Abstract
Acetate conversion pathways of methanogenic consortia in acetate-fed chemostats at dilution rates of 0.025 and 0.6 day−1 were investigated by using 13C-labeled acetates, followed by gas chromatography-mass spectrometry (GC-MS) analysis of the CH4 and CO2 produced. Nonaceticlastic syntrophic oxidation by acetate-oxidizing syntrophs and hydrogenotrophic methanogens was suggested to occupy a primary pathway (approximately 62 to 90%) in total methanogenesis at the low dilution rate. In contrast, aceticlastic cleavage of acetate by aceticlastic methanogens was suggested to occupy a primary pathway (approximately 95 to 99%) in total methanogenesis at the high dilution rate. Phylogenetic analyses of transcripts of the methyl coenzyme M reductase gene (mcrA) confirmed that a significant number of transcripts of the genera Methanoculleus (hydrogenotrophic methanogens) and Methanosarcina (aceticlastic methanogens) were present in the chemostats at the low and high dilution rates, respectively. The mcrA transcripts of the genus Methanosaeta (aceticlastic methanogens), which dominated the population in a previous study (T. Shigematsu, Y. Tang, H. Kawaguchi, K. Ninomiya, J. Kijima, T. Kobayashi, S. Morimura, and K. Kida, J. Biosci. Bioeng. 96:547-558, 2003), were poorly detected at both dilution rates due to the limited coverage of the primers used. These results demonstrated that the dilution rate could cause a shift in the primary pathway of acetate conversion to methane in acetate-fed chemostats.
Under methanogenic conditions, acetate is quantitatively the most dominant intermediate of anaerobic degradation of organic matter. It is estimated that approximately 70 to 80% of methane is derived from acetate in anoxic environments (11, 13, 14). Two processes by which acetate is converted to methane have been described. The acetate-utilizing methanogens, the genera Methanosaeta and Methanosarcina, use the aceticlastic cleavage pathway in which the methyl group of acetate is converted to methane, while the carboxyl group is converted to CO2 (4). The second process includes the syntrophic oxidation of acetate to CO2 and hydrogen by one organism and the subsequent reduction of carbon dioxide to methane by a hydrogenotrophic methanogen. Two thermophilic bacteria, strain AOR and Thermacetogenium phaeum, and one mesophilic bacterium, Clostridium ultunense, were demonstrated to be capable of acetate oxidation in cocultures with hydrogenotrophic methanogens (7, 10, 19). The net reaction is the same as the reaction for aceticlastic cleavage of acetate in the syntrophic acetate oxidation process, but 14C-labeled substrates have been used to differentiate between the two processes (16, 18, 24). The quantitative information for these two acetate conversion pathways in total methanogenic microbial communities has been limited to date.
In a previous study, chemostat cultures of mesophilic acetate-degrading methanogenic consortia were constructed (8). The relative concentration of coenzyme F420, which is involved in hydrogenotrophic methanogenesis, was much higher at low dilution rates than that at high dilution rates. Microbial community structure analysis of the chemostat cultures at dilution rates of 0.025 and 0.6 day−1 revealed that a significant number of cells of the genus Methanoculleus, which is a hydrogenotrophic methanogen, were detected only at the low dilution rate, although larger populations of aceticlastic methanogens affiliated with the genera Methanosaeta and Methanosarcina were detected at both dilution rates (21). The detection of hydrogenotrophic methanogens and higher F420 concentrations in the chemostat cultures at the low dilution rate suggests that a significant proportion of methanogenesis occurs by syntrophic acetate oxidation rather than by aceticlastic cleavage of acetate. In this study, we analyzed the acetate conversion pathways of methanogenic consortia using 13C-labeled substrates and gas chromatography-mass spectrometry (GC-MS) analysis of the CH4 and CO2 produced. We also performed a phylogenetic analysis of transcripts of the mcrA gene, which encodes the α-subunit of methyl coenzyme M reductase I (MCR I), at both dilution rates.
MATERIALS AND METHODS
Operation of acetate-fed chemostats.
Two anaerobic chemostats were operated for more than 2 years at 37°C with acetate as the only added carbon source and electron donor at dilution rates of 0.025 and 0.6 day−1. Completely stirred tank reactors, each with a working volume of 1.7 liters, were used as the chemostats. The inoculum for these reactors was digested sludge acclimatized in our laboratory for 6 months. Detailed operation methods have been described previously (8). Under steady-state conditions, the acetate concentrations in the chemostats were approximately 10 and 250 mg/liter at dilution rates of 0.025 and 0.6 day−1, respectively.
Batchwise gas evolution test with 13C-labeled substrate.
A 10-ml sample of culture broth was taken from a chemostat and centrifuged at 25,000 × g for 15 min. The precipitate was washed with 30 ml of synthetic wastewater (8) containing no carbon source and was resuspended in 10 ml of synthetic wastewater containing no carbon source. The precipitate was transferred to vials and supplemented with [1-13C]sodium acetate, [2-13C]sodium acetate, or [1,2-13C]sodium acetate to give a final concentration of 100 mM. Washing and suspension of the pellet were performed by using an anaerobic glove box (model 1025; Forma Scientific, Marietta, Ohio) supplemented with helium gas. The vials were immersed in a thermostat-controlled water bath at 37°C. After 12 h of incubation with mixing, the CH4 and CO2 in the headspace were analyzed by using a GCMS-QP5000 GC-MS (Shimadzu, Kyoto, Japan) equipped with a GS-GasPro column (30 m by 0.32 mm; J & W Scientific, Folsom, Calif.). Helium was used as carrier gas at a flow rate of 1.7 ml/min. The column temperature was 30°C. The peaks at m/z 15 and 17 in the mass spectrum, which were derived from a retention time of 1.2 min in the gas chromatogram, were regarded as the fragment ion for 12CH4 and the molecular ion for 13CH4, respectively. The peaks at m/z 44 and 45, which were derived from a retention time of 1.5 min, were regarded as the molecular ions for 12CO2 and 13CO2, respectively.
RT-PCR amplification and cloning.
Total nucleic acids from the culture broth in a chemostat were extracted by a method described previously (21). RNA was then extracted by the method of Griffiths et al. (5) and purified with an RNeasy kit (QIAGEN, Hilden, Germany). Reverse transcription (RT) reactions were performed with a Gene Amp Gold RNA PCR reagent kit (Applied Biosystems, Foster City, Calif.) by using 500 and 100 ng of the template RNA extracted from the chemostats at dilution rates of 0.025 and 0.6 day−1, respectively, and the reverse primer ME2 (5′-TCAT(G/T)GC(A/G)TAGTT(A/G/T)GG(A/G)TAGT-3′) (6). The resulting cDNA was purified with a MicroSpin S-400 HR column (Amersham Biosciences, Piscataway, N.J.) and was used as the template for amplification of mcrA with AmpliTaq (Applied Biosystems, Foster City, Calif.) used according to the manufacturer's instructions (1× PCR buffer, 2.5 U of AmpliTaq DNA polymerase, each deoxynucleoside triphosphate at a concentration of 250 μM, and 40 pmol of each primer in a 100-μl reaction mixture). The PCR primers used in the amplification were the forward primer ME1 (5′-GC(A/C)ATGCA(A/G)AT(A/C/T)GG(A/T)ATGTC-3′) (6) and the reverse primer ME2b (5′-TCCTG(G/C)AGGTCG(A/T)A(A/G)CCGAAGAA-3′). Reactions were performed with a GeneAmp PCR System 2400 (Applied Biosystems) with the following cycle conditions: preincubation at 95°C for 2 min; 25 cycles of 95°C for 1 min, 50°C for 1 min, and 72°C for 1 min; and a final extension at 72°C for 7 min. The amplified mcrA fragments were cloned into a plasmid pT7Blue T vector (Novagen Inc., Madison, Wis.) by using a DNA ligation kit (version 2; Takara, Kyoto, Japan).
Sequencing and phylogenetic analysis.
Cloned RT-PCR products were prepared from randomly selected recombinants and used as templates for sequencing. Sequencing was performed by using a DNA sequencer (CEQ8000; Beckman Coulter, Fullerton, Calif.) with a CEQ Quick Start Master Mix kit (Beckman Coulter). DNA and deduced amino acid sequences were analyzed with the GENETYX-WIN software package (version 5.1; Software Development, Tokyo, Japan). The search for homologous proteins was conducted with the BLAST program (1). Multiple alignments were run by using the Clustal X program, version 1.8 (22). Phylogenetic analyses were conducted with MEGA, version 2.1 (9). Identical sequences (100% similarity) were recognized by matrix analysis and manual comparison and were used in subsequent analyses as an operational taxonomic unit (OTU). The OTUs were designated ALM01 to ALM08 for clones from the culture broth at a dilution rate of 0.025 day−1 and AHM01 to AHM08 for clones at a dilution rate of 0.6 day−1.
Quantitative RT-PCR of mcrA transcripts.
Real-time quantitative RT-PCR experiments were conducted to quantify mcrA transcripts of different taxonomic groups by using the TaqMan fluorogenic PCR system. The RT reaction was performed with a Gene Amp Gold RNA PCR reagent kit (Applied Biosystems) by using 10 μg of the extracted RNA as a template and the reverse primer ME2 in a 100-μl reaction mixture. The resulting cDNA was precipitated by ethanol precipitation and then vacuum dried and resuspended in 20 μl of Tris-EDTA buffer (pH 7.4). The cDNA was purified by using a MicroSpin S-400 HR column (Amersham Biosciences) and was then used as a template for a quantitative PCR. The quantitative PCR was carried out by using primers ME1 and ME2b and a genus-specific TaqMan probe. Three TaqMan probes, SAE716TAQ (5′-AGGCCTTCCCCACTCTGCTTGAGGAT-3′), SAR716TAQ (5′-AGAAATTCCCAACAGCCCTTGAAGAC-3′), and MCU716TAQ (5′-AGCAGTACCCGACCATGATGGAGGAC-3′), were used for detection of the mcrA gene products of the genera Methanosaeta, Methanosarcina, and Methanoculleus, respectively. The specificities of these probes for the target mcrA genes were confirmed by manual comparison of the nucleotide sequences of mcrA genes in the DDBJ/EMBL/GenBank database. All TaqMan probes were 5′ end labeled with 6-carboxyfluorescein and 3′ end labeled with 6-carboxytetramethyl rhodamine, obtained from Applied Biosystems. In order to evaluate the selectivity of the primer-probe sets, three clones, ALM07, AHM01, and ALM01, were used as controls. By using the three sets of primers and TaqMan probes, fluorescence signal monitoring was performed with the GeneAmp 5700 sequence detection system (Applied Biosystems). Reaction mixtures for fluorogenic PCR in which the concentrations of both the primer and the TaqMan probe were optimized (300 and 200 nM, respectively) were prepared. The concentration of control DNA varied between 15.63 and 625 pg per 50 μl of reaction mixture. The TaqMan Universal PCR Master Mix (Applied Biosystems) was used with the following cycle conditions: an initial step of 50°C for 2 min and then 95°C for 10 min; and two-step cycles of 95°C for 15 s and 60°C for 1 min. All assays were performed at least in duplicate. Post-PCR analysis was performed by using GeneAmp 5700 SDS software.
Nucleotide sequence accession numbers.
The DDBJ/EMBL/GenBank accession numbers for the sequences of clones ALM01 to ALM08 and AHM01 to AHM08 are AB158524 to AB158539.
RESULTS
Batchwise gas evolution test with 13C-labeled substrates.
Two acetate-fed mesophilic anaerobic chemostats were operated for more than 2 years at dilution rates of 0.025 and 0.6 day−1. Under steady-state conditions, the acetate concentrations in the chemostats were approximately 0.2 and 4 mM at dilution rates of 0.025 and 0.6 day−1, respectively. To evaluate the acetate conversion pathway in the two chemostats, batchwise cultivation of culture broth extracted from the chemostats was carried out by using forms of 13C-labeled sodium acetate as the substrates. If the culture broth used aceticlastic cleavage of acetate, the methyl and carboxyl groups of acetate would have been converted to methane and CO2, respectively (4), but if the culture broth used syntrophic acetate oxidation, 2 mol of CO2 would have been produced from 1 mol of acetate, while 1 mol of the CO2 produced would have been concurrently reduced to methane (24). In the latter case, the methyl and carboxyl groups of acetate would have been converted to equal amounts of methane and CO2.
When the culture broth of the chemostat at a dilution rate of 0.025 day−1 was used for batch cultivation, 33 and 45% of the methane were considered to be derived from the carboxyl group of acetate when [2-13C]sodium acetate and [1-13C]sodium acetate, respectively, were used as the substrates (Table 1). For CO2, 31 and 32% were considered to be derived from the methyl group when [2-13C]sodium acetate and [1-13C]sodium acetate, respectively, were used as the substrates (Table 2). On the other hand, only about 2% of the methane and 6% of the CO2 were considered to be derived from the carboxyl and methyl groups of acetate, respectively, when the culture broth of the chemostat at a dilution rate of 0.6 day−1 was used. These results suggested that the syntrophic oxidation pathway accounted for approximately 62 to 90% of the total methanogenesis in the chemostat at the low dilution rate. In contrast, at the high dilution rate, the aceticlastic cleavage of acetate was suggested to account for 95 to 99% of total methanogenesis. Because we used batchwise cultivation for the 13C-labeled substrate assay, the results might not precisely reflect the in situ activities of the microorganisms in the chemostats. But the culture broth at the low dilution rate had an obviously larger potential for syntrophic acetate oxidation than for aceticlastic cleavage of acetate, whereas the culture broth at the high dilution rate had a larger potential for aceticlastic cleavage than for syntrophic acetate oxidation.
TABLE 1.
GC-MS analysis of CH4 produced from 13C-labeled acetate
| Dilution rate (day−1) | Substrate | Peak intensities
|
CH4 from carboxyl base/total CH4 | CH4 from methyl base/total CH4 | ||
|---|---|---|---|---|---|---|
|
m/z 15 (12CH4)
|
m/z 17(13CH4) (actual) | |||||
| Actual | Background subtracteda | |||||
| 0.025 | 13CH312COONa | 2,695b | 2,695 | 5,474 | 0.33 | 0.67 |
| 12CH313COONa | 1,139 | 1,139 | 928 | 0.45 | 0.55 | |
| 13CH313COONa | 0 | 1,361 | ||||
| 0.6 | 13CH312COONa | 81,973 | 14,066 | 614,114 | 0.022 | 0.98 |
| 12CH313COONa | 636,401 | 562,089 | 13,860 | 0.024 | 0.98 | |
| 13CH313COONa | 74,312 | 588,433 | ||||
The peak intensities at an m/z value of 15 from 13CH313COONa were regarded as the background.
All values are averages for duplicate experiments.
TABLE 2.
GC-MS analysis of CO2 produced from 13C-labeled acetate
| Dilution rate (day−1) | Substrate | Peak intensities
|
CO2 from methyl base/total CO2 | CO2 from carboxyl base/total CO2 | ||
|---|---|---|---|---|---|---|
|
m/z 44 (12CO2)
|
m/z 45(13CO2) (actual) | |||||
| Actual | Background subtracteda | |||||
| 0.025 | 13CH312COONa | 4,041b | 2,635 | 1,196 | 0.31 | 0.69 |
| 12CH313COONa | 3,424 | 2,018 | 4,296 | 0.32 | 0.68 | |
| 13CH313COONa | 1,406 | 3,380 | ||||
| 0.6 | 13CH312COONa | 87,919 | 79,915 | 5,302 | 0.062 | 0.94 |
| 12CH313COONa | 12,951 | 4,947 | 80,702 | 0.058 | 0.94 | |
| 13CH313COONa | 8,004 | 73,096 | ||||
The peak intensities at an m/z value of 44 from 13CH313COONa were regarded as the background.
All values are averages for duplicate experiments.
Analysis of mcrA gene transcripts from the chemostats.
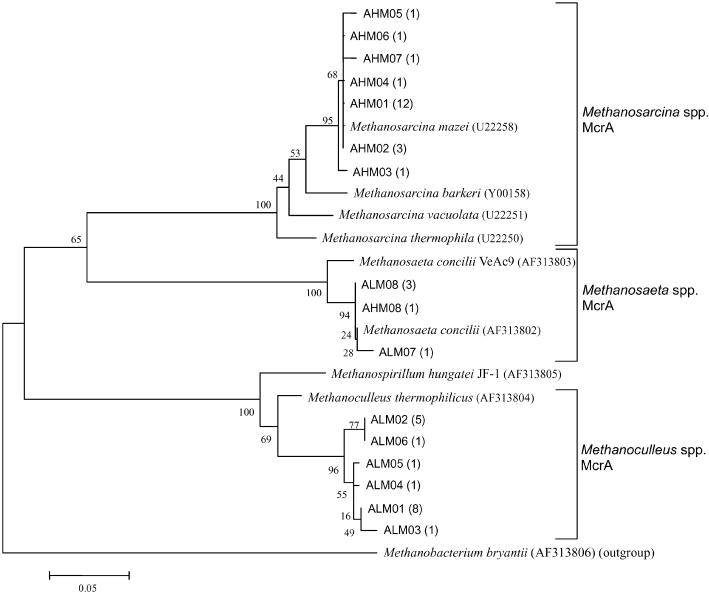
MCR I appears to be unique to methanogens and to be present in all methanogens. The mcrA gene encoding the α-subunit of MCR I has been used a marker gene for the specific detection of methanogens in various environments (6, 12). The mcrA transcripts in community RNA extracted from the acetate-fed chemostats were amplified by RT-PCR and were used to construct two clone libraries, designated ALM (mcrA transcripts from the chemostat at a dilution rate of 0.025 day−1) and AHM (mcrA transcripts from the chemostat at a dilution rate of 0.6 day−1). Twenty-one clones from each library were randomly selected and sequenced. All nucleotide sequences and deduced amino acid sequences showed significant similarities with sequences of known mcrA genes and McrAs, respectively. In the ALM library, eight different sequences (OTUs) were obtained. Six OTUs (ALM01 to ALM06, 17 clones) were closely related to the mcrA gene of Methanoculleus thermophilicus (Fig. 1) and were regarded as mcrA genes of the genus Methanoculleus (Table 3). The other two OTUs (ALM07 and ALM08, four clones) were closely related to the mcrA gene of Methanosaeta concilii and were regarded as mcrA genes of the genus Methanosaeta. In the AHM library, eight OTUs were obtained. Seven OTUs (AHM01 to AHM07, 20 clones) were closely related to the mcrA gene of Methanosarcina mazei and were regarded as mcrA genes of the genus Methanosarcina. Another OTU (AHM08, one clone) was closely related to mcrA of M. concilii and was regarded as an mcrA gene of the genus Methanosaeta.
FIG. 1.
Phylogenetic relationships of deduced amino acid sequences of α-subunits of MCR I (McrA). The tree was constructed from phylogenetic distances obtained by the neighbor-joining method (17). ALM and AHM indicate clones from cultivation at the low (0.025 day−1) and high (0.6 day−1) dilution rates, respectively. The numbers of clones that had identical sequences are shown in parentheses. Bar = 5 amino acid substitutions per 100 amino acids. Bootstrap probabilities (3) are indicated at branch nodes. The DDBJ/EMBL/GenBank accession numbers for reference strains are shown in parentheses. The tree was rooted by using McrA of Methanobacterium bryantii as the outgroup.
TABLE 3.
Composition of and quantification of mcrA transcripts of three taxonomic groups
| Phylogenetic affiliationa | Dilution rate of 0.025 day−1
|
Dilution rate of 0.6 day−1
|
||
|---|---|---|---|---|
| No. of clones | Quantitative RT-PCR (copies/μg of total RNA)b | No. of clones | Quantitative RT-PCR (copies/μg of total RNA)b | |
| Methanosaeta spp. mcrA | 4 | NDc | 1 | ND |
| Methanosarcina spp. mcrA | 0 | ND | 20 | 4.15 × 107 |
| Methanoculleus spp. mcrA | 17 | 9.06 × 106 | 0 | ND |
Primers ME1 and ME2 used in this study were reported not to be suitable for amplification of the mcrA genes of the genus Methanosaeta (12).
The values for quantitative RT-PCR are averages for duplicate experiments.
ND, not detected.
Real-time quantitative RT-PCR experiments were conducted to quantify mcrA transcripts of different taxonomic groups. mcrA transcripts of the genus Methanosaeta could not be detected in the RNA from the chemostat at either dilution rate (Table 3). mcrA transcripts of the genus Methanosarcina could be detected only in the RNA from the chemostat at a dilution rate of 0.6 day−1. In contrast, mcrA transcripts of the genus Methanoculleus could be detected only in the RNA from the chemostat at a dilution rate of 0.025 day−1. These results agreed with our clonal sequence analysis which showed that the mcrA transcripts of the genera Methanoculleus and Methanosarcina were dominant in RNA from the chemostats at the low dilution rate and the high dilution rate, respectively. Because the ME1 and ME2 primers were reported not to be suitable for amplification of the mcrA genes of the genus Methanosaeta (12), the selectivity of the primer sets should be considered. However, our results still indicated that the levels of mcrA transcription activity of the genera Methanoculleus and Methanosarcina were significant in the chemostats at the low and high dilution rates, respectively.
DISCUSSION
We used a stable-isotope technique to analyze the primary pathway of acetate conversion to methane in acetate-fed chemostats. To our knowledge, this was the first application of 13C-labeled substrates combined with GC-MS analysis of the methane and CO2 produced to analyze acetate conversion pathways to methane. This technique is more useful and convenient for determining which groups of acetate are converted to methane and CO2 than the conventional technique using 14C-labeled substrates. The results obtained in the RT-PCR experiment targeting mcrA transcripts supported the findings obtained by using 13C-labeled substrates, although the limited coverage of the primers used requires further consideration.
The specific growth rate of the mesophilic acetate-oxidizing syntroph C. ultunense cocultured with a hydrogenotrophic methanogen by using acetate as a substrate was reported to be 0.027 to 0.035 day−1 (20). The specific growth rates of the mesophilic aceticlastic methanogens M. concilii and M. mazei were 0.24 to 0.28 and 0.98 day−1, respectively (2). At the high dilution rate (0.6 day−1), the cells affiliated with the genus Methanosarcina, which were able to grow rapidly, would have been predominant in the chemostat and engaged in aceticlastic cleavage of acetate to methane as the primary pathway. On the other hand, the low dilution rate (0.025 day−1) was sufficiently low for growth of the three acetate-utilizing members, acetate-oxidizing syntrophs, Methanosaeta, and Methanosarcina. In this case, competition among the substrate affinities of the three acetate-utilizing members would have been decisive for dominance. The apparent Km for acetate of a thermophilic acetate-oxidizing syntroph was reported to be 0.65 mM (16), although no Km value of a mesophilic acetate-oxidizing syntroph is currently available. The apparent Km values for acetate of the genera Methanosaeta and Methanosarcina were approximately 0.8 to 0.9 and 3 to 5 mM, respectively (23). The acetate-oxidizing syntroph associated with Methanoculleus, according to the high substrate affinity, was better adapted to convert acetate primarily in the chemostat at a low dilution rate. The genus Methanosaeta, whose population was previously shown to be the largest among the three members by previous ribosomal DNA analyses (21), played a secondary role for acetate conversion by aceticlastic cleavage in the chemostat at the low dilution rate. It is possible that even cells of a dominant population could not show a dominant metabolic function in a consortium because of their lower metabolic activity. The correlation between the population dominance of the genus Methanosaeta and its lower metabolic activity for acetate conversion at the low dilution rate still requires further analysis in terms of quantification and comparison of mcrA transcripts of the genera Methanosaeta and Methanoculleus with more universal primers. We have no direct evidence of which bacteria are responsible for acetate oxidation in our chemostats. However, the dominance of bacteria belonging to the phylum Firmicutes, with which C. ultunense and T. phaeum are affiliated, was shown by 16S ribosomal DNA clonal sequence analysis (21). Some members of this phylum, which is related to the known acetate-oxidizing syntrophs, may contribute to the syntrophic acetate oxidation in the chemostat at the low dilution rate.
The results described above, combined with previous findings (8, 21) demonstrate that the dilution rate could cause a shift in the primary pathway of acetate conversion to methane in acetate-fed chemostats. At the low dilution rate, the acetate-oxidizing syntrophs, associated with hydrogenotrphic methanogens, could metabolically overcome the aceticlastic methanogens and play a primary role in the conversion of acetate to methane. Recently, Nüsslein et al. reported that a large proportion of methanogenesis in lake sediment occurs by syntrophic acetate oxidation rather than by aceticlastic cleavage of acetate (15). Most natural environments fulfill the conditions of low dilution rate and low acetate concentration that were present in our chemostat. Syntrophic acetate oxidation might be a common mechanism in natural methanogenic environments.
Acknowledgments
This study was financially supported by a grant-in-aid for scientific research (project 14580593) from the Japan Society for the Promotion of Science.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boone, D. R., W. B. Whitman, and Y. Koga. 2001. Order III. Methanosarcinales ord. nov., p. 268-294. In D. R. Boone and R. W. Castenholz (ed.), Bergey's manual of systematic bacteriology, 2nd ed., vol. 1. The Archaea and the deeply branching and phototrophic Bacteria. Springer-Verlag, New York, N.Y.
- 3.Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783-791. [DOI] [PubMed] [Google Scholar]
- 4.Ferry, J. G. 1993. Fermentation of acetate, p. 304-334. In J. G. Ferry (ed.), Methanogenesis—ecology, physiology, biochemistry & genetics. Chapman & Hall, New York, N.Y.
- 5.Griffiths, R. I., A. S. Whiteley, A. G. O'Donnell, and M. J. Bailey. 2000. Rapid method for coextraction of DNA and RNA from natural environments for analysis of ribosomal DNA- and rRNA-based microbial community composition. Appl. Environ. Microbiol. 66:5488-5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hales, B. A., C. Edwards, D. A. Ritchie, G. Hall, R. W. Pickup, and J. R. Saunders. 1996. Isolation and identification of methanogen-specific DNA from blanket bog peat by PCR amplification and sequence analysis. Appl. Environ. Microbiol. 62:668-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hattori, S., Y. Kamagata, S. Hanada, and H. Shoun. 2000. Thermoacetogenium phaeum gen. nov., sp. nov., a strictly anaerobic, thermophilic, syntrophic acetate-oxidizing bacterium. Int. J. Syst. Evol. Microbiol. 50:1601-1609. [DOI] [PubMed] [Google Scholar]
- 8.Kida, K., T. Shigematsu, J. Kijima, M. Numaguchi, Y. Mochinaga, N. Abe, and S. Morimura. 2001. Influence of Ni2+ and Co2+ on methanogenic activity and the amounts of coenzymes involved in methanogenesis. J. Biosci. Bioeng. 91:590-595. [DOI] [PubMed] [Google Scholar]
- 9.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 10.Lee, M. J., and S. H. Zinder. 1988. Isolation and characterization of a thermophilic bacterium which oxidizes acetate in syntrophic association with a methanogen and which grows acetogenically on H2-CO2. Appl. Environ. Microbiol. 54:124-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lovley, D. R., and M. J. Klug. 1982. Intermediary metabolism of organic matter in the sediments of a eutrophic lake. Appl. Environ. Microbiol. 43:552-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lueders, T., K.-J. Chin, R. Conrad, and M. Friedrich. 2001. Molecular analyses of methyl-coenzyme M reductase α-subunit (mcrA) genes in rice field soil and enrichment cultures reveal the methanogenic phenotype of a novel archaeal lineage. Environ. Microbiol. 3:194-204. [DOI] [PubMed] [Google Scholar]
- 13.Mackie, R. I., and M. P. Bryant. 1981. Metabolic activity of fatty acid-oxidizing bacteria and the contribution of acetate, propionate, butyrate, and CO2 to methanogenesis in cattle waste at 40 and 60°C. Appl. Environ. Microbiol. 41:1363-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mountfort, D. O., and R. A. Asher. 1978. Changes in proportions of acetate and carbon dioxide used as methane precursors during the anaerobic digestion of bovine waste. Appl. Environ. Microbiol. 35:648-654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nüsslein, B., K.-J. Chin, W. Eckert, and R. Conrad. 2001. Evidence for anaerobic syntrophic acetate oxidation during methane production in the profundal sediment of subtropical Lake Kinneret (Israel). Environ. Microbiol. 3:460-470. [DOI] [PubMed] [Google Scholar]
- 16.Petersen, S. P., and B. K. Ahring. 1991. Acetate oxidation in a thermophilic anaerobic sewage-sludge digestor: the importance of non-aceticlastic methanogenesis from acetate. FEMS Microbiol. Ecol. 86:149-158. [Google Scholar]
- 17.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 18.Schnürer, A., F. P. Houwen, and B. H. Svensson. 1994. Mesophilic syntrophic acetate oxidation during methane formation by a triculture at high ammonium concentration. Arch. Microbiol. 162:70-74. [Google Scholar]
- 19.Schnürer, A., B. Schink, and B. H. Svensson. 1996. Clostridium ultunense sp. nov., a mesophilic bacterium oxidizing acetate in syntrophic association with a hydrogenotrophic methanogenic bacterium. Int. J. Syst. Bacteriol. 46:1145-1152. [DOI] [PubMed] [Google Scholar]
- 20.Schnürer, A., B. H. Svensson, and B. Schink. 1997. Enzyme activities in and energetics of acetate metabolism by the mesophilic syntrophically acetate-oxidizing anaerobe Clostridium ultunense. FEMS Microbiol. Lett. 154:331-336. [Google Scholar]
- 21.Shigematsu, T., Y. Tang, H. Kawaguchi, K. Ninomiya, J. Kijima, T. Kobayashi, S. Morimura, and K. Kida. 2003. Effect of dilution rate on structure of a mesophilic acetate-degrading methanogenic community during continuous cultivation. J. Biosci. Bioeng. 96:547-558. [DOI] [PubMed] [Google Scholar]
- 22.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zinder, S. H. 1993. Physiological ecology of methanogens, p. 128-206. In J. G. Ferry (ed.), Methnaogenesis—ecology, physiology, biochemistry & genetics. Chapman & Hall, New York, N.Y.
- 24.Zinder, S. H., and M. Koch. 1984. Non-aceticlastic methanogenesis from acetate: acetate oxidation by a thermophilic syntrophic coculture. Arch. Microbiol. 138:263-272. [Google Scholar]