Abstract
We have developed a system for rapid and reliable assessment of gene essentiality in Haemophilus influenzae Rd strain KW20. We constructed two “suicide” complementation vectors (pASK5 and pASK6) containing 5′ and 3′ regions of the nonessential ompP1 gene flanking a multiple cloning site and a selectable marker (a chloramphenicol resistance gene or a tetracycline resistance cassette). Transformation of H. influenzae with the complementation constructs directs chromosomal integration of a gene of interest into the ompP1 locus, where the strong, constitutive ompP1 promoter drives its expression. This single-copy, chromosome-based complementation system is useful for confirming the essentiality of disrupted genes of interest. It allows genetic analysis in a background free of interference from any upstream or downstream genetic elements and enables conclusive assignment of essentiality. We validated this system by using the riboflavin synthase gene (ribC), a component of the riboflavin biosynthetic pathway. Our results confirmed the essentiality of ribC for survival of H. influenzae Rd strain KW20 and demonstrated that a complementing copy of ribC placed under control of the ompP1 promoter reverses the lethal phenotype of a strain with ribC deleted.
Haemophilus influenzae is a nonmotile, aerobic, gram-negative coccobacillus that colonizes the human upper respiratory tract as a normal commensal organism. H. influenzae is one of the most common agents of community-acquired pneumonia and causes a number of other diseases ranging in severity from otitis media and sinusitis to meningitis (10). Increasing emergence of virulent, antibiotic-resistant strains of H. influenzae and other important bacterial pathogens is driving the need for better-targeted pharmaceutical therapies (6, 7, 21).
The bacterial genomic era began in 1995, when the first complete genome of a free-living organism, H. influenzae, was published (5). Since then, genomics has played an increasingly significant role in the identification and validation of novel targets for antibacterial drug discovery (9). In this sense, “target validation” refers to experimental confirmation that a specific gene product is essential for the viability of an organism during growth and infection (9). Although the H. influenzae genome sequence was completed several years ago, the function and essentiality of many of its annotated genes remain unknown. Genome scale studies have been performed with H. influenzae in order to identify genes essential for growth and survival (1, 13). These studies focused on the identification of nonessential genes by performing transposon mutagenesis so that essential genes (EGs) could be deduced by mutant exclusion and zero-time analysis (1, 13). Akerley et al. (1) estimated that H. influenzae carries 478 EGs (no insertions) of which greater than 50% (259 genes) had no ascribed function (1). This is an efficient way to generate an inventory of potentially EGs; however, more directed studies are required to fully validate targets for antimicrobial drug discovery.
It is critical to develop genetic tools that will allow one to conclusively define gene essentiality, as well as address questions related to physiological functions. In many cases, a gene disruption can impact neighboring genes (e.g., operon structure), leading to pleiotropic effects and false interpretations of experimental data. Therefore, it is equally important to complement the disrupted gene in trans with a second copy of the gene to restore, as closely as possible, the original phenotype of the strain. Here we report the development of an efficient complementation system useful in assessing gene essentiality and function in H. influenzae.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains used in this study are listed in Table 1. Chemically competent Escherichia coli TOP10 cells (Invitrogen Corp., Carlsbad, Calif.) were used for cloning and propagation of plasmid constructs. Cells were maintained and selected on Luria-Bertani agar and in Luria-Bertani broth in the presence of the appropriate antibiotics. Ampicillin was used at 100 μg/ml, chloramphenicol was used at 50 μg/ml, and tetracycline was used at 20 μg/ml for selection in E. coli. Transformation of E. coli cells was performed as described by the manufacturer (Invitrogen).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Reference |
|---|---|---|
| E. coli TOP10 | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 deoR recA1 araD139 Δ(ara-leu) 7697 galU galK rpsL (Strr) endA1 nupG | Invitrogen |
| H. influenzae | ||
| Rd KW20 | WT genome sequence strain | 19 |
| SM1 | Rd(ΔribC::Kan) Kanr | This study |
| ASK1 | Rd(ΔompP1::cat) Camr | This study |
| ASK2 | Rd(ΔompP1::tetA tetR) Tetr | This study |
| ASK3 | Rd(ΔompP1::ribC+cat) Camr | This study |
| ASK4 | Rd(ΔompP1::ribC+tetA tetR) Tetr | This study |
| ASK5 | Rd(ΔribC::Kan ΔompP1::ribC+cat) Camr Kanr | This study |
| SM2 | Rd(ΔribC::Kan ΔompP1::ribC+tetA tetR) Tetr Kanr | This study |
| Plasmids | ||
| pUC18K | Kmr Apr; pUC18 containing nonpolar aph-A3 cassette | 8 |
| pUC19 | Apr; cloning vector | 20 |
| pGEM-T | Apr; TA cloning vector | Promega |
| pCMR | Cmr; 1.3-kb Cmr cassette from pACYC184 in pCR2.1 | 18 |
| pGESYII | Tcr; 2.8-kb Tcr cassette from pGJB103 in pACYC177 | 14 |
| pSM1 | Apr; pGEM-T 2,039-bp DNA fragment containing ribC and flanking region | This study |
| pSM2 | Kmr Apr; pSM1 with 442-bp deletion in ribC replaced by aph-A3 | This study |
| pSM3 | Apr Tcr; pGEM-T::tetA tetR | This study |
| pASK1 | Apr; pGEM-T, 645-bp ompP1 5′ homology sequence | This study |
| pASK2 | Apr; pGEM-T, 606-bp ompP1 3′ homology sequence | This study |
| pASK3 | Apr; pUC19 with ompP1 5′ 3′ homology sequence | This study |
| pASK4 | Apr Cmr; pGEM-T, 1238-bp Cmr cassette from pCMR flanked by SmaI, BamHI, XbaI, SalI, and PstI sites | This study |
| pASK5 | Apr Cmr; pUC19, ompP1 5′, 3′ homology sequence | This study |
| pASK6 | Apr Tcr; pUC19, ompP1 5′, 3′ homology sequence | This study |
| pASK7 | Apr; pGEM-T::ribC | This study |
| pASK8 | Apr Cmr; pUC19, ompP1 5′, 3′ homology sequence; ribC | This study |
| pASK9 | Apr Tcr; pUC19, ompP1 5′, 3′ homology sequence; ribC | This study |
H. influenzae Rd strain KW20 (5, 19) was grown and maintained at 37°C on brain heart infusion agar (Difco Laboratories) supplemented with 10 μg of βNAD per ml and 12 μg of hemin per ml (sBHI; Sigma, St. Louis, Mo.) (4). H. influenzae transformants were selected on sBHI containing the appropriate concentration of antibiotics and other supplements. In this case, chloramphenicol was used at 1.5 μg/ml, kanamycin was used at 10 μg/ml, tetracycline was used at 5 μg/ml, and riboflavin was used at 100 μg/ml. Transformation of H. influenzae was performed as previously described (4, 16).
H. influenzae competent cell preparation and transformation.
Competent cells were prepared and transformed as previously described (4). Briefly, H. influenzae cells were grown overnight on sBHI agar. Riboflavin and kanamycin were added to the medium for the ribC KO strain. Cells were scraped from the plates, suspended in brucella broth, and used to inoculate 100 ml of sBHI broth to a starting optical density at 600 nm of about 0.07 to 0.08. Cells were grown with shaking (160 rpm) at 37°C to an optical density at 600 nm of about 0.3, at which time they were centrifuged (15 min at 3,000 × g), and the pellet was washed by gentle pipetting in 50 ml of freshly prepared MIV medium (4). The washed cells were centrifuged as described above, resuspended in 80 ml of MIV medium, and grown with shaking (100 rpm) for 100 min at 37°C for competence development. Competent cells were frozen in 1-ml aliquots containing 20% (vol/vol) glycerol at −80°C.
One-milliliter aliquots of frozen cells were thawed on ice and transferred into 15-ml screw-cap tubes. Cells were pelleted, and each aliquot was resuspended in 1 ml of fresh MIV medium. For each transformation, approximately 1 μg of DNA was added to the cells and incubated at 37°C. After 30 min, 5 ml of sBHI broth was added to each transformation and incubation was continued for an additional 3 h. Finally, the cells were pelleted, resuspended in 100 μl of brucella broth, and plated on sBHI agar plates containing the appropriate supplements. Transformants were obtained after overnight incubation of the plates at 37°C.
Molecular biology procedures.
H. influenzae chromosomal DNA was prepared with a Wizard genomic DNA purification kit, and plasmid DNA was prepared with a Wizard Plus plasmid miniprep kit (Promega, Madison, Wis.). Restriction enzymes were obtained from New England Biolabs (Beverly, Mass.). Ligations were performed with a Rapid DNA ligation kit (Roche Diagnostics Corp., Indianapolis, Ind.). PCR Supermix High Fidelity (Invitrogen) was used to generate DNA fragments, and the reactions were purified with a QuickStep PCR purification kit (Edge Biosystems, Gaithersburg, Md.). All PCR-generated clones and selected PCR-generated DNA fragments were sequenced with an ABI Prism 3100 Genetic Analyzer after preparing ABI Prism BigDye Terminator Cycle Sequencing v.2.0 Ready Reactions (PE Biosystems, Foster City, Calif.). The resulting DNA sequence chromatographs were assembled and analyzed with Sequencher software v.4.0.5 (Gene Codes Corporation, Ann Arbor, Mich.). Oligonucleotide primers used for PCR and sequencing were synthesized at Invitrogen (Table 2). DNA was extracted from preparative agarose gels with a QIAEX II kit (QIAGEN Inc., Valencia, Calif.). The cloning vectors used were pGEM-T (Promega) and pUC19 (20).
TABLE 2.
Primers used for plasmid constructions
| Primer | Sequence (5′→3′) |
|---|---|
| CAT1 | CCCGGGGATCCTCTAGAGTCGACCTGCAGCCTCCACGGGGAGAGCCTGA |
| CAT2 | CCCGGGGATCCTCTAGAGTCGACCTGCAGCCACCCCGTCAGTAGCTGAACAGGAGGG |
| CAT3 | CATTGGGATATATCAACGGTGG |
| CAT4 | GATTCAGGTTCATCATGCC |
| TetRF | TGCTGCAGTTAAGACCCACTTTCACATTTAAGTTGTTTTTCTAATCCGCAAATGATCAATTCAAGGCC |
| TetAF | TAACCCGGGAAGCTTGCATGCCTGCAGACATCATTAATTCCTAATTTTTGTTGAC |
| TetAR | TTGCCCGGGAAGCTTGCATGCTGCAGGCTATTTACCGCGGCTTTTTATTGAGC |
| ompP1 | CGTACGGACGTCAAGCGTATTCAGCAAGCG |
| ompP2 | TTTTTTCATATGGAACCCTTTGTTATAAATAAAAATCTGGG |
| ompP3 | CCTTACCTGCAGGTATTGCTTACGATCAGGC |
| ompP4 | CTCGCAAAGCTTGCCAAATGGCTTGTTGAAATGCG |
| 5′ ompP1 | GAAATTGAGCTACATTCAGACGG |
| 3′ ompP1 | GCTCTCCATAAGTTGAAAGCTCG |
| HI-1613-1F | CGAAAATTAATCAGCATCAAATAAGC |
| HI-1613-1R | CAAGGCGAGGCTTTAGAAATTTCGCC |
| HI-1613-2F | CCACAAACCCGGGCTATTGTAGATACGGTAG |
| HI-1613-2R | ATTTCCCGGGCTTTACGCATTTCAGGTAGTAATTTTACC |
| HI-1613-3F | GGTAATGACCATCGCAGGCTTGG |
| HI-1613-3R | CGGCTCAAATGTTATTTGCGC |
| K1 | CTCCCACCAGCTTATATACCTTAGC |
| K2 | TTGCCTTCTGCGTCCGGTCGATCAGG |
| ribCNdeF | AAAATTCATATGTTTACTGGAATTGTACAAGGC |
| ribCSalR | TTTTAAGTCGACCTAGAAATTTTTTGACTGCAAATAATTTTCTACCG |
Plasmid constructions.
The plasmids used in this study are listed in Table 1. To generate an H. influenzae riboflavin synthase (ribC gene number HI1613) disruption plasmid, a 2,039-bp PCR fragment containing the ribC gene plus 498 bp upstream of the ATG start codon and 926 bp downstream of the TAG stop codon, was PCR amplified with primers HI-1613-1F and -1R and subsequently cloned into pGEM-T to create pSM1 (Fig. 1a). Next, primers HI-1613-2F and -2R (with a SmaI site) were used to perform an inverse PCR around pSM1, creating a defined 442-bp deletion (out of 615 bp) in ribC. The resulting PCR product was digested with SmaI. An 840-bp DNA fragment containing a promoterless aphA-3 kanamycin resistance cassette was excised from pUC18K with SmaI and ligated in frame in place of the 442-bp deletion in ribC to make pSM2 (8). In addition, a 2,039-bp linear version of the disruption construct was prepared by digesting pSM2 with PstI and SacII and gel purifying the DNA fragment containing the disrupted ribC gene (Fig. 1b). This plasmid and linear DNA fragment were used to make the strains with ribC disrupted that are described in Results.
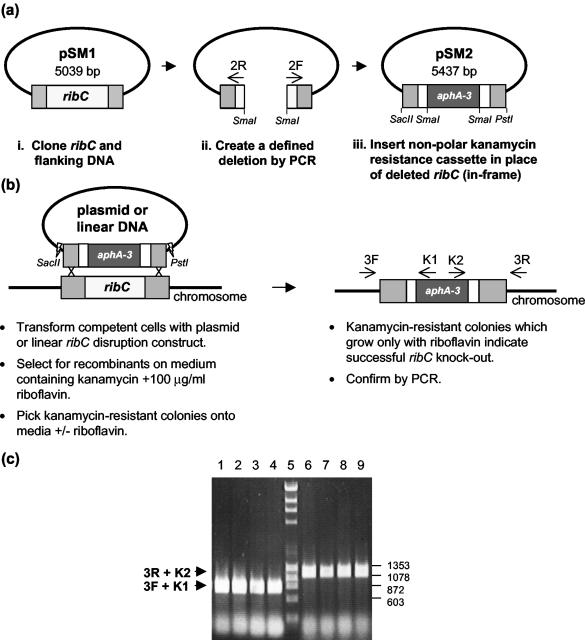
FIG. 1.
(a) Construction of ribC disruption plasmid pSM2. (i) ribC (615 bp) and flanking DNA sequences (498 bp upstream of the ATG start codon and 926 bp downstream of the TAG stop codon) were cloned as a 2,039-bp PCR product into pGEM-T. (ii) A defined 442-bp deletion was created in the ribC coding sequence by inverse PCR with primers 2F and 2R, generating a 4,597-bp product that was then digested with SmaI. (iii) An 840-bp SmaI-digested DNA fragment containing a promoterless, nonpolar kanamycin resistance cassette (aphA-3) was ligated, in frame, in place of the deleted ribC sequence to make pSM2. (b) pSM2 or a linear ribC disruption construct was transformed into H. influenzae to generate a ribC disruption strain (SM1). The linear ribC disruption construct was generated by digestion of pSM2 with SacII and PstI, followed by gel purification. Primers 3R and K2 and primers 3F and K1 were used to verify the disruption of ribC by PCR amplification. 3F and 3R bind outside of the originally cloned 2,039-bp DNA fragment of pSM1 (c) PCR products obtained from four representative Kanr colonies with primers 3R and K2 (lanes 1 to 4) and primers 3F and K1 (lanes 6 to 9) were analyzed by agarose gel electrophoresis. Only transformants grown in the presence of riboflavin carried a disrupted copy of ribC as determined by visualization of the expected 740-bp bands in lanes 1 to 4 (crossover at the 5′ end) and 1,195-bp bands in lanes 6 to 9 (crossover at the 3′ end). DNA size standards are shown in lane 5 with corresponding relevant sizes (base pairs) at the right.
Two complementation vectors were constructed to enable complementation analysis of EG disruptions in H. influenzae. The nonessential ompP1 locus (HI-0401) was used as the target for homologous recombination of complementing DNA into the chromosome. First, primers ompP1 and ompP2 (containing engineered AatII and NdeI sites, respectively) were used to amplify a 645-bp region of the Rd KW20 chromosome 5′ to and including the ompP1 start (ATG) codon (5′ ompP1). This fragment was cloned into pGEM-T to make pASK1. Primers ompP3 (with a PstI site) and ompP4 (with an HindIII site) were used to generate a 606-bp fragment at the 3′ end of the gene (3′ ompP1) that was also cloned into pGEM-T to make pASK2. The 5′ ompP1 and 3′ ompP1 DNAs were subsequently excised from pASK1 (AatII-NdeI fragment) and pASK2 (PstI-HindIII fragment) and directionally cloned into pUC19 to make pASK3. Both DNA fragments contained the 9-bp core DNA uptake sequence (AAGTGCGGT) required for efficient transformation of H. influenzae (15). Next, antibiotic resistance cassettes were amplified for cloning into pASK3. A 1,238-bp chloramphenicol resistance cassette containing the chloramphenicol acetyltransferase gene (cat) was amplified from pCMR (18) with the CAT1 and CAT2 primers, which were designed to introduce SmaI, BamHI, XbaI, SalI, and PstI restriction sites flanking the cat gene. This PCR product was cloned into pGEM-T to make pASK4. The chloramphenicol resistance cassette was then excised from pASK4 with PstI and ligated into pASK3 to make pASK5 (Fig. 2). A tetracycline resistance cassette (2,064 bp) comprising tetA and tetR was amplified with primers TetRF and TetAR from pGESYII (14) and cloned into pGEM-T to make pSM3. The cassette was modified to remove an internal NdeI site in tetR and incorporate flanking PstI restriction sites for subcloning. The tetracycline resistance cassette was excised from pSM3 with PstI and ligated into pASK3 to make pASK6 (Fig. 2).
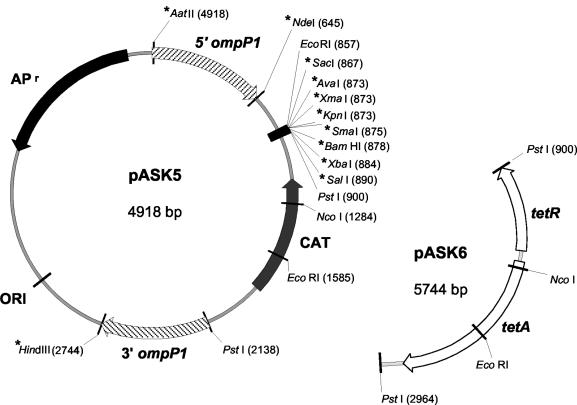
FIG. 2.
Complementation vectors developed for H. influenzae. pUC19-based vectors pASK5 (4,918 bp) and pASK6 (5,744 bp) were constructed as described in Materials and Methods. Genomic sequences found flanking the ompP1 coding region (645 bp upstream and 606 bp downstream) were inserted into both vectors. An NdeI site located 3′ to the 645-bp upstream ompP1 sequence fragment allows in-frame insertion of the gene of interest. A multiple cloning site allows directional cloning. pASK5 and pASK6 contain resistance genes for chloramphenicol (cat) and tetracycline (tetA and tetR), respectively, to allow selection of integrants. An asterisk indicates a unique restriction enzyme site.
The ribC gene (615 bp) was PCR amplified from H. influenzae Rd KW20 genomic DNA with primers ribCNdeF and ribCSalR and cloned into pGEM-T to make pASK7. The primers were designed so that the resultant PCR product would contain a 5′ NdeI site and a 3′ SalI site to facilitate subcloning into complementation vectors pASK5 and pASK6. ribC was then subcloned into both complementation vectors to make pASK8 and pASK9.
In all cases, both strands of each clone, including the junctions, were sequenced.
RESULTS
The basic strategy for complementation of EG disruptions described in this report is outlined in Fig. 3. The first step involves transformation of wild-type (WT) H. influenzae cells with a complementation vector carrying a functional copy of an EG and a unique antibiotic resistance gene(s) (cat or tetA tetR). We tested our complementation system with the conditionally essential ribC gene. ribC was selected to validate the complementation plasmids because a ribC disruption mutant is expected to be auxotrophic and therefore should be viable when the recombinant strain is grown in the presence of excess riboflavin. This allows us to study ribC as a nonessential gene. Additionally, when a strain with ribC disrupted is deprived of riboflavin, it should not be viable and would therefore allow us to study ribC as an EG for the sake of EG complementation.
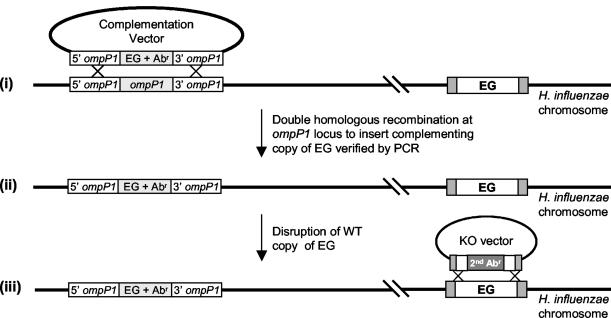
FIG. 3.
Chromosomal complementation strategy for assessing gene essentiality in H. influenzae. (i) WT H. influenzae cells are transformed with a suicide complementation vector containing a copy of a putative EG together with an antibiotic resistance (Abr) gene for selection. Integration of this copy of the EG is targeted to the nonessential ompP1 locus facilitated by ompP1 5′ and 3′ sequences flanking the EG-plus-Abr gene sequence. (ii) Transformants carrying the second copy of the EG, now placed under control of the ompP1 promoter, are selected on medium containing the appropriate antibiotic. Proper integration is verified by PCR analysis. (iii) A suicide disruption vector containing the EG disrupted by a nonpolar Abr gene is transformed into the strain created in step ii. KO, knockout.
Construction of a strain with ribC disrupted.
Since ribC was our gene of choice for validating the complementation system, we first created an H. influenzae strain with ribC disrupted to verify that loss of this gene function would result in a distinguishable phenotype that could subsequently be complemented in accordance with the strategy described in Fig. 3. A schematic of the gene disruption procedure is illustrated in Fig. 1. WT H. influenzae strain KW20 was first transformed with ribC disruption plasmid pSM2, and transformants were selected on sBHI agar containing kanamycin, with and without riboflavin (Table 3). Since all of the plasmids used for transformation of H. influenzae in this study contain a ColE1 origin, they are unable to autonomously replicate and must integrate into the chromosome to survive antibiotic selection. After transformation and selection, kanamycin-resistant colonies that grew in the presence or absence of riboflavin were analyzed by genomic PCR. PCRs indicative of double homologous recombination at the ribC locus were prepared with primers 3F and K1 (recombination at the 5′ end of ribC) or 3R and K2 (recombination at the 3′ end) (Fig. 1b). When constructing an EG disruption, it is important to delete most of the EG of interest and provide substantial 5′ and 3′ flanking DNA in order to favor recombination at the original site of the EG and avoid recombination at the site of complementation.
TABLE 3.
Transformation efficiencya of ribC disruption constructs
| DNA constructb | With riboflavinc | Without riboflavind | Ratioe |
|---|---|---|---|
| Plasmid DNA (pSM2) | 4.9 × 105 | 7.1 × 103 | 69 |
| Linear DNA | 2.2 × 106 | 1 × 102 | 22,000 |
Transformation efficiency refers to the number of kanamycin-resistant colonies recovered per microgram of DNA. These data represent a single representative (reproducible) experiment.
ribC disruption constructs as referred to in Materials and Methods and Fig. 1b.
Number of kanamycin-resistant colonies recovered when selection is carried out in the presence of 100 μg of riboflavin per ml.
Number of kanamycin-resistant colonies recovered when selection is carried out in the absence of riboflavin.
Ratio refers to the number of kanamycin-resistant colonies obtained in the presence of riboflavin divided by the number of kanamycin-resistant colonies obtained in the absence of riboflavin.
PCR analysis of the transformants obtained in the presence of riboflavin indicated that ribC was disrupted because of double recombination at the ribC locus. The resulting size of the PCR products corresponded to those expected upon proper integration of the ribC disruption construct at the WT ribC locus (Fig. 1c). Also, when these colonies were subcultured onto medium without riboflavin they did not grow, as expected given that the ribC gene is essential for riboflavin synthesis. In contrast, PCR analysis of the transformants obtained in the absence of riboflavin generated only a single PCR product from either the 3′ or the 5′ PCR but not both. This result indicated that the entire plasmid, pSM2, had integrated via single homologous recombination at the 3′ end or the 5′ end of ribC and that the strain contained a disrupted ribC gene, as well as an adjacent functional WT ribC gene (data not shown).
Since transformation with pSM2 yielded a large number of false-positive (putatively disrupted ribC) colonies when ribC was treated as an EG (selection without riboflavin), we tested linear DNA (containing the disrupted ribC gene) excised from pSM2 with PstI and SacII and gel purified. Molar equivalent amounts of linear and plasmid (pSM2) DNAs were transformed into H. influenzae. Compared to transformation with plasmid DNA, transformation with linear DNA yielded much fewer background colonies when ribC was treated as an EG (selection without riboflavin; Table 3). When selection for Kanr was done in the presence of riboflavin, similar transformation numbers were obtained for linear and circular DNAs (Table 3). Therefore, when testing the essentiality of a gene it appears that transformation with linear DNA significantly reduces potential false positives due to single-crossover recombination events (plasmid integration) since the linear DNA must integrate via double homologous recombination to yield a kanamycin-resistant phenotype. The magnitude of the difference in recovering false-positive colonies when transforming with linear versus plasmid is shown in Table 3.
Follow-up experiments revealed that the linear DNA version of the ribC disruption construct could be prepared by PCR amplification from pSM2 with primers 1F and 1R to achieve the same result as with restriction enzyme-generated linear DNA (data not shown).
Selection of a chromosomal site for complementation.
In order to identify a nonessential gene that would provide a suitable site for integrating a second copy of a gene of interest, we reviewed the available literature for well-characterized, nonessential H. influenzae genes. We identified 16 candidates of which the outer membrane protein OmpP1 was the most suitable for our purposes. The ompP1 gene is under control of a strong, constitutively active promoter that has been shown also to be active in E. coli (12). The ability to place a gene of interest under the control of this promoter would ensure robust expression in vivo, allowing effective screening in the complementation analysis. Further, the nonessential nature of this gene was confirmed by Tn5 mutagenesis in H. influenzae type b and biogroup aegyptius, where OmpP1 has been extensively studied to assess its potential as a vaccine candidate (11). Strains with ompP1 disrupted were observed to grow normally and did not exhibit any detectable phenotypic differences from the WT parent strain (11).
Complementation with pASK8 and pASK9 of an H. influenzae strain with ribC disrupted.
Competent WT KW20 cells were transformed with either pASK8 or pASK9 (both carry ribC). Chloramphenicol (pASK8)- and tetracycline (pASK9)-resistant colonies were checked for specific homologous recombination by PCR with the primer pairs indicated in Fig. 4 and Table 2. For chloramphenicol-resistant transformants (pASK8 derived), proper integration at the 5′ and 3′ ends was verified in individual reaction mixtures containing primers 5′ompP1 and Cmr3 or primers Cmr4 and 3′ompP1, respectively. Agarose gel electrophoresis of the PCR products confirmed the presence of the 977-bp (3′ end) and 1,683-bp (5′ end) fragments expected upon integration of the complementing construct at the ompP1 locus (Fig. 4a, lanes 1 and 2). Tetracycline-resistant integrants (pASK9 derived) were similarly analyzed with primers 5′ompP1 and HI-1613-2R (5′ end) or primers TetAF and 3′ompP1 (3′ end). Recombination at both the 3′ and 5′ ends of ompP1 was verified upon agarose gel visualization of 2,151- and 803-bp DNA fragments, respectively (Fig. 4b, lanes 1 and 2). All of the resistant colonies analyzed exhibited double homologous recombination of pASK8 and pASK9 at the ompP1 locus. One H. influenzae colony each from the pASK8 (strain ASK3) and pASK9 (strain ASK4) transformations described above was subcultured, and competent cells were made for transformation with ribC disruption construct pSM2.
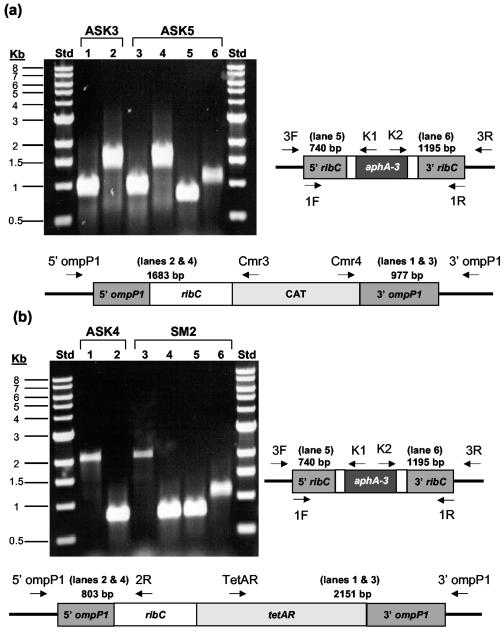
FIG. 4.
Genetic complementation with complementation vectors pASK8 (a) and pASK9 (b) of strains with ribC disrupted. Site-specific integration of the ribC gene supplied on complementation vectors pASK8 and pASK9 into the ompP1 locus to create strains ASK3 and ASK4 was verified by PCR analysis after transformation of WT strain KW20. Subsequently, targeted disruption of the authentic copy of ribC (but not the complementing copy at the ompP1 locus) by transformation with pSM2 was also verified by PCR. This disruption resulted in the creation of H. influenzae strains ASK5 and SM2. The specific primer pairs shown in the schematics were used for PCR amplifications from chromosomal DNAs prepared from H. influenzae strains ASK3, ASK4, ASK5, and SM2. Schematic representations of the gene arrangements resulting from the transformations described above and the resulting agarose gel electrophoresis patterns for the PCR products obtained are shown. In lanes 1 and 3, the 977-bp ASK3 and ASK5 (a) and the 2,151-bp ASK4 and SM2 (b) products are indicative of integration of ribC at the 3′ end of ompP1, while in lanes 2 and 4, the 1,683-bp ASK3 and ASK5 (a) and the 803-bp ASK4 and SM2 (b) fragments result from recombination at the 5′ end of the ompP1 locus. The 740- and 1,195-bp PCR products in lanes 5 and 6 of both gels verify disruption of the WT copy of the ribC gene. Std = size standards in kilobase pairs (Kb) are shown. Lane numbers are bracketed.
PCR-based analysis of genomic DNAs prepared from the transformants was used to verify disruption of the WT ribC gene. Disruption of the WT ribC locus was confirmed in individual PCRs set up with genomic DNAs isolated from ASK5 and SM2 (Fig. 4a and b, lanes 5 and 6). In both pASK8- and pASK9-derived strains transformed with the ribC disruption construct, 100% of the transformants obtained in the absence of riboflavin were confirmed as having a disrupted copy of ribC.
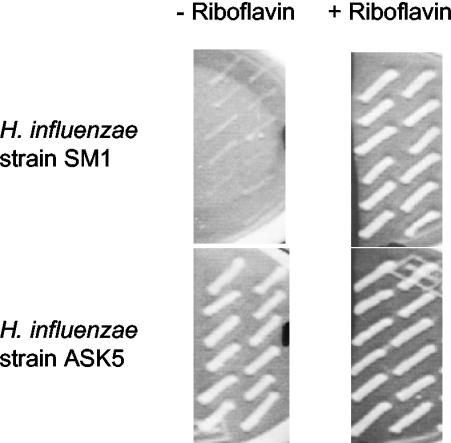
The ability of the ribC gene placed under control of the ompP1 promoter to complement the ribC disruption was further characterized by growth on media with and without riboflavin. H. influenzae strain SM1 grew well on medium supplemented with riboflavin but was entirely unable to grow in the absence of riboflavin (Fig. 5, top). This conditional lethal phenotype was reversed when a complementing copy of the ribC gene, supplied on complementation vector pASK8 or pASK9, recombined at the nonessential ompP1 locus, indicating that expression of ribC was being driven by the ompP1 promoter (Fig. 5, bottom). ribC can therefore be said to be essential for H. influenzae because of its participation in the essential riboflavin biosynthesis pathway.
FIG. 5.
Growth behavior of complemented versus noncomplemented H. influenzae strains with ribC disrupted. The growth behavior of WT H. influenzae cells transformed with complementation vector pASK8 and/or ribC disruption plasmid pSM2 was recorded in the presence and absence of riboflavin. Twelve colonies from each transformation were patched in duplicate onto media with and without riboflavin. pSM2-mediated disruption of the ribC gene rendered the cells dependent on an exogenous supply of riboflavin for survival (strain SM1). Introduction of a functional ribC gene at the ompP1 locus as a result of transformation with the complementation vector successfully restored the WT phenotype and allowed cells to grow equally well both in the presence and in the absence of riboflavin in the growth medium (strain ASK5).
DISCUSSION
The riboflavin synthase α subunit gene, ribC, was identified as a model gene to test the efficiency and robustness of our system. Riboflavin, a precursor molecule of the coenzymes flavin adenine dinucleotide and flavin mononucleotide, is essential for basic cellular metabolism (2). It is produced endogenously in plants and most microbes but must be acquired exogenously by higher animals (2). Riboflavin biosynthesis has been proposed as a potential target for chemotherapy of gram-negative bacterial infections because E. coli and other gram-negative pathogens lack a transport system for riboflavin, and as a result, riboflavin auxotrophs only survive if grown in the presence of nonphysiologically high concentrations of the vitamin (3, 17). Little is known about riboflavin biosynthesis in H. influenzae other than what can be construed on the basis of homology to other tested systems.
H. influenzae is an important human pathogen. It is amenable to DNA manipulations and therefore an attractive model organism. Its genome has been completely sequenced, greatly facilitating cloning and expression of most genes of interest (5). It is naturally competent for DNA uptake and integration (15), and strain KW20 is highly sensitive to most antibiotics for which resistance cassettes are available (14, 18). The gene disruption and complementation system reported here should serve as a useful tool in evaluating gene function and essentiality in H. influenzae. It should find applications in the identification of genes as potential targets for drug discovery. A large number of genes can be screened, and their associated effects and functions can be studied. The use of linear versus plasmid disruption constructs greatly improves the yield of the correct double recombinants. In vivo expression of the complementing gene allows a clearer interpretation of the resultant phenotype without contending with issues of plasmid copy number, polar effects, and interference from or of other overlapping genetic elements (operon effects). We have validated the system with the conditionally essential ribC gene and found that our method allows efficient evaluation of putative EGs.
Acknowledgments
We thank Paul Manning for critical reading of the manuscript, T. L. Stull and D. J. Morton for the generous gift of the plasmids pCMR and pGESYII, and K. MacCormack for DNA sequencing.
The AstraZeneca R&D Boston summer internship program supported this work.
REFERENCES
- 1.Akerley, B. J., E. J. Rubin, V. L. Novick, K. Amaya, N. Judson, and J. J. Mekalanos. 2002. A genome-scale analysis for identification of genes required for growth or survival of Haemophilus influenzae. Proc. Natl. Acad. Sci. USA 99:966-971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bacher, A., S. Eberhardt, and G. Richter. 1996. Biosynthesis of riboflavin, p. 657-664. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coil and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 3.Bandrin, S. V., M. Y. Beburov, P. M. Rabinovich, and A. I. Stepanov. 1979. Riboflavin auxotrophs of Escherichia coli. Genetika 15:2063-2065. [PubMed] [Google Scholar]
- 4.Barcak, G. J., M. S. Chandler, R. J. Redfield, and J. F. Tomb. 1991. Genetic systems in Haemophilus influenzae. Methods Enzymol. 204:321-342. [DOI] [PubMed] [Google Scholar]
- 5.Fleischmann, R. D., M. D. Adams, O. White, R. A. Clayton, E. F. Kirkness, A. R. Kerlavage, et al. 1995. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269:496-512. [DOI] [PubMed] [Google Scholar]
- 6.Georgiou, M., R. Munoz, F. Roman, R. Canton, R. Gomez-Lus, J. Campos, and A. G. De La Campa. 1996. Ciprofloxacin-resistant Haemophilus influenzae strains possess mutations in analogous positions of GyrA and ParC. Antimicrob. Agents Chemother. 40:1741-1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jacobs, M. R., D. Felmingham, P. C. Appelbaum, and R. N. Gruneberg. 2003. The Alexander Project 1998-2000: susceptibility of pathogens isolated from community-acquired respiratory tract infection to commonly used antimicrobial agents. J. Antimicrob. Chemother. 52:229-246. [DOI] [PubMed] [Google Scholar]
- 8.Menard, R., P. J. Sansonetti, and C. Parsot. 1993. Nonpolar mutagenesis of the ipa genes defines IpaB, IpaC, and IpaD as effectors of Shigella flexneri entry into epithelial cells. J. Bacteriol. 175:5899-5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mills, S. D. 2003. The role of genomics in antimicrobial discovery. J. Antimicrob. Chemother. 51:749-752. [DOI] [PubMed] [Google Scholar]
- 10.Moxon, E. R. 1995. Haemophilus influenzae, p. 2039-2045. In G. L. Mandell, J. E. Bennet, and R. Dolin. (ed.), Principles and practice of infectious diseases, vol. 2. Churchill Livingstone, New York, N.Y. [Google Scholar]
- 11.Munson, R., Jr., and A. Hunt. 1989. Isolation and characterization of a mutant of Haemophilus influenzae type b deficient in outer membrane protein P1. Infect. Immun. 57:1002-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Munson, R., Jr., and S. Grass. 1988. Purification, cloning, and sequence of outer membrane protein P1 of Haemophilus influenzae type b. Infect. Immun. 56:2235-2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reich, K. A., L. Chovan, and P. Hessler. 1999. Genome scanning in Haemophilus influenzae for identification of essential genes. J. Bacteriol. 181:4961-4968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ren, Z., H. Jin, D. J. Morton, and T. L. Stull. 1998. hgpB, a gene encoding a second Haemophilus influenzae hemoglobin- and hemoglobin-haptoglobin-binding protein. Infect. Immun. 66:4733-4741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith, H. O., J. F. Tomb, B. A. Dougherty, R. D. Fleischmann, and J. C. Venter. 1995. Frequency and distribution of DNA uptake signal sequences in the Haemophilus influenzae Rd genome. Science 269:538-540. [DOI] [PubMed] [Google Scholar]
- 16.Spencer, H. T., and R. M. Herriott. 1965. Development of competence of Haemophilus influenzae. J. Bacteriol. 90:911-920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vitreschak, A. G., D. A. Rodionov, A. A. Mironov, and M. S. Gelfand. 2002. Regulation of riboflavin biosynthesis and transport genes in bacteria by transcriptional and translational attenuation. Nucleic Acids Res. 30:3141-3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Whitby, P. W., D. J. Morton, and T. L. Stull. 1998. Construction of antibiotic resistance cassettes with multiple paired restriction sites for insertional mutagenesis of Haemophilus influenzae. FEMS Microbiol. Lett. 158:57-60. [DOI] [PubMed] [Google Scholar]
- 19.Wilcox, K. W., and H. O. Smith. 1975. Isolation and characterization of mutants of Haemophilus influenzae deficient in an adenosine 5′-triphosphate-dependent deoxyribonuclease activity. J. Bacteriol. 122:443-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 21.Zhanel, G. G., J. A. Karlowsky, D. E. Low, and D. J. Hoban. 2000. Antibiotic resistance in respiratory tract isolates of Haemophilus influenzae and Moraxella catarrhalis collected from across Canada in 1997-1998. J. Antimicrob. Chemother. 45:655-662. [DOI] [PubMed] [Google Scholar]