional machinery (45) and recruiting HDACs. TLE corepressors lack a DNA-binding domain but are tethered to cis-acting regulatory elements via protein-protein interactions to transcription factors such as Homeobox gene expressed in ES cells, TCF: T-cell factor/lymphoid enhancer-binding factor, and SIX family members. TLEs also act as corepressors for other transcription factors during early pituitary development (46–48). Groucho-related genes (GRG4 and GRG5) interact with mouse SIX3 and SIX6 (30). SIX3-mediated autorepression in eye development requires its interaction with members of the Groucho-related family of corepressors (30). Investigation of TLE mRNA expression using qPCR and quantitative RNA-sequencing analysis in the gonadotrope cell lines revealed the expression patterns of six TLE mRNAs (28). Therefore, we tested the function of SIX6 and its corepressor by cis-mutation of the Six6 eh1 domain. The abolishment of transcriptional regulation by the μ-eh1 demonstrates that the recruitment of TLEs by SIX6 is required for transcriptional regulation of Cga, GnRHR, LHb, and FSHb (Figure 4C).
The pituitary-specific homeobox protein, PITX1, is expressed throughout the pituitary during development and plays a critical role in activation of a number of pituitary genes (31, 32, 49, 50). The Pitx1 and Pitx2 genes start to be expressed at approximately e9.5 and participate in the differentiation of the central nervous system and pituitary organogenesis (49). PITX1 transactivates Cga, FSHb, and LHb (31). PITX1 and PITX2 also collaborate in thyrotrope differentiation by acting synergistically with Cga and TSHb transactivation (34). Synergistic interactions between steroidogenic factor 1 (SF-1), early growth response 1 (EGR1), and PITX1 are essential for GnRH induction of LHb gene expression (33, 51). PITX1 is also necessary for maintaining corticotrope-specific transcription (31). PITX1 deletion in mice affects anterior pituitary development, leading to a reduction in the number of gonadotropes (among other cells). All 4 gonadotrope specific genes, Cga, GnRHR, FSHb, and LHb, have Pitx1-response elements within their promoters. We find that Six3- and Six6-responsive elements are located in similar regions of these promoters. Mutations within the Pitx-response elements and EMSA studies indicate that SIX6 may compete with the binding of the activator PITX1 and thus repress transcription of GnRHR, LHb, and FSHb. These results, combined with overexpression and siRNA knockdown experiments, lead to the conclusion that transcription of the gonadotrope-specific genes is negatively regulated by SIX6 and that this repression may occur by displacing or competing with PITX1 or other HD activators in the promoters of the Cga, GnRHR, LHb, and FSHb genes. PITX1 may not be the only transcription factor that can be interfered with by SIX6. Other HD transcription factors, such as ISL1, that also share similar consensus ATTA-binding elements, may also compete for binding to these sites by SIX6. Therefore, the molecular mechanism involved in SIX-mediated transcriptional repression may reflect competition with other HD activators. One intriguing question is why mature gonadotrope cells might retain a transcriptional repressor for LH and FSH. Based on the competition between SIX6 and other HD proteins, and with consideration of the stimulatory effect of SIX6 on Cga and GnRHR, we propose that SIX6 transcriptional regulation focuses on balancing and modulating, rather than only repressing gene expression.
Interestingly, in LβT2 cells, overexpression of Six6 inhibits the mRNA expression of FSHb (Figure 4), knockdown of Six6 induces FSHb (Figure 6), whereas FSHb mRNA increases in Six6-KO embryonic pituitary, but not in adult (Figure 1). However, Six6-KO males have reduced serum FSH levels, although females have normal levels. In contrast, we observe no changes in LHb gene expression in vivo or after siRNA knockdown, whereas overexpression of either SIX3 or SIX6 represses the LHb promoter in LβT2 cells. For GnRHR, absence of SIX6 in embryonic pituitary causes increased expression, whereas overexpression of SIX6 or SIX3 in αT3-1 cells or SIX3 in LβT2 cells causes repression. However, we could not observe changes to GnRHR in the siRNA knockdown experiments and in LβT2 cells, SIX6 overexpression induced GnRHR. Possible explanations for these differences may include induction of Six3 in vivo due to absence of Six6 allowing compensation, or that manipulation of gene expression in the transfection studies by overexpression or knockdown may interrupt the balancing role of SIX proteins with other HD proteins and emphasize the role of SIX6 itself. Furthermore, differences between the overexpression data in LβT2 cells vs the αT3-1 cells could be due to the enormous difference in endogenous expression of Six6 between these 2 cell lines. Therefore, these contradictory findings may further support our hypothesis that SIX6 functions as a balancing transcription factor instead of simply a repressor.
To further understand the physiologic context of SIX3 and SIX6 in adult pituitary, we investigated whether pituitary SIX3 and SIX6 respond to feedback by steroid hormones. Our findings show that pituitary Six6 was decreased during the LH surge in OVX+E2 females, whereas neither Six3 nor Six6 were affected in CX+T males. This suggests that specifically Six6 is regulated by sex hormones in the female. Interestingly, we determined that both Six3 and Six6 were expressed in a circadian manner in the pituitary with higher levels in the morning than in the evening. These results are comparable with those we previously described for Six3 and Six6 in the hypothalamus (2), indicating these homeoproteins are circadian regulated. These data identify a circadian expression of Six3 and Six6 in the pituitary, and that Six6 is strongly regulated by estrogen. Surprisingly, T and DHT were both unable to modulate Six3 or Six6 transcript in CX males, identifying a sexually dimorphic regulation by sex hormones in the pituitary.
The studies presented herein further support the role of SIX3 and SIX6 as critical transcriptional regulators necessary for anterior pituitary development, specifically for gonadotrope-specific gene programming. The molecular mechanism involved in this regulation may be due to competition with the HD transcription factor PITX1. In vitro siRNA knockdown and Six6-KO mice have been used to confirm regulation by SIX3 and SIX6 of gonadotrope-specific genes, including Cga, GnRHR, LHb, and FSHb. More importantly, our studies have also shown that, although Six3 and Six6 are expressed during gonadotrope development and play distinct roles, their functions are overlapping and each can compensate the loss of its closely related subfamily protein.
Acknowledgments
We thank María Inés Pérez-Millán and Sally A. Camper (University of Michigan) for the αGSU-iCre mice, Dr Xue Li (Children's Hospital of Boston, Harvard Medical School, Boston, MA) for the Six6-null mice, and Paul S. Amieux for the RiboTag mice (University Washington, Seattle). We also thank Dr Varykina Thackray for the HDBE plasmid and Varykina Thackray and Erica Schoeller for many helpful comments on the manuscript.
Present address for S.W.D.: Department of Biology, University of South Carolina 29208.
Present address for R.L.: Wellcome Trust-MRC Institute of Metabolic Science, University of Cambridge, Cambridge, CB2 0QQ, United Kingdom.
This work was supported by National Institutes of Health (NIH) Grants R01 DK044838, R01 HD072754, R01 HD082567, and R01 HD020377 (to P.L.M.), by National Institute of Child Health and Human Development/NIH through a cooperative agreement (U54 HD012303) as part of the Specialized Cooperative Centers Program in Reproduction and Infertility Research (to P.L.M.), and NIH Grants R01 HD034283 and R01 HD030428 (to Sally Camper). P.L.M. was partially supported by NIH Grants P30 DK063491, P42 ES101337, and P30 CA023100; H.X. was partially supported by NIH Grants T32 HD007203 and F32 HD070579; and C.M. and C.T. were partially supported by a The Endocrine Society Student Summer Research Fellowship. The University of Virginia, Center for Research in Reproduction, Ligand Assay and Analysis Core is supported by the Eunice Kennedy Shriver National Institute of Child Health and Human Development/NIH (Specialized Cooperative Centers Program in Reproduction and Infertility Research) Grant U54 HD28934.
Disclosure Summary: The authors have nothing to disclose.
Footnotes
- CGA
- α-subunit of the glycoprotein hormones
- CMV
- cytomegalovirus
- CX
- castrated
- DHT
- dihydrotestosterone
- e
- embryonic day
- E2
- estradiol
- μ-eh1
- mutation of the eh1 domain of Six6
- GnRHR
- GnRH receptor
- GSU
- glycoprotein hormones, α-subunit
- HD
- homeodomain
- HDBE
- HD-binding element
- ISL1
- ISL1 transcription factor
- KO
- knockout
- OVX
- ovariectomy
- PITX1
- pituitary homeobox 1
- qPCR
- quantitative polymerase chain reaction
- siRNA
- small interfering RNA
- SIX
- sine oculis-related homeobox
- Sry
- sex determining region of Chr Y
- TLE
- transducin-like enhancer of split
- WT
- wild-type.
References
- 1. Larder R, Clark DD, Miller NL, Mellon PL. Hypothalamic dysregulation and infertility in mice lacking the homeodomain protein Six6. J Neurosci. 2011;31:426–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Clark DD, Gorman MR, Hatori M, Meadows JD, Panda S, Mellon PL. Aberrant development of the suprachiasmatic nucleus and circadian rhythms in mice lacking the homeodomain protein six6. J Biol Rhythms. 2013;28:15–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Japon MA, Rubinstein M, Low MJ. In situ hybridization analysis of anterior pituitary hormone gene expression during fetal mouse development. J Histochem Cytochem. 1994;42:1117–1125. [DOI] [PubMed] [Google Scholar]
- 4. Windle JJ, Weiner RI, Mellon PL. Cell lines of the pituitary gonadotrope lineage derived by targeted oncogenesis in transgenic mice. Mol Endocrinol. 1990;4:597–603. [DOI] [PubMed] [Google Scholar]
- 5. Alarid ET, Windle JJ, Whyte DB, Mellon PL. Immortalization of pituitary cells at discrete stages of development by directed oncogenesis in transgenic mice. Development. 1996;122:3319–3329. [DOI] [PubMed] [Google Scholar]
- 6. Lew D, Brady H, Klausing K, et al. GHF-1 promoter-targeted immortalization of a somatotropic progenitor cell results in dwarfism in transgenic mice. Genes Dev. 1993;7:683–693. [DOI] [PubMed] [Google Scholar]
- 7. Pernasetti F, Spady TJ, Hall SB, et al. Pituitary tumorigenesis targeted by the ovine follicle-stimulating hormone β-subunit gene regulatory region in transgenic mice. Mol Cell Endocrinol. 2003;203:169–183. [DOI] [PubMed] [Google Scholar]
- 8. Yusta B, Alarid ET, Gordon DF, Ridgway EC, Mellon PL. The thyrotropin β-subunit gene is repressed by thyroid hormone in a novel thryrotrope cell line, mouse TαT1 cells. Endocrinology. 1998;139:4476–4482. [DOI] [PubMed] [Google Scholar]
- 9. Pernasetti F, Vasilyev VV, Rosenberg SB, et al. Cell-specific transcriptional regulation of FSHβ by activin and GnRH in the LβT2 pituitary gonadotrope cell model. Endocrinology. 2001;142:2284–2295. [DOI] [PubMed] [Google Scholar]
- 10. Graham KE, Nusser KD, Low MJ. LbT2 gonadotroph cells secrete follicle stimulating hormone (FSH) in response to activin A. J Endocrinol. 1999;162:R1–R5. [DOI] [PubMed] [Google Scholar]
- 11. Jean D, Bernier G, Gruss P. Six6 (Optx2) is a novel murine Six3-related homeobox gene that demarcates the presumptive pituitary/hypothalamic axis and the ventral optic stalk. Mech Dev. 1999;84:31–40. [DOI] [PubMed] [Google Scholar]
- 12. Oliver G, Mailhos A, Wehr R, Copeland NG, Jenkins NA, Gruss P. Six3, a murine homologue of the sine oculis gene, demarcates the most anterior border of the developing neural plate and is expressed during eye development. Development. 1995;121:4045–4055. [DOI] [PubMed] [Google Scholar]
- 13. Conte I, Morcillo J, Bovolenta P. Comparative analysis of Six 3 and Six 6 distribution in the developing and adult mouse brain. Dev Dyn. 2005;234:718–725. [DOI] [PubMed] [Google Scholar]
- 14. Aijaz S, Allen J, Tregidgo R, van Heyningen V, Hanson I, Clark BJ. Expression analysis of SIX3 and SIX6 in human tissues reveals differences in expression and a novel correlation between the expression of SIX3 and the genes encoding isocitrate dehyhrogenase and cadherin 18. Genomics. 2005;86:86–99. [DOI] [PubMed] [Google Scholar]
- 15. Li X, Perissi V, Liu F, Rose DW, Rosenfeld MG. Tissue-specific regulation of retinal and pituitary precursor cell proliferation. Science. 2002;297:1180–1183. [DOI] [PubMed] [Google Scholar]
- 16. Brinkmeier ML, Potok MA, Davis SW, Camper SA. TCF4 deficiency expands ventral diencephalon signaling and increases induction of pituitary progenitors. Dev Biol. 2007;311:396–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lagutin OV, Zhu CC, Kobayashi D, et al. Six3 repression of Wnt signaling in the anterior neuroectoderm is essential for vertebrate forebrain development. Genes Dev. 2003;17:368–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Thackray VG, McGillivray SM, Mellon PL. Androgens, progestins and glucocorticoids induce follicle-stimulating hormone β-subunit gene expression at the level of the gonadotrope. Mol Endocrinol. 2006;20:2062–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Duval DL, Nelson SE, Clay CM. The tripartite basal enhancer of the gonadotropin-releasing hormone (GnRH) receptor gene promoter regulates cell-specific expression through a novel GnRH receptor activating sequence. Mol Endocrinol. 1997;11:1814–1821. [DOI] [PubMed] [Google Scholar]
- 20. Sasson R, Luu SH, Thackray VG, Mellon PL. Glucocorticoids induce human glycoprotein hormone α-subunit gene expression in the gonadotrope. Endocrinology. 2008;149:3643–3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rosenberg SB, Mellon PL. An Otx-related homeodomain protein binds an LHb promoter element important for activation during gonadotrope maturation. Mol Endocrinol. 2002;16:1280–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Skarra DV, Arriola DJ, Benson CA, Thackray VG. Forkhead box O1 is a repressor of basal and GnRH-induced Fshb transcription in gonadotropes. Mol Endocrinol. 2013;27:1825–1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hoffmann HM, Tamrazian A, Xie H, Perez-Millan MI, Kauffman AS, Mellon PL. Heterozygous deletion of ventral anterior homeobox (Vax1) causes subfertility in mice. Endocrinology. 2014;155:4043–4053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sanz E, Yang L, Su T, Morris DR, McKnight GS, Amieux PS. Cell-type-specific isolation of ribosome-associated mRNA from complex tissues. Proc Natl Acad Sci USA. 2009;106:13939–13944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Perez-Millan MI, Zeidler MG, Saunders TL, Camper SA, Davis SW. Efficient, specific, developmentally appropriate cre-mediated recombination in anterior pituitary gonadotropes and thyrotropes. Genesis. 2013;51:785–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cherrington BD, Bailey JS, Diaz AL, Mellon PL. NeuroD1 and Mash1 temporally regulate GnRH receptor gene expression in immortalized mouse gonadotrope cells. Mol Cell Endocrinol. 2008;295:106–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Givens ML, Rave-Harel N, Goonewardena VD, et al. Developmental regulation of gonadotropin-releasing hormone gene expression by the MSX and DLX homeodomain protein families. J Biol Chem. 2005;280:19156–19165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Xie H, Cherrington BD, Meadows JD, Witham EA, Mellon PL. Msx1 homeodomain protein represses the αGSU and GnRH receptor genes during gonadotrope development. Mol Endocrinol. 2013;27:422–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lopez-Rios J, Tessmar K, Loosli F, Wittbrodt J, Bovolenta P. Six3 and Six6 activity is modulated by members of the groucho family. Development. 2003;130:185–195. [DOI] [PubMed] [Google Scholar]
- 30. Zhu CC, Dyer MA, Uchikawa M, Kondoh H, Lagutin OV, Oliver G. Six3-mediated auto repression and eye development requires its interaction with members of the Groucho-related family of co-repressors. Development. 2002;129:2835–2849. [DOI] [PubMed] [Google Scholar]
- 31. Tremblay JJ, Lanctôt C, Drouin J. The pan-pituitary activator of transcription, Ptx1 (pituitary homeobox 1), acts in synergy with SF-1 and Pit1 and is an upstream regulator of the Lim-homeodomain gene Lim3/Lhx3. Mol Endocrinol. 1998;12:428–441. [DOI] [PubMed] [Google Scholar]
- 32. Suh H, Gage PJ, Drouin J, Camper SA. Pitx2 is required at multiple stages of pituitary organogenesis: pituitary primordium formation and cell specification. Development. 2002;129:329–337. [DOI] [PubMed] [Google Scholar]
- 33. Tremblay JJ, Marcil A, Gauthier Y, Drouin J. Ptx1 regulates SF-1 activity by an interaction that mimics the role of the ligand-binding domain. EMBO J. 1999;18:3431–3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Drouin J, Lamolet B, Lamonerie T, Lanctôt C, Tremblay JJ. The PTX family of homeodomain transcription factors during pituitary development. Mol Cell Endocrinol. 1998;140:31–36. [DOI] [PubMed] [Google Scholar]
- 35. Wu Y, Luo H, Liu J, Kang D, McNeilly AS, Cui S. LIM homeodomain transcription factor Isl-1 enhances follicle stimulating hormone-β and luteinizing hormone-β gene expression and mediates the activation of leptin on gonadotropin synthesis. Endocrinology. 2010;151:4787–4800. [DOI] [PubMed] [Google Scholar]
- 36. Jiang Q, Jeong KH, Horton CD, Halvorson LM. Pituitary homeobox 1 (Pitx1) stimulates rat LHβ gene expression via two functional DNA-regulatory regions. J Mol Endocrinol. 2005;35:145–158. [DOI] [PubMed] [Google Scholar]
- 37. DiMattia GE, Rhodes SJ, Krones A, et al. The Pit-1 gene is regulated by distinct early and late pituitary-specific enhancers. Dev Biol. 1997;182:180–190. [DOI] [PubMed] [Google Scholar]
- 38. McGillivray SM, Bailey JS, Ramezani R, Kirkwood BJ, Mellon PL. Mouse GnRH receptor gene expression is mediated by the LHX3 homeodomain protein. Endocrinology. 2005;146:2180–2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Citation for the Richard E. Weitzman Memorial Award of the Endocrine Society to Pamela L. Mellon. Mol Endocrinol. 1989;3:1333–1334. [PubMed] [Google Scholar]
- 40. Lein ES, Hawrylycz MJ, Ao N, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. [DOI] [PubMed] [Google Scholar]
- 41. Koop KE, MacDonald LM, Lobe CG. Transcripts of Grg4, a murine groucho-related gene, are detected in adjacent tissues to other murine neurogenic gene homologues during embryonic development. Mech Dev. 1996;59:73–87. [DOI] [PubMed] [Google Scholar]
- 42. Leon C, Lobe CG. Grg3, a murine Groucho-related gene, is expressed in the developing nervous system and in mesenchyme-induced epithelial structures. Dev Dyn. 1997;208:11–24. [DOI] [PubMed] [Google Scholar]
- 43. Dehni G, Liu Y, Husain J, Stifani S. TLE expression correlates with mouse embryonic segmentation, neurogenesis, and epithelial determination. Mech Dev. 1995;53:369–381. [DOI] [PubMed] [Google Scholar]
- 44. Mallo M, Franco del Amo F, Gridley T. Cloning and developmental expression of Grg, a mouse gene related to the groucho transcript of the Drosophila enhancer of split complex. Mech Dev. 1993;42:67–76. [DOI] [PubMed] [Google Scholar]
- 45. Yu X, Li P, Roeder RG, Wang Z. Inhibition of androgen receptor-mediated transcription by amino-terminal enhancer of split. Mol Cell Biol. 2001;21:4614–4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mallo M, Gendron-Maguire M, Harbison ML, Gridley T. Protein characterization and targeted disruption of Grg, a mouse gene related to the groucho transcript of the Drosophila enhancer of split complex. Dev Dyn. 1995;204:338–347. [DOI] [PubMed] [Google Scholar]
- 47. Brinkmeier ML, Potok MA, Cha KB, et al. TCF and Groucho-related genes influence pituitary growth and development. Mol Endocrinol. 2003;17:2152–2161. [DOI] [PubMed] [Google Scholar]
- 48. Carvalho LR, Brinkmeier ML, Castinetti F, Ellsworth BS, Camper SA. Corepressors TLE1 and TLE3 interact with HESX1 and PROP1. Mol Endocrinol. 2010;24:754–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tremblay JJ, Goodyer CG, Drouin J. Transcriptional properties of Ptx1 and Ptx2 isoforms. Neuroendocrinology. 2000;71:277–286. [DOI] [PubMed] [Google Scholar]
- 50. Pulichino AM, Lamolet B, Vallette-Kasic S, et al. Tpit−/−NeuroD1−/− mice reveal novel aspects of corticotroph development. Endocr Res. 2004;30:551–552. [DOI] [PubMed] [Google Scholar]
- 51. Dorn C, Ou Q, Svaren J, Crawford PA, Sadovsky Y. Activation of luteinizing hormone β gene by gonadotropin-releasing hormone requires the synergy of early growth response-1 and steroidogenic factor-1. J Biol Chem. 1999;274:13870–13876. [DOI] [PubMed] [Google Scholar]