Abstract
Progesterone, acting through the progesterone receptors (PGRs), is one of the most critical regulators of endometrial differentiation, known as decidualization, which is a key step toward the establishment of pregnancy. Yet a long-standing unresolved issue in uterine biology is the precise roles played by the major PGR isoforms, PGR-A and PGR-B, during decidualization in the human. Our approach, expressing PGR-A and PGR-B individually after silencing endogenous PGRs in human endometrial stromal cells (HESCs), enabled the analysis of the roles of these isoforms separately as well as jointly. Chromatin immunoprecipitation-sequencing in combination with gene expression profiling revealed that PGR-B controls a substantially larger cistrome and transcriptome than PGR-A during HESC differentiation. Interestingly, PGR-B directly regulates the expression of PGR-A. De novo motif analysis indicated that, although the 2 isoforms bind to the same DNA sequence motif, there are both common and unique neighboring motifs where other transcription factors, such as FOSL1/2, JUN, C/EBPβ, and STAT3, bind and dictate the transcriptional activities of these isoforms. We found that PGR-A and PGR-B regulate overlapping as well as distinct sets of genes, many of which are known to be critical for decidualization and establishment of pregnancy. When PGR-A and PGR-B were coexpressed during HESC differentiation, PGR-B played a predominant role, although both isoforms influenced each other's transcriptional activity. This study revealed the gene networks that operate downstream of each PGR isoform to mediate critical functions, such as regulation of the cell cycle, angiogenesis, lysosomal activation, insulin receptor signaling, and apoptosis, during decidualization in the human.
Decidualization, the differentiation of endometrium into a supportive tissue for the implanting embryo, is one of the most critical processes during the establishment of pregnancy. Each step of this process is regulated by the concerted actions of many transcription factors and signaling molecules. Progesterone (P), secreted from the newly formed corpus luteum after ovulation, is one of the earliest and most important regulators of endometrial differentiation. It is well established that P, acting through the nuclear P receptors (PGRs) in endometrial cells, is required for the precise and timely regulation of this process (1–6). Several in vitro and in vivo studies, including characterization of the Pgr knockout mouse, have established a key role for PGR during decidualization in mice and humans (1–8).
PGR exists as 2 isoforms, PGR-A and PGR-B, which are transcribed from the same gene (9). Despite having the same DNA binding and ligand binding domains, PGR-A and PGR-B often display very different transcriptional activities in the cell. The dissimilarities in their activities originate from the differences in their amino-terminal regions, as PGR-B has an additional transactivation domain that interacts with adjacent PGR-B dimers, different transcription factors, and coactivators (10). A number of cell-based reporter assays suggested that PGR-A could act as a repressor of PGR-B transcriptional activity at certain promoters (11, 12). Studies in breast cancer cell lines showed that PGR-A and PGR-B regulate different gene networks, and their relative expression levels, which change in some breast cancer types, might affect downstream gene expression (13, 14).
Both PGR isoforms are expressed in human endometrial stroma, and their relative levels change throughout the menstrual cycle (15). In the early proliferative stage, PGR-A levels are relatively higher than those of PGR-B. During the periovulatory period, there is a robust induction of PGR-B, making its level comparable to that of PGR-A. By the late secretory phase, expression of both PGR-A and PGR-B starts to decline, and their levels return to those seen in the early proliferative phase (15, 16). Although several PGR-regulated pathways have been identified in the endometrium (5, 7, 17), a comprehensive analysis of the primary gene targets of each isoform during human decidualization has not been performed.
To achieve this goal, we used a well-established in vitro system in which human primary endometrial stromal cells (HESCs) undergo differentiation in response to steroid hormones and cAMP. We used adenoviral vectors to express PGR-A and PGR-B isoforms individually or in combination in these cells after removal of the endogenous PGR upon treatment with small interfering RNA (siRNA). This strategy allowed the identification of genome-wide binding sites and downstream gene networks of each isoform during endometrial differentiation. Our results offer unique insights into the roles of the PGR isoforms in human uterine biology.
Materials and Methods
Primary HESC culture
Our studies involving human endometrial biopsies and endometrial cell cultures adhere to the regulations set forth for the protection of human subjects participating in clinical research and are approved by the institutional review boards of Emory University, Wake Forest University, and the University of Illinois at Urbana-Champaign. Endometrial samples from the early proliferative stage of the menstrual cycle were obtained by Pipelle biopsy at Emory University and Wake Forest Medical Centers from fertile, regularly cycling women under anesthesia before laparoscopy as described previously (18). They had no signs of endometriosis or other endometrial pathological conditions and provided written informed consent. The subjects ranged in age from 28 to 42 years and in parity from 1 to 2.
These cells were cultured in DMEM/F-12 medium (Invitrogen) supplemented with 5% (v/v) fetal bovine serum (HyClone), 50 μg/mL penicillin, and 50 μg/mL streptomycin (Invitrogen). For in vitro differentiation, the cells were treated with differentiation cocktail composed of 0.5 mM 8-bromo-adenosine-3′,5′-cyclic monophosphate (8-Br-cAMP) (Sigma-Aldrich), 1 μM P (Sigma-Aldrich), and 10 nM 17β-estradiol (Sigma-Aldrich) in DMEM/F-12 medium (Invitrogen) supplemented with 2% (v/v) charcoal dextran–stripped fetal bovine serum. The medium/differentiation cocktail was refreshed every 48 hours.
siRNA transfection and adenovirus transduction
HESCs were transfected with siRNA targeting the 3′-untranslated region (UTR) of PGR mRNA (Dharmacon) or control scrambled siRNA (Dharmacon) following the manufacturer's protocol (siLentFect; Bio-Rad Laboratories). In brief, a final concentration of 50 nM siRNA was mixed with siLentFect to transfect the cells. At 24 hours after siRNA treatment, cells were transduced with the adenovirus expressing flag-tagged PGR-A or PGR-B for 24 hours. The adenovirus was kindly provided by Dr Dean Edwards of Baylor College of Medicine (Houston, Texas). At 48 hours of siRNA transfection and 24 hours of concurrent adenovirus transduction, medium was removed, and cells were washed with PBS and treated with differentiation cocktail for the time periods indicated in the figure legends.
Western blot analysis
Primary HESCs were subjected to different treatments for the indicated periods of time as described in the figure legends. Whole-cell extracts were prepared, equal amounts of proteins were analyzed by SDS-PAGE and transferred to polyvinylidene difluoride membrane, and the specific proteins were detected by Western blotting using the antibody against calnexin (Santa Cruz Biotechnology) and PGR. The PGR antibody was kindly provided by Dr Dean Edwards of Baylor College of Medicine.
Chromatin immunoprecipitation (ChIP) and ChIP-sequencing (ChIP-seq) analysis
HESCs were seeded on 150-mm dishes. After the cells attached, they were placed in medium containing 2% (v/v) charcoal dextran–stripped fetal bovine serum. When the cells reached 70% to 80% confluence, they were transfected with siRNA and after 24 hours were transduced with PGR-A or PGR-B adenovirus or control virus for 24 hours. On the next day, the cells were treated with a hormonal cocktail containing estrogen (E), P, and 8-Br-cAMP for 2 hours. ChIP assays were performed using the EZ-ChIP kit (Millipore), according to the manufacturer's instructions with minor modifications. In brief, cells were fixed with 1% formaldehyde for 10 minutes, and excess formaldehyde was quenched with 0.25 M glycine for 5 minutes. Cells were then washed twice with cold PBS, scraped and collected in PBS containing protease inhibitors in a conical tube, and centrifuged. Cell pellets were resuspended in lysis buffer containing protease inhibitors for 15 minutes to lyse the cells. Next, chromatin was sonicated using 4 15-second pulses on power 4 with cooling between pulses (model 100 sonic dismembrator; Fisher Scientific). One percent of the cell lysate was used for input control. The remainder of the lysate was incubated with anti-flag M2 affinity gel (A2220; Sigma-Aldrich) overnight at 4°C to isolate the immune complexes (flag-PGR-A and flag-PGR-B). For additional ChIP experiments, antibodies against CCAAT/enhancer-binding protein β (C/EBPβ) (sc-150; Santa Cruz Technology) and signal transducer and activator of transcription 3 (STAT3) (sc-8019 and sc-482; Santa Cruz Technology) were used. The immunoprecipitates were washed consecutively with buffers containing low salt, high salt, and LiCl and twice with Tris-EDTA (TE) buffer. For PGR isoforms, the immunocomplexes were eluted from the beads using flag peptide and for C/EBPβ and STAT3 ChIP, immunocomplexes were eluted in SDS elution buffer. Elutes were heated at 65°C for 6 hours to reverse the cross-linking. After digestions with RNase A and proteinase K, DNA fragments were purified using a QIAquick PCR purification kit (QIAGEN). For real-time PCR experiments, primers were designed to amplify potential chromatin binding sites. The resulting signals were normalized to input DNA, and relative fold enrichment over that of negative control ChIP was calculated.
Purified chromatin samples were sequenced using the Illumina platform. For mapping the Illumina sequencing reads to the human genome, we used the BOWTIE software package (19). Mapped reads were then analyzed with MACS version 1.4.2 (20). To identify peaks enriched in PGR-A or PGR-B, ChIP samples were compared with genomic input. The ChIP-sequencing (ChIP-seq) data can be accessed by Gene Expression Omnibus (GEO) via accession number GSE62475.
Microarray analysis
HESCs were transfected with siRNA targeting PGR mRNA (Dharmacon) or control scrambled siRNA (Dharmacon) as explained above. At 24 hours after siRNA treatment, cells were transduced with the adenovirus expressing flag-tagged PGR-A or PGR-B or both PGR-A and PGR-B for 24 hours. On the next day, cells were washed with PBS and treated with the differentiation cocktail for 6 hours. We performed 2 independent microarray experiment using 2 different clinical samples. Total RNA and protein were isolated by TRIzol reagent (Invitrogen). Protein samples were used to confirm expression of comparable levels of PGR-A and PGR-B (data not shown). Total RNA was purified using RNeasy columns (QIAGEN) following the manufacturer's instructions. RNA samples were processed at the Biotechnology Center of the University of Illinois at Urbana-Champaign. RNA integrity was verified using an Agilent 2100 bioanalyzer. Each RNA sample was processed to generate labeled cRNA following the established protocols for hybridization to the GeneChip Human Genome U133A 2.0 arrays. The resulting data files were first normalized and analyzed by Affymetrix GeneChip Expression Console software using the RMA (robust multiarray average) algorithm. Genes with a minimum 1.2-fold change in the same direction in duplicate microarrays were considered as differentially regulated genes. The array data can be accessed in GEO via accession number GSE48853.
RNA isolation and real-time PCR analysis
Total RNA was extracted from cultured HESCs using TRIzol (Invitrogen) according to the manufacturer's instructions. Total RNA was converted to cDNA using the Stratagene cDNA reverse transcription kit, following the manufacturer's instructions. cDNA was subjected to quantitative PCR analysis using gene-specific primers. Sequences of real-time PCR primers are available upon request. 36B4 was used as the reference gene. For a given sample, threshold cycle (Ct) and SD were calculated as an average of individual Ct values from 3 replicates. The normalized mean Ct (ΔCt) was calculated by subtracting the mean Ct of the reference gene 36B4 from the mean Ct of a target gene. The ΔΔCt was then calculated for each gene as a difference in ΔCt values between the control and the experimental sample. Fold change in gene expression was then computed as 2−ΔΔCt.
Statistical analysis
Validations of the ChIP-seq and microarray results were performed using at least 3 independent clinical samples. When one experimental sample was compared with the control sample, statistical significance between the control and experimental sample was determined using the Student t test. The statistical significance within a group of experimental samples was determined using within-subjects ANOVA followed by the Tukey honestly significant difference (HSD) comparison test. A P value of ≤.05 was considered to be significant.
Results
A strategy to analyze the roles of PGR-A and PGR-B during HESC differentiation
Undifferentiated HESCs expressed both PGR-A and PGR-B (Figure 1A, lane 1). Then 24 hours after the addition of a differentiation cocktail containing E, P, and 8-bromo-cAMP, a cAMP analog, the levels of both isoforms rose significantly (Figure 1A, lane 2). The increase in the level of PGR-B was more pronounced than that of PGR-A (Figure 1A), such that both isoforms are expressed at comparable levels in differentiating HESCs. To analyze the roles of each isoform separately, we used a strategy to generate HESCs expressing only one or the other of the isoforms. Both endogenous PGR isoforms were effectively depleted in HESCs by transfecting the cells with siRNA targeted to the 3′-UTR of the PGR mRNA (Figure 1B, lanes 1 and 2). Next, the cells were transduced with recombinant adenovirus harboring cDNA encoding flag-tagged PGR-A or PGR-B or a control adenovirus carrying the empty vector. The exogenous PGR expression constructs lack 3′-UTR sequences, allowing the expression of the corresponding transcripts and proteins even in the presence of the siRNA, thereby permitting the reconstitution of PGR-A or PGR-B. To ascertain that the protein levels of exogenously expressed isoforms are comparable to their endogenous levels, we used Western blot analysis with an antibody that recognizes both PGR-A and PGR-B. We optimized the amount of adenovirus to express each isoform based on endogenous protein levels (Figure 1B, lanes 3 and 4).
Figure 1.
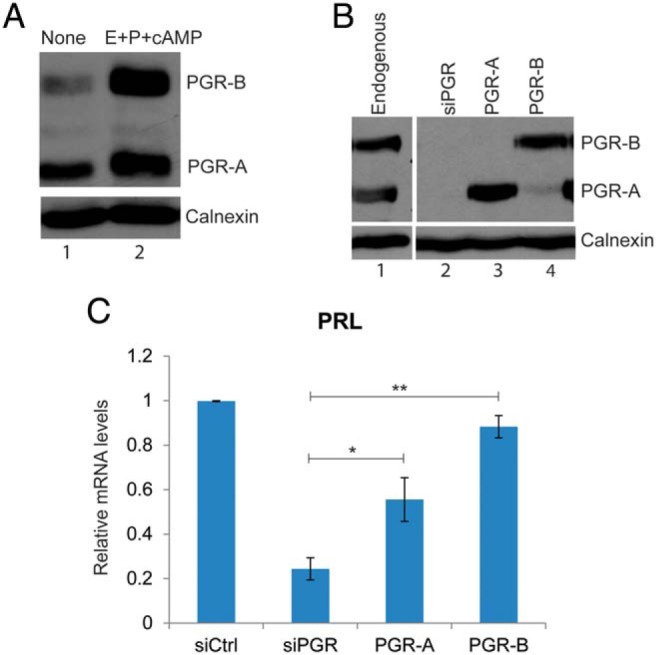
Analysis of individual roles of PGR-A and PGR-B in differentiating HESCs. A, HESCs were either untreated (lane 1) or treated with a differentiation cocktail composed of E, P, and cAMP (lane 2) as indicated. Cells were lysed at 24 hours after treatment, analyzed by SDS-PAGE, and subjected to Western blotting using a monoclonal antibody that recognizes both PGR-A and PGR-B and a calnexin antibody. Calnexin immunostaining served as a loading control. Data are representative of 5 independent experiments. B, HESCs were treated with siRNA targeting the 3′-UTR of PGR mRNA. On the next day, cells were transduced with empty vector (lane 2), adenovirus harboring flag-tagged PGR-A (lane 3), or PGR-B (lane 4) for 24 hours. Lane 1 represents the endogenous PGR levels in untransfected cells at 24 hours of differentiation. Whole-cell lysates were prepared from these cells. For each sample, whole-cell lysates was analyzed by Western blotting using a monoclonal antibody that recognizes both PGR-A and PGR-B. Immunostaining of calnexin was used as a loading control. C, HESCs were treated with 50 nM control siRNA or siRNA targeting the 3′-UTR of PGR mRNA. Next, cells were transduced with adenovirus harboring flag-tagged PGR-A or PGR-B or control virus for 24 hours. Then, siRNA and adenovirus were removed, and cells were treated with differentiation cocktail. Total RNA was isolated 60 hours later, and cDNAs were prepared. Real-time PCR was performed using gene-specific primers for 36B4 and PRL. PCR data were normalized to 36B4 mRNA levels. Relative fold changes were normalized with respect to the control siRNA treatment. Graphs represent means ± SEM, n = 2–3.
We next tested the biological activities of the flag-tagged PGR-A and flag-tagged PGR-B by assessing their ability to rescue differentiation of HESCs depleted of endogenous PGR. For this purpose, we monitored the expression of transcripts of decidual prolactin (PRL), a well-known marker of stromal decidualization. In response to siRNA-mediated knockdown of endogenous PGR-A and PGR-B, the expression of PRL was strongly down-regulated. Reconstitution of PGR-B expression fully restored the PRL mRNA expression, whereas the rescue of the expression of this mRNA by PGR-A was less effective (Figure 1C). Similar results were obtained with other markers of decidualization, such as IGFBP1 and WNT4 (Supplemental Figure 1). These results support our view that adenovirus-induced flag-PGR-A and flag-PGR-B are biologically active and able to restore stromal differentiation, although they may differ subtly in their inherent capacities to control this process.
Genome-wide binding sites of PGR-A and PGR-B in HESCs during decidualization
To understand the mechanisms by which the PGR isoforms control the decidualization process, we sought to identify the primary genomic binding sites of these isoforms in differentiating HESCs. To map these binding sites, we used ChIP followed by high-throughput sequencing (ChIP-seq). HESCs depleted of endogenous PGR isoforms and expressing adenovirus-induced flag-PGR-A or flag-PGR-B were treated with differentiation cocktail for 2 hours. ChIP assays were performed using an antibody specific for the flag epitope. Purified chromatin samples were sequenced using the Illumina platform as described in Materials and Methods.
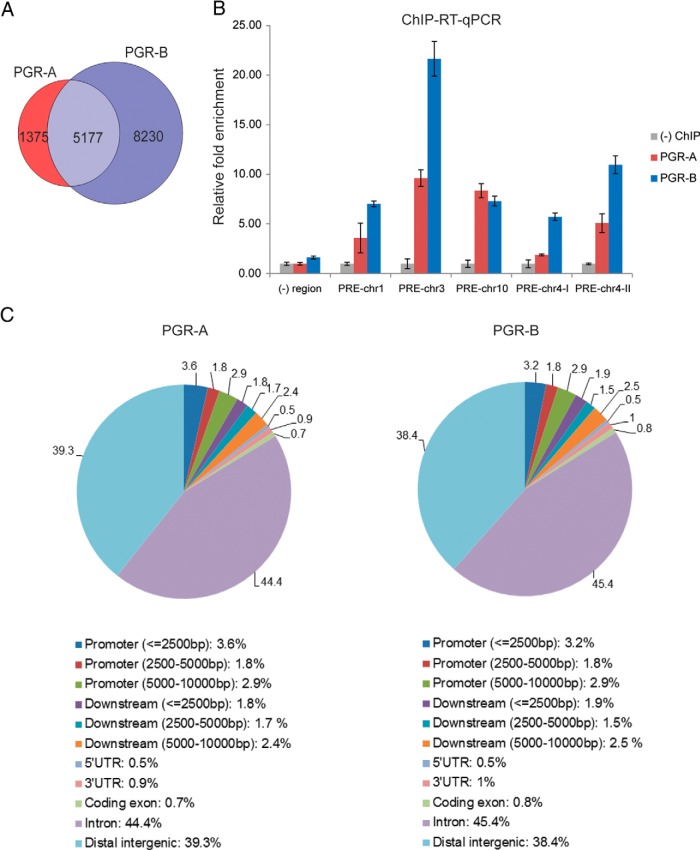
To identify genomic binding sites for each isoform, we used genomic input DNA comparison and for further analysis we considered sequences with a false discovery rate of ≤5% and fold enrichment of ≥4.5. We identified 6552 genome-wide binding sites for PGR-A and 13 407 sites for PGR-B (GSE62475). Most PGR-A binding sites were common to PGR-B, whereas PGR-B also occupied many unique binding sites (Figure 2A). A subset of target sequences common to both PGR-A and PGR-B ChIP-seq data were selected for further validation by ChIP followed by real-time RT-PCR (Figure 2B).
Figure 2.
Genome-wide PGR-A and PGR-B binding locations in differentiating HESCs. A, ChIP-seq–derived PGR-A and PGR-B binding sites. Venn diagrams show overlap of PGR-A vs PGR-B binding sites with a false discovery rate of <5% cutoff. B, A subset of PGR binding sites identified in the ChIP-seq experiment was selected for validation. ChIP was performed as described in Materials and Methods. Chromatin enrichment was quantified by real-time PCR using primers flanking these potential binding sites on different chromosomes as indicated in the figure. The negative (−) region was a PRE-deficient region. To calculate the enrichment of chromatin, the resulting signals were normalized to 1% input DNA. Data are presented as fold enrichment over that of the negative control ChIP and represent 3 independent experiments. C, Genomic region associations of genome-wide PGR-A and PGR-B binding sites. CEAS (a chromosome enrichment annotation tool of Cistrome) was used to analyze the distribution of binding sites with the following parameters: promoter/downstream lower interval, 2500 bp; middle interval, 5000 bp; and upper interval, 10 000 bp. In brief, regions between 0 and 2500 bp, 2500 and 5000 bp, and 5000 and 10 000 bp upstream of TSSs were selected as promoter intervals. Similarly, regions between 0 and 2500 bp, 2500 and 5000 bp, and 5000 and 10 000 bp downstream of a transcript were defined as different downstream intervals.
To further characterize the binding sites, we mapped the enriched regions identified by PGR-A and PGR-B ChIP-seq with custom defined genomic regions using the CEAS (cis-regulatory element annotation system) tool of Cistrome (21, 22). Most of the binding sites for the PGR isoforms were localized at great distances from the transcription start site (TSS) of the target genes, and many were present in introns or intergenic regions (Figure 2C). Similar observations were reported previously for E receptors (23, 24). A comparison of our data with ENCODE data assembled from several human cell lines (25) further revealed that about 80% of all binding regions overlapped with either enhancer- or promoter-associated histone marks, such as histone H3 lysine 4 methylation (H3K4Me1), histone H3 lysine 4 trimethylation (H3K4Me3), and nuclease hypersensitive sites, resembling distant enhancers (data not shown). It is conceivable that, in uterine tissue, both PGR-A and PGR-B regulate the expression of most their target genes through such long-range enhancer interactions rather than binding to proximal promoter regions of the target genes.
PGR-B regulates PGR-A expression
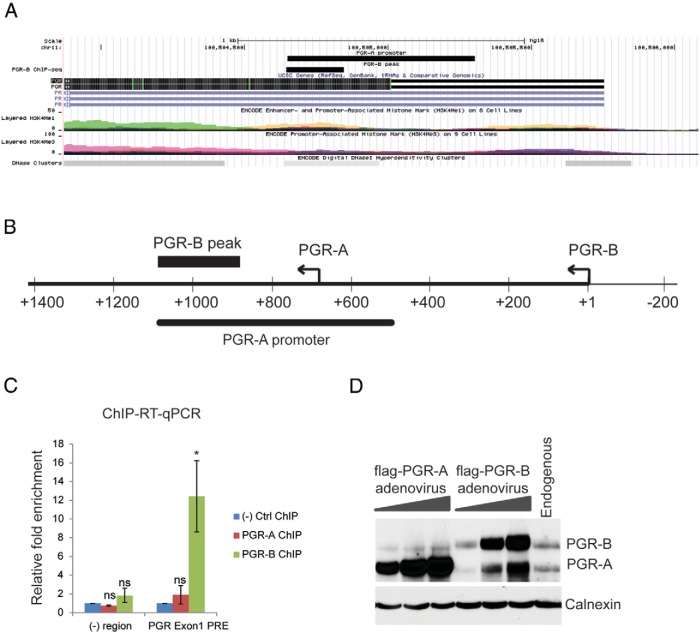
We identified 2 PGR binding sites in the PGR gene itself. Interestingly, these sites were bound by PGR-B but not PGR-A. The binding site in the first exon of the PGR gene aligned with the sequence previously reported to be a PGR-A promoter (Figure 3, A and B) (9), suggesting that PGR-B is a potential regulator of PGR-A expression. We validated PGR-B binding to this region by ChIP (Figure 3C). To assess whether PGR-B controls the expression of PGR-A, we examined the regulation of one isoform by the other at the protein level. With increasing expression of PGR-A, no noticeable increase in PGR-B protein was detected. In contrast, we observed a considerable increase in PGR-A protein levels in response to increasing levels of exogenously expressed PGR-B (Figure 3D), suggesting that PGR-B regulates PGR-A expression by directly binding to its promoter.
Figure 3.
PGR-B regulates the expression of PGR-A. A, UCSC Genome Browser illustration localized a PGR-B binding site in the first exon of the PGR gene. B, Schematic diagram showing the location of the previously identified PGR-A promoter with respect to the PGR-B binding site. C, ChIP-RT-qPCR validation of PGR-B binding in the first exon of the PGR gene. ChIP was performed as described in Materials and Methods. Chromatin enrichment was quantified by RT-PCR using primers flanking the PGR-B binding site in the PGR-A promoter region. The negative region was a PRE-deficient region. To calculate the enrichment of chromatin, the resulting signals were normalized to 1% input DNA. Data are presented as fold enrichment of PGR-A or PGR-B binding over that of the negative control ChIP. The graph represents the mean ± SEM from 4 independent experiments. Within-subjects ANOVA followed by a Tukey HSD test was used for statistical analysis: *, P ≤ .05; ns, not significant. D, HESCs were transduced with increasing multiplicity of infections of adenovirus harboring PGR-A or PGR-B for 24 hours; a control sample was not transduced. Next, cells were treated with differentiation cocktail for 6 hours. Whole-cell protein extracts were prepared from these cells and analyzed by Western blotting using a monoclonal antibody that recognizes both PGR-A and PGR-B. Immunostaining of calnexin was used as a loading control.
Analysis of transcription factor binding motifs associated with PGR-A and PGR-B binding sites
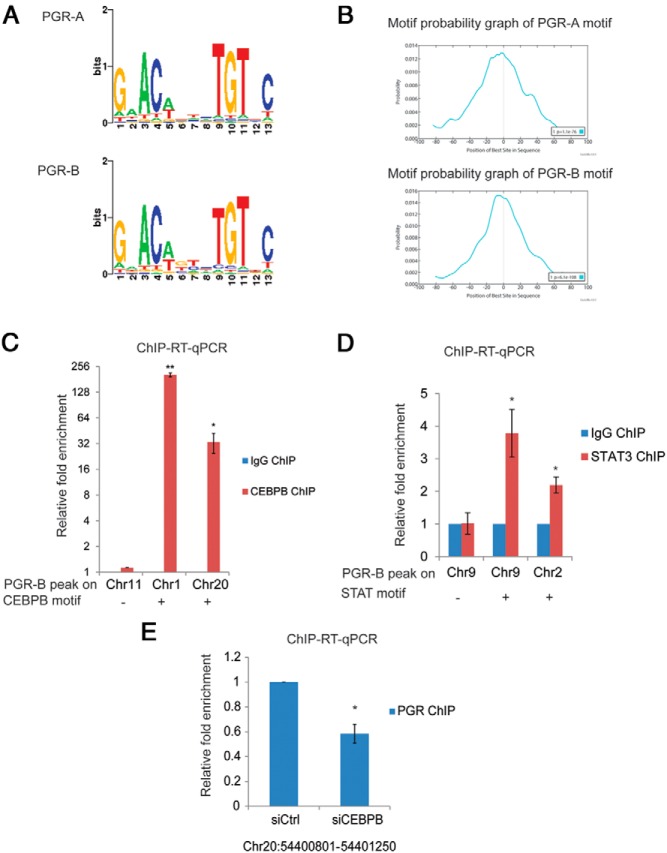
To identify enriched transcription factor binding motifs in the PGR binding sites identified by ChIP-seq, we applied the de novo motif detection tools MEME (26) and SeqPos (22) to analyze sequences within ±100 bp of the summits of PGR-A and PGR-B binding sites. Motif analysis of both PGR-A and PGR-B binding sites identified G.ACA… TGT.C as the major motif, which did not differ between the 2 isoforms (Figure 4A). This motif, which is identical to the P response element (PRE) described previously (17), was located centrally within the 200-bp sequence spanning the summit (Figure 4B). Descriptions of various other motifs observed in the 200-nucleotide sequence around the summits of PGR-A and PGR-B binding sites are listed in Supplemental Tables 1 and 2. We found several motifs common for both PGR isoforms. These common motifs represented the binding sites of various transcription factors, such as JUN, FOSL1/2, SOX10, the Ets domain family (ELK4), C/EBPα, C/EBPβ, FOXP1, HIF1A/B, REST, PAX1, and MTF1. The AP-1 (FOS/JUN) family motif was one of the most abundant motifs in both PGR-A and PGR-B target sequences. It was also identified as a prominent motif in the mouse PGR cistrome (17). Interestingly, JUN and FOS motifs were found more frequently in PGR-A than in PGR-B bound sites. Motifs unique to PGR-A included BACHs. Motifs unique to PGR-B included BCL6, STATs, TRP63, and RARA. It is conceivable that these nearby motifs serve as binding sites of cognate transcription factors that interact with PGR-A and PGR-B to induce different transcriptional outcomes.
Figure 4.
Motif analysis. A, PGR binding motif for PGR-A and PGR-B identified by de novo motif analysis using the SeqPos tool of Cistrome (22). B, Motif probability graphs showing the relative positions of PGR-A and PGR-B binding motifs within the summits of the top 1000 binding sites of PGR-A and PGR-B were generated using MEME-ChIP (21). The center of the sequences corresponds to the vertical line in the center of the graph. C, HESCs were subjected to treatment with differentiation cocktail for 48 hours. C/EBPβ ChIP was performed as described in Materials and Methods. Chromatin enrichment was quantified by real-time PCR using primers flanking the PGR-B binding sites, which contain a C/EBPβ motif within 100 bp (on chromosomes 1 and 20) and a C/EBPβ motif–negative PGR-B binding site (chromosome 11) as indicated in the figure. To calculate the enrichment of chromatin, the resulting signals were normalized to 1% input DNA. Data are presented as fold enrichment of C/EBPβ binding over that of the rabbit IgG ChIP. Graphs represent means ± SEM from 3 independent experiments. The Student t test was used for statistical analysis: **, P ≤ .01; *, P ≤ .05. D, HESCs were subjected to treatment with differentiation cocktail for 72 hours. STAT3 ChIP was performed as described in Materials and Methods. Chromatin enrichment was quantified by real-time PCR using primers flanking the PGR-B binding sites that contain a STAT motif within 100 bp (on chromosomes 9 and 2) and a STAT motif–negative PGR-B binding site (on chromosome 9) as indicated in the figure. To calculate the enrichment of chromatin, the resulting signals were normalized to 1% input DNA. Data are presented as fold enrichment of STAT3 binding over that of the IgG ChIP. Graphs represent the means ± SEM from 3 experiments. The Student t test was used for statistical analysis: *, P ≤ .05. E, HESCs were transfected with siRNA targeting C/EBPβ. At 48 hours after transfection, cells were treated with differentiation cocktail for 48 hours. PGR ChIP was performed. Chromatin enrichment was quantified by real-time PCR using primers flanking the PGR-B binding site on chr20: 54400801 to 54401250. To calculate the enrichment of chromatin, the resulting signals were normalized to 1% input DNA. Data are presented as fold enrichment of PGR enrichment in C/EBPβ siRNA-treated cells over that in control siRNA-treated cells. Graphs represent means ± SEM from 3 independent experiments. The Student t test was used for statistical analysis: *, P ≤ .05.
Previous studies in our laboratory have demonstrated that C/EBPβ and STAT3 play essential roles in HESC decidualization (27). To assess whether the PGR-B binding sites that contained a C/EBP or STAT motif are actually occupied by the corresponding transcription factors, we analyzed by ChIP the chromatin enrichment of C/EBPβ and STAT3 at selected sites. As shown in Figure 4C, sequences that had a C/EBPβ motif were significantly enriched, whereas there was no enrichment of a control sequence lacking a C/EBPβ motif. We obtained similar results with STAT3 ChIP (Figure 4D). We further examined whether depletion of C/EBPβ alters the binding of PGR-B to the Chr20 site shown in Figure 4C. This site, co-occupied by C/EBPβ during decidualization, is located upstream of the TSS of AURKA, a PGR-regulated gene identified by our microarray analysis. Depletion of C/EBPβ using siRNA led to a significant decrease in PGR enrichment at this site (Figure 4E). In the reciprocal experiment, when PGR was depleted by siRNA, C/EBPβ enrichment at that site was decreased (data not shown). These results established the interdependency of C/EBPβ and PGR binding to the PGR regulatory region of the AURKA gene.
Genes regulated by PGR isoforms in differentiating HESCs
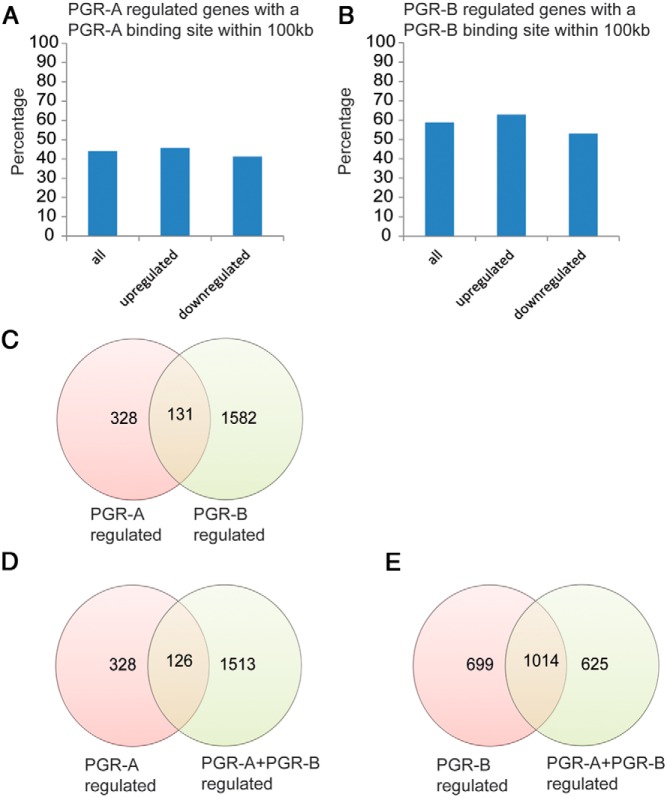
We next performed gene expression profiling to examine whether the occupancy of the genomic sites by PGR-A and PGR-B can be correlated with the regulation of expression of the corresponding genes. For the microarray analysis, we used HESCs expressing either flag-tagged PGR-A or PGR-B as described in Figure 1. Importantly, we also coexpressed PGR-A and PGR-B to see whether the presence of one isoform influences the transcriptional activity of the other. Our analysis yielded 451, 1724, and 1636 differentially regulated genes in HESCs expressing PGR-A alone, PGR-B alone, and PGR-A plus PGR-B, respectively, compared with HESCs in which endogenous PGR is depleted (GSE48853). We assessed the percentage of up-regulated and down-regulated genes that had at least 1 PGR-A or PGR-B-binding site within ±100 kb of their TSS. About 40% of the genes regulated by PGR-A had a PGR-A binding site, while 60% of genes regulated by PGR-B contained a PGR-B binding site (Figure 5, A and B). Several genes, such as BMP2, which were previously reported to be P-regulated (18), were bound by both PGR-A and PGR-B.
Figure 5.
Gene networks downstream of PGR-A, PGR-B, or PGR-A and PGR-B coexpressing HESCs. A, Graph showing the percentage of the PGR-A up-regulated and down-regulated genes containing at least 1 PGR-A binding site within ±100 kb from their TSS. B, Graph showing the percentage of the PGR-B up-regulated and down-regulated genes containing at least 1 PGR-B binding site within ±100 kb from their TSS. C, Venn diagram representing the number and overlap of the PGR-A regulated genes with the PGR-B regulated genes. D, Venn diagram representing the number and overlap of the PGR-A regulated genes with the PGR-A and PGR-B coregulated genes. E, Venn diagram representing the number and overlap of the PGR-B regulated genes with the PGR-A and PGR-B coregulated genes.
To have a better understanding of the key biological functions associated with the gene pathways downstream of PGR-A and PGR-B, we classified them into various biological categories, using DAVID (28, 29) and PANTHER (30) annotation tools. We noted that about 25% of genes regulated by PGR-A overlapped with those regulated by PGR-B (Figure 5C). These common pathways included genes that regulate blood vessel development and responses to hypoxia. PGR-A-regulated genes also encoded several protein kinases and transcription factors involved in the regulation of apoptosis (including TNFSF12, BCL6, CFLAR, MCL1, and BIRC5), and factors involved in blood vessel development (including CYR61 and VEGFA) (Supplemental Table 3).
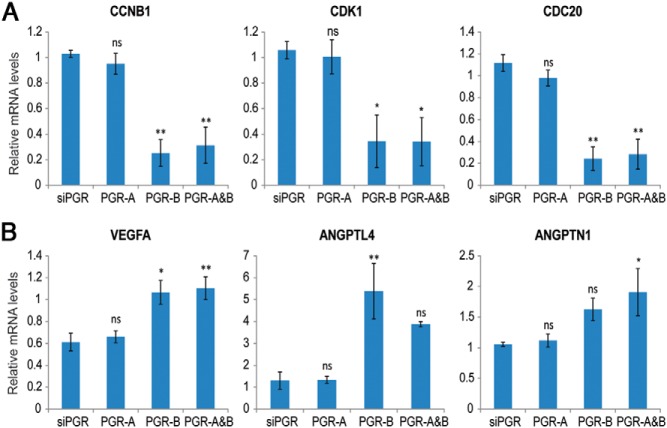
Our analysis highlighted the predominant roles of PGR-B in the regulation of the cell cycle, angiogenesis, lysosomal activity, and secretion of chemokines and cytokines (Supplemental Table 4). Genes up-regulated by PGR-B included several signaling molecules involved in angiogenesis, apoptosis, and cell adhesion and numerous solute carrier family members. Interestingly, many genes regulating the mitotic phase of the cell cycle, including CCNB1, CCNB2, CDC20, CDC25A, CDK1, and BUB1, were suppressed by PGR-B, suggesting that the PGR-B isoform may be playing a critical role in suppression of cell proliferation during the differentiation of HESCs. We validated the down-regulation of some of these genes by PGR-B using real-time PCR (Figure 6A). To further assess the effects of the PGR isoforms on HESC proliferation, endogenous PGR expression was silenced, and PGR-A and PGR-B were individually reexpressed. HESCs were incubated in the differentiation medium for 24 hours, and the percentages of proliferating cells were quantified using Ki-67 immunostaining as a marker of cell proliferation. Compared with the PGR-depleted control, there was a significant decrease in the percentage of Ki-67–positive HESCs upon PGR-B reexpression, whereas there was no significant change in cell proliferation when PGR-A was reexpressed (Supplemental Figure 2, A and B). These results are consistent with our view that PGR-B, but not PGR-A, suppresses proliferation of the HESCs as they enter the differentiation program.
Figure 6.
Validation of PGR-B regulation of cell cycle– and angiogenesis-related pathways. A, HESCs were treated with 50 nM control siRNA or siRNA targeting the 3′-UTR of PGR mRNA. Next, cells were transduced with adenovirus harboring flag-tagged PGR-A or PGR-B or control virus for 24 hours. Then, siRNA and adenovirus were removed, and cells were treated with differentiation cocktail for 6 hours. Real-time PCR analysis was performed using gene-specific primers for CCNB1, CDK1, CDC20, and 36B4. PCR data were normalized to 36B4 mRNA levels. Relative fold changes were normalized with respect to control siRNA treatment. Graphs represent means ± SEM from 3 independent experiments. Within-subjects ANOVA followed by the Tukey HSD test was used for statistical analysis: *, P ≤ .05; **, P ≤ .01. B, The same samples were used for real time PCR with gene-specific primers for VEGFA, ANGPTNL4, and ANGPTN1. Graphs represent means ± SEM from 3 independent experiments. Within-subjects ANOVA followed by the Tukey HSD test was used for statistical analysis: *, P ≤ .05; **, P ≤ .01.
Angiogenesis appears to be another biological process regulated by PGR-B. We found several genes, including VEGFA, ANGPT2, ANGPTL4, EPAS1, EREG, TGFB2, EGFR, FGF2, and TNFAIP2 that are up regulated by PGR-B. Many of these genes, such as VEGFA, TGFB2, ANGPT2, ANGPTL4, and EGFR, harbor PGR-B binding sites, indicating that they may be direct targets of PGR-B. We validated the induction of some of these genes by PGR-B using real-time PCR (Figure 6B).
When both isoforms were coexpressed, differentially regulated genes were more commonly associated with PGR-B (Supplemental Table 5 and Figure 5, D and E). However, PGR-A clearly modulated PGR-B activity as the magnitudes of expression of several genes were altered when both isoforms were coexpressed. These genes included numerous glycoproteins involved in cell signaling and immunoregulation, ion channels, peroxisome-related genes, and certain peptidases involved in lysosomal processes.
IRS2, a PGR-regulated metabolic factor, controls decidualization
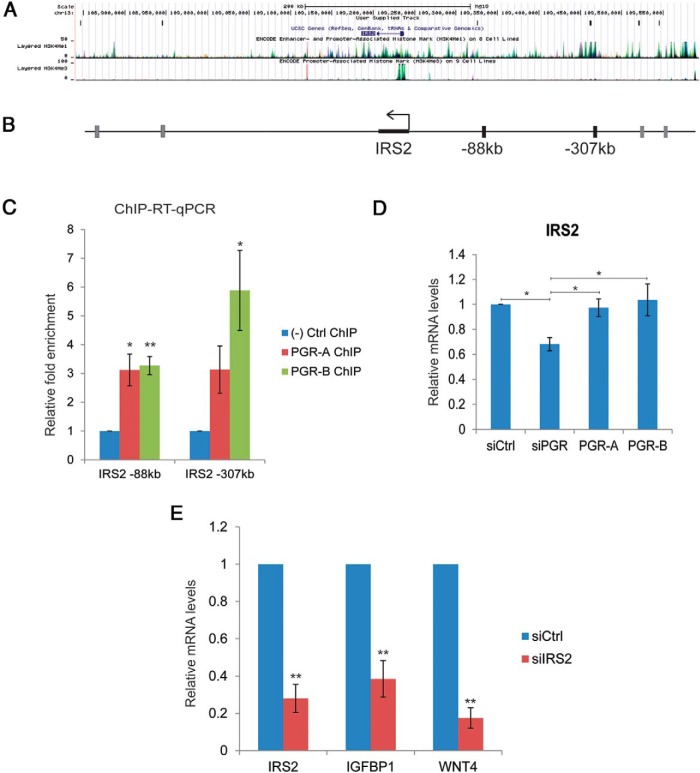
We identified several components of the insulin signaling pathway, including insulin receptor substrate 2 (IRS2), SHC-transforming proteins 1 and 3 (SHC1 and SHC3), GRB10, and SOS1 downstream of PGR-B. Among these potential target genes, IRS2 contained binding sites for both PGR-A and PGR-B at 88 kb upstream of its TSS, in addition to several other binding sites further (−307 kb) upstream or downstream of its open reading frame (Figure 7, A and B). We monitored IRS2 expression in primary cultures of differentiating HESCs. Addition of the differentiation cocktail led to an increase in IRS2 mRNA levels within 24 hours (data not shown). To establish IRS2 as a direct target of PGR, we validated the binding of PGR-A and PGR-B to the −88 kb upstream region and an additional region further upstream. Both PGR-A and PGR-B were recruited to these regions (Figure 7C). Next, we confirmed transcriptional regulation of IRS2 by PGR-A and PGR-B after 24 hours of HESC differentiation. When endogenous PGR was silenced by siRNA, IRS2 expression was down-regulated. Reconstitution of either PGR-A or PGR-B expression restored IRS2 mRNA expression (Figure 7D). To test the possibility of a potential role of IRS2 in HESC differentiation, we examined the effect of its down-regulation on the differentiation process. As shown in Figure 7E, siRNA-mediated suppression of IRS2 transcripts led to a significant decrease in the expression of mRNAs corresponding to IGFBP1 and WNT4, well-known biochemical markers of decidualization in the human, indicating a lack of stromal differentiation in the absence of IRS2. Therefore, we concluded that IRS2, a potential direct target of PGR, is critical for the differentiation of HESCs.
Figure 7.
IRS2 is regulated by PGR-A and PGR-B and is critical for stromal differentiation. A, UCSC Genome Browser illustration localized PGR-A and PGR-B binding sites around the IRS2 gene. B, Schematic diagram showing the locations of binding sites common to PGR-A and PGR-B around the IRS2 gene. Sites used for validation by ChIP in panel C are marked as black blocks. C, HESCs were transduced with or without recombinant adenovirus harboring flag-tagged PGR-A or PGR-B for 24 hours. Then, samples were subjected to treatment with differentiation cocktail for 2 hours. ChIP was performed as described in Materials and Methods. Chromatin enrichment was quantified by real-time PCR using primers flanking potential PGR-A and PGR-B binding sites in the 5′-flanking region of IRS2 at −88 and −307 kb. Graphs represent means ± SEM; n = 4. Within-subjects ANOVA followed by the Tukey HSD test was used for statistical analysis: *, P ≤ .05; **, P ≤ .01. D, HESCs were treated with 50 nM control siRNA or siRNA targeting the 3′-UTR of PGR mRNA. Next, cells were transduced with adenovirus harboring flag-tagged PGR-A or PGR-B or control virus for 24 hours. Then, siRNA and adenovirus were removed, and cells were treated with differentiation cocktail for 24 hours. Prepared cDNAs were subjected to real-time PCR using primers for IRS2. PCR data were normalized to 36B4 mRNA levels. Relative fold changes were normalized with respect to those of the control nontargeting siRNA treatment. Graphs represent means ± SEM from 3 independent experiments. Within-subjects ANOVA followed by the Tukey HSD test was used for statistical analysis: *, P ≤ .05. E, HESCs were transfected with siRNA targeting IRS2 or scrambled siRNA (control). At 48 hours after transfection, HESCs were treated with differentiation cocktail to initiate in vitro differentiation. Cells were harvested at 72 hours after differentiation cocktail treatment. Total RNA was isolated, and cDNAs were prepared. cDNAs were subjected to real-time PCR using gene-specific primers for IRS2, IGFBP1, and WNT4. PCR data were normalized to 36B4 mRNA levels. Relative fold changes were normalized with respect to those for the control nontargeting siRNA treatment. Graphs represent mean ± SEM from 3 independent experiments. The Student t test was used for statistical analysis: **, P ≤ .01.
Discussion
The present study is unique in terms of identifying the genomic binding sites of the human PGR-A and PGR-B isoforms, which are expressed at comparable levels in endometrial stromal cells during decidualization. Using an unbiased genome-wide approach and parallel gene expression analyses, we identified the transcriptomes of PGR-A and PGR-B at an early time point during in vitro decidualization. We found that PGR-B had a larger cistrome and transcriptome than PGR-A (Figures 2A and 5C). Although the reason for this is unclear, it is possible that cooperative interactions between PGR-B homodimers occupying multiple adjacent binding sites may stabilize its binding to DNA, facilitating transcription of target genes. Furthermore, interactions with other transcription factors that bind to nearby motifs may also stabilize PGR-B binding to the genome more than that of PGR-A.
When we analyzed the motifs within a ±100-bp window from the summits of target sequences of PGR-A and PGR-B, we identified motifs that were unique to PGR-A (BACHs) and PGR-B (BCL6, STATs, TRP63, and RARA) (Supplemental Tables 1 and 2). In addition, PGR-A and PGR-B had common nearby motifs, including JUN, C/EBPs, SOXs, REST, and ESR1, some of which had been observed previously in the mouse PGR cistrome (17). We speculate that the transcription factors that bind to these motifs facilitate binding of PGR-A or PGR-B to the PRE and vice versa. Consistent with this view, it has been proposed that PGR can act as a pioneering factor that modulates chromatin availability for other transcription factors (31). Alternatively, DNA-bound PGR-A or PGR-B may interact with these transcription factors to cooperatively regulate target gene transcription. Whatever the mechanism is, it adds another level of complexity to the pattern of temporal regulation of PGR target genes, because activation and nuclear localization of some of these transcription factors occur hours to days after the initial differentiation stimulus. Previous studies suggested that PGR undergoes functional interactions with FOXO1 (forkhead box protein O1), STAT3, and C/EBPβ to regulate the expression of target genes in differentiating stromal cells (32–34). The present study demonstrates a significant enrichment of C/EBPβ and STAT3 adjacent to several PGR-B binding sites (Figure 4, C and D). Interestingly, when we grouped genes downstream of PGR-B according to their direction of regulation, the REST (RE1-silencing transcription factor) motif was enriched more in repressed genes, whereas the C/EBPβ, HIF1, ELKx, and ETVx motifs were found more in up-regulated genes, suggesting that the presence of specific transcription factors near a PGR binding site may also dictate the direction of regulation of the target gene. Collectively, these results support the concept that coordinated gene regulation involving PGR and these transcription factors is critical to execute the decidualization program.
Our gene expression analysis, using HESCs expressing PGR-A, PGR-B, or a combination of PGR-A plus PGR-B, provided a comprehensive view of the pathways regulated by P signaling during decidualization. Almost half of the genes found downstream of PGR-A and PGR-B at 6 hours of differentiation contained at least 1 PGR-A or PGR-B binding site within 100 kb of its TSS (Figure 5, A and B), suggesting that some of these genes may be direct targets of P signaling. Several of these genes, such as BMP2, STAT3, WNT4, HOXA10, and HAND2 are critical for HESC differentiation (6, 35). Our findings indicated that PGR-B was a more potent transcription factor than PGR-A in human stromal cell differentiation, in terms of the extent to which it interacts with genomic binding sites and regulates downstream genes. We also found that when PGR-A and PGR-B were coexpressed, PGR-B was the dominant isoform, although PGR-A was able to influence the transcriptional activity of PGR-B in some cases (Figure 5, D and E). Many of the potential direct targets of both PGR-A and PGR-B were found to be involved in regulation of angiogenesis, the immune response, the cell cycle, apoptosis, and cell-matrix interactions (Supplemental Tables 3 and 4), biological events considered critical for decidualization and maintenance of pregnancy.
We observed that many genes involved in the mitotic phase of the cell cycle, such as CCNB1, CDK1, and CDC20, were repressed in differentiating HESCs expressing PGR-B (Figure 6A). It is generally observed that the cell cycle exit is needed for differentiation (36). We previously reported that HESC proliferation ceases with the addition of the differentiation cocktail to the primary cultures (27). Based on these findings, we propose that the induction of PGR-B contributes to the transition of HESCs from a proliferating state to a differentiating state by suppressing progression to mitosis. The biological relevance of this is found in the observed increase in PGR expression in HESCs at the periovulatory period as they begin to differentiate and transition from a proliferative to a secretory phase of the cycle (37).
Angiogenesis is essential for normal endometrial development and placentation. New blood vessels must be formed in the decidua during pregnancy to support the developing embryo. Many diseases of pregnancy, such as preeclampsia and fetal growth restriction, have been associated with abnormal angiogenesis (38). Our gene expression–profiling analysis showed that, although both PGR-A and PGR-B regulate several genes involved in angiogenesis, PGR-B was the predominant regulator of this process. Angiogenesis-related genes found downstream of PGR-B included vascular endothelial growth factor (VEGF) A, angiopoietin 2 (ANGPT2), angiopoietin-like 4 (ANGPTL4), and fibroblast growth factor 2 (FGF2). Among them, ANGPT2, ANGPTL4, and FGF2 all contained PGR-B binding sites upstream of their TSS, suggesting that they are direct targets of PGR-B. VEGFA, a key angiogenic factor in human endometrium, was previously shown to be regulated by P in the uterus (39). In our study, we found VEGFA transcripts to be induced by PGR-B at 6 hours after the initiation of differentiation (Figure 6B). Consistent with this observation, we identified several PGR-B binding sites within a ±200-kb window of the VEGF TSS. We speculate that these regions might be bound by PGR-B, which then regulates VEGFA expression through a looping mechanism. Further research is necessary to understand regulation of VEGFA by PGR-B.
One of the most prominent functional categories of genes up-regulated by PGR-B were the lysosomal proteins, including acid phosphatases, cathepsins, arylsulfatase, and lysosome-associated membrane proteins. Changes in lysosomal enzyme activities, including increases in acid phosphatase and arylsulfatase B activity during the initial stages of decidualization, were reported previously (40). P tends to stabilize lysosomal membranes, maintaining intralysosomal contents during the secretory phase. However, P withdrawal during menstruation results in the release of these enzymes (41). Future studies will help us understand the roles of these lysosomal enzymes and membrane proteins during decidualization and pregnancy.
According to our gene expression analysis, apoptosis is one of the most prominent biological processes regulated by PGR-A. During the menstrual cycle, there is little to no apoptosis in the proliferative phase, but by the late secretory phase, marked stromal apoptosis occurs, and it progresses through most of the functional layer of endometrium (42). Compared with undifferentiated HESCs, differentiating cells were reported to be resistant to oxidative stress–induced apoptosis (43), suggesting that resistance to apoptosis might be an acquired feature of decidualized HESCs and part of the differentiation process itself. Interestingly, we found both negative (BIRC5 and CFLAR) and positive (SMAD3 and TGFB2) regulators of apoptosis downstream of PGR-A, suggesting that PGR-A might be involved in regulating the critical balance of these factors to control apoptosis appropriately in response to various internal and external signals.
We also found several solute carrier proteins, including monoamine transporters, glucose transporters, and ion exchangers regulated in PGR-B and PGR-A plus PGR-B coexpressing cells. These molecules may be critical for monoamine metabolism, which is important for regulation of blood flow and capillary permeability (44) and nutrient/ion transport. Further research is necessary to determine the biological roles of these transporter proteins in decidualization and reproductive physiology.
Using ChIP-seq and gene expression analyses, we identified IRS2 as a direct target of both PGR-A and PGR-B (Figure 7). These findings are consistent with previous reports indicating a role of P in regulating IRS2 expression, which was further linked to IGF signaling (45–47). IRS2, together with IRS1 and SHC1, is a key mediator for the downstream signaling pathways of both insulin and IGF-1. The expression and phosphorylation of IRS2 increased in endometrial stromal cells during in vitro decidualization, suggesting that it is involved in the signaling mechanisms controlling this process (48). In the present study, we showed that silencing of IRS2 expression impairs HESC differentiation (Figure 7E). Aberrant IRS2 signaling has been implicated in reproductive disorders associated with insulin metabolism (49). Critical endometrial functions, such as proliferation, differentiation, and apoptosis are energy-demanding processes. Growing evidence suggests that the regulation of energy metabolism is essential for the establishment of successful pregnancy (50). We propose that IRS2 is a critical link between PGR signaling and energy metabolism during stromal differentiation. IRS2 may be involved in localization of glucose transporters to the cell membrane to enable the endometrial stromal cells to take up enough glucose to meet the increased energy needs during their proliferation and differentiation. IRS2 may also be involved in the regulation of genes controlling glucose metabolism (51). Future studies, addressing the level and localization of glucose transporters in stromal cells lacking IRS2 would be useful to test this possibility. It would also be useful to investigate whether IRS2 regulates glycolytic enzymes, such as hexokinase, phosphofructokinase, and pyruvate kinase during decidualization.
It is of interest to compare our ChIP-seq and gene expression results with those of the DeMayo group (17) who recently used an ovariectomized mouse model to identify PGR-regulated genes induced by acute P treatment. Although their experimental design did not allow dissection of the individual functions of the PGR isoforms, several P-regulated genes, such as BMP2, HAND2, WNT7A, LIFR, and VEGFA, were common in both studies. As shown in the mouse cistrome, several circadian rhythm genes, such as CLOCK, NPAS2, CRY1, PER1, NR1D2, and RORC, also contained binding sites for human PGR-B and PGR-A. In addition, several common neighboring motifs representing the binding sites of JUN, FOSL, C/EBPβ, STAT3, the Ets1 family, ESR1, ESR2, and a number of SOX family proteins were identified by both studies. However, there were also interesting differences between the 2 data sets. For example, in contrast to our finding that several cell cycle regulatory genes, such as CDK1 and CCNB1, were suppressed by PGR-B, these genes were up-regulated in ovariectomized mouse uterus upon treatment with P. Furthermore, the IRS2 gene, a direct target of PGR in both mouse and human uteri, was suppressed in the mouse uterus in response to P. These differences most likely arose from the fact that the mouse stromal cells do not undergo decidualization under the hormone treatment conditions used by the DeMayo group, whereas our study addressed the roles of human PGR isoforms during decidualization. This comparison indicated that, although certain aspects of gene regulation by the PGRs are similar between the undifferentiated and differentiated states of the uterine stromal cells and are conserved between mice and humans, important differences in regulatory outcomes do exist based on the biological context.
In summary, our study provided a molecular understanding of several of the known roles of P, such as cell cycle regulation, angiogenesis, and apoptosis, during human endometrial decidualization. We were also able to identify unique target genes, such as IRS2, downstream of P signaling that are likely to play a critical role in pregnancy. This new knowledge will help us to better understand normal PGR biology and its dysfunction in various clinical endometrial disorders.
Acknowledgments
This work was supported by the Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health, through cooperative agreement U54 HD055787 as part of the Specialized Cooperative Centers Program in Reproduction and Infertility Research (R.N.T., I.C.B., and M.K.B.).
Disclosure Summary: The authors have nothing to disclose.
Footnotes
- 8-Br-cAMP
- 8-bromo-adenosine-3′,5′-cyclic monophosphate
- C/EBPβ
- CCAAT/enhancer-binding protein β
- ChIP
- chromatin immunoprecipitation
- ChIP-seq
- ChIP-sequencing
- E
- estrogen
- FGF2
- fibroblast growth factor 2
- GEO
- Gene Expression Omnibus
- HESC
- human primary endometrial stromal cell
- HSD
- honestly significant difference
- IRS2
- insulin receptor substrate 2
- P
- progesterone
- PGR
- P receptor
- PRE
- P response element
- PRL
- prolactin
- siRNA
- small interfering RNA
- STAT3
- signal transducer and activator of transcription 3
- TSS
- transcription start site
- UTR
- untranslated region.
References
- 1. Lydon JP, DeMayo FJ, Funk CR, et al. Mice lacking progesterone receptor exhibit pleiotropic reproductive abnormalities. Genes Dev. 1995;9:2266–2278. [DOI] [PubMed] [Google Scholar]
- 2. Franco HL, Jeong JW, Tsai SY, Lydon JP, DeMayo FJ. In vivo analysis of progesterone receptor action in the uterus during embryo implantation. Semin Cell Dev Biol. 2008;19:178–186. [DOI] [PubMed] [Google Scholar]
- 3. Brosens JJ, Gellersen B. Death or survival—progesterone-dependent cell fate decisions in the human endometrial stroma. J Mol Endocrinol. 2006;36:389–398. [DOI] [PubMed] [Google Scholar]
- 4. Conneely OM, Mulac-Jericevic B, Lydon JP, De Mayo FJ. Reproductive functions of the progesterone receptor isoforms: lessons from knock-out mice. Mol Cell Endocrinol. 2001;179:97–103. [DOI] [PubMed] [Google Scholar]
- 5. Popovici RM, Kao LC, Giudice LC. Discovery of new inducible genes in in vitro decidualized human endometrial stromal cells using microarray technology. Endocrinology. 2000;141:3510–3513. [DOI] [PubMed] [Google Scholar]
- 6. Ramathal CY, Bagchi IC, Taylor RN, Bagchi MK. Endometrial decidualization: of mice and men. Semin Reprod Med. 2010;28:17–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cloke B, Huhtinen K, Fusi L, et al. The androgen and progesterone receptors regulate distinct gene networks and cellular functions in decidualizing endometrium. Endocrinology. 2008;149:4462–4474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brosens JJ, Hayashi N, White JO. Progesterone receptor regulates decidual prolactin expression in differentiating human endometrial stromal cells. Endocrinology. 1999;140:4809–4820. [DOI] [PubMed] [Google Scholar]
- 9. Kastner P, Krust A, Turcotte B, et al. Two distinct estrogen-regulated promoters generate transcripts encoding the two functionally different human progesterone receptor forms A and B. EMBO J. 1990;9:1603–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Conneely OM, Mulac-Jericevic B, Lydon JP. Progesterone-dependent regulation of female reproductive activity by two distinct progesterone receptor isoforms. Steroids. 2003;68:771–778. [DOI] [PubMed] [Google Scholar]
- 11. Vegeto E, Shahbaz MM, Wen DX, Goldman ME, O'Malley BW, McDonnell DP. Human progesterone receptor A form is a cell- and promoter-specific repressor of human progesterone receptor B function. Mol Endocrinol. 1993;7:1244–1255. [DOI] [PubMed] [Google Scholar]
- 12. Wen DX, Xu YF, Mais DE, Goldman ME, McDonnell DP. The A and B isoforms of the human progesterone receptor operate through distinct signaling pathways within target cells. Mol Cell Biol. 1994;14:8356–8364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Khan JA, Bellance C, Guiochon-Mantel A, Lombes M, Loosfelt H. Differential regulation of breast cancer-associated genes by progesterone receptor isoforms PRA and PRB in a new bi-inducible breast cancer cell line. PLoS One. 2012;7:e45993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Richer JK, Jacobsen BM, Manning NG, Abel MG, Wolf DM, Horwitz KB. Differential gene regulation by the two progesterone receptor isoforms in human breast cancer cells. J Biol Chem. 2002;277:5209–5218. [DOI] [PubMed] [Google Scholar]
- 15. Mote PA, Balleine RL, McGowan EM, Clarke CL. Colocalization of progesterone receptors A and B by dual immunofluorescent histochemistry in human endometrium during the menstrual cycle. J Clin Endocrinol Metab. 1999;84:2963–2971. [DOI] [PubMed] [Google Scholar]
- 16. Mangal RK, Wiehle RD, Poindexter AN, 3rd, Weigel NL. Differential expression of uterine progesterone receptor forms A and B during the menstrual cycle. J Steroid Biochem Mol Biol. 1997;63:195–202. [DOI] [PubMed] [Google Scholar]
- 17. Rubel CA, Lanz RB, Kommagani R, Franco HL, Lydon JP, DeMayo FJ. Research resource: Genome-wide profiling of progesterone receptor binding in the mouse uterus. Mol Endocrinol. 2012;26:1428–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li Q, Kannan A, Wang W, et al. Bone morphogenetic protein 2 functions via a conserved signaling pathway involving Wnt4 to regulate uterine decidualization in the mouse and the human. J Biol Chem. 2007;282:31725–31732. [DOI] [PubMed] [Google Scholar]
- 19. Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang Y, Liu T, Meyer CA, et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008;9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shin H, Liu T, Manrai AK, Liu XS. CEAS: Cis-regulatory element annotation system. Bioinformatics. 2009;25:2605–2606. [DOI] [PubMed] [Google Scholar]
- 22. Liu T, Ortiz JA, Taing L, et al. Cistrome: an integrative platform for transcriptional regulation studies. Genome Biol. 2011;12:R83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Welboren WJ, van Driel MA, Janssen-Megens EM, et al. ChIP-Seq of ERα and RNA polymerase II defines genes differentially responding to ligands. EMBO J. 2009;28:1418–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Carroll JS, Meyer CA, Song J, et al. Genome-wide analysis of estrogen receptor binding sites. Nat Genet. 2006;38:1289–1297. [DOI] [PubMed] [Google Scholar]
- 25. Rosenbloom KR, Sloan CA, Malladi VS, et al. ENCODE data in the UCSC Genome Browser: year 5 update. Nucleic Acids Res. 2013;41:D56–D63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bailey TL, Boden M, Buske FA, et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang W, Taylor RN, Bagchi IC, Bagchi MK. Regulation of human endometrial stromal proliferation and differentiation by C/EBPβ involves cyclin E-cdk2 and STAT3. Mol Endocrinol. 2012;26:2016–2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. [DOI] [PubMed] [Google Scholar]
- 30. Mi H, Muruganujan A, Thomas PD. PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 2013;41:D377–D386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ballaré C, Castellano G, Gaveglia L, et al. Nucleosome-driven transcription factor binding and gene regulation. Mol Cell. 2013;49:67–79. [DOI] [PubMed] [Google Scholar]
- 32. Takano M, Lu Z, Goto T, et al. Transcriptional cross talk between the forkhead transcription factor forkhead box O1A and the progesterone receptor coordinates cell cycle regulation and differentiation in human endometrial stromal cells. Mol Endocrinol. 2007;21:2334–2349. [DOI] [PubMed] [Google Scholar]
- 33. Lee JH, Kim TH, Oh SJ, et al. Signal transducer and activator of transcription-3 (Stat3) plays a critical role in implantation via progesterone receptor in uterus. FASEB J. 2013;27:2553–2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Christian M, Pohnke Y, Kempf R, Gellersen B, Brosens JJ. Functional association of PR and CCAAT/enhancer-binding protein beta isoforms: promoter-dependent cooperation between PR-B and liver-enriched inhibitory protein, or liver-enriched activatory protein and PR-A in human endometrial stromal cells. Mol Endocrinol. 2002;16:141–154. [DOI] [PubMed] [Google Scholar]
- 35. Li Q, Kannan A, Das A, et al. WNT4 acts downstream of BMP2 and functions via β-catenin signaling pathway to regulate human endometrial stromal cell differentiation. Endocrinology. 2013;154:446–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Miller JP, Yeh N, Vidal A, Koff A. Interweaving the cell cycle machinery with cell differentiation. Cell Cycle. 2007;6:2932–2938. [DOI] [PubMed] [Google Scholar]
- 37. Fung HY, Wong YL, Wong FW, Rogers MS. Study of oestrogen and progesterone receptors in normal human endometrium during the menstrual cycle by immunocytochemical analysis. Gynecol Obstet Invest. 1994;38:186–190. [DOI] [PubMed] [Google Scholar]
- 38. Reynolds LP, Killilea SD, Redmer DA. Angiogenesis in the female reproductive system. FASEB J. 1992;6:886–892. [PubMed] [Google Scholar]
- 39. Ancelin M, Buteau-Lozano H, Meduri G, et al. A dynamic shift of VEGF isoforms with a transient and selective progesterone-induced expression of VEGF189 regulates angiogenesis and vascular permeability in human uterus. Proc Natl Acad Sci USA. 2002;99:6023–6028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Moulton BC, Koenig BB, Borkan SC. Uterine lysosomal enzyme activity during ovum implantation and early decidualization. Biol Reprod. 1978;19:167–170. [DOI] [PubMed] [Google Scholar]
- 41. Fritz MA, Speroff L. Clinical Gynecologic Endocrinology and Infertility. 8th ed Philadelphia, PA: Wolters Kluwer Health/Lippincott Williams & Wilkins; 2010. [Google Scholar]
- 42. Shikone T, Kokawa K, Yamoto M, Nakano R. Apoptosis of human ovary and uterine endometrium during the menstrual cycle. Horm Res. 1997;48(Suppl 3):27–34. [DOI] [PubMed] [Google Scholar]
- 43. Kajihara T, Uchino S, Suzuki M, Itakura A, Brosens JJ, Ishihara O. Human chorionic gonadotropin confers resistance to oxidative stress-induced apoptosis in decidualizing human endometrial stromal cells. Fertil Steril. 2011;95:1302–1307. [DOI] [PubMed] [Google Scholar]
- 44. Hansson SR, Bottalico B, Noskova V, Casslén B. Monoamine transporters in human endometrium and decidua. Hum Reprod Update. 2009;15:249–260. [DOI] [PubMed] [Google Scholar]
- 45. Vassen L, Wegrzyn W, Klein-Hitpass L. Human insulin receptor substrate-2 (IRS-2) is a primary progesterone response gene. Mol Endocrinol. 1999;13:485–494. [DOI] [PubMed] [Google Scholar]
- 46. Cui X, Lazard Z, Zhang P, Hopp TA, Lee AV. Progesterone crosstalks with insulin-like growth factor signaling in breast cancer cells via induction of insulin receptor substrate-2. Oncogene. 2003;22:6937–6941. [DOI] [PubMed] [Google Scholar]
- 47. Yudt MR, Berrodin TJ, Jelinsky SA, et al. Selective and opposing actions of progesterone receptor isoforms in human endometrial stromal cells. Mol Cell Endocrinol. 2006;247:116–126. [DOI] [PubMed] [Google Scholar]
- 48. Ganeff C, Chatel G, Munaut C, Frankenne F, Foidart JM, Winkler R. The IGF system in in-vitro human decidualization. Mol Hum Reprod. 2009;15:27–38. [DOI] [PubMed] [Google Scholar]
- 49. Burks DJ, Font de Mora J, Schubert M, et al. IRS-2 pathways integrate female reproduction and energy homeostasis. Nature. 2000;407:377–382. [DOI] [PubMed] [Google Scholar]
- 50. Frolova AI, Moley KH. Glucose transporters in the uterus: an analysis of tissue distribution and proposed physiological roles. Reproduction. 2011;142:211–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Roncero I, Alvarez E, Acosta C, et al. Insulin-receptor substrate-2 (IRS-2) is required for maintaining glucokinase and glucokinase regulatory protein expression in mouse liver. PLoS One. 2013;8:e58797. [DOI] [PMC free article] [PubMed] [Google Scholar]