Abstract
U.S. Environmental Protection Agency method 1623 is widely used to monitor source waters and drinking water supplies for Cryptosporidium oocysts. Matrix spikes, used to determine the effect of the environmental matrix on the method's recovery efficiency for the target organism, require the collection and analysis of two environmental samples, one for analysis of endemic oocysts and the other for analysis of recovery efficiency. A new product, ColorSeed, enables the analyst to determine recovery efficiency by using modified seeded oocysts that can be differentiated from endemic organisms in a single sample. Twenty-nine stream water samples and one untreated effluent sample from a cattle feedlot were collected in triplicate to compare modified seeding procedures to conventional seeding procedures that use viable, unmodified oocysts. Significant negative correlations were found between the average oocyst recovery and turbidity or suspended sediment; this was especially apparent in samples with turbidities greater than 100 nephelometric turbidity units and suspended sediment concentrations greater than 100 mg/liter. Cryptosporidium oocysts were found in 16.7% of the unseeded environmental samples, and concentrations, adjusted for recoveries, ranged from 4 to 80 oocysts per 10 liters. Determining recovery efficiency also provided data to calculate detection limits; these ranged from <2 to <215 oocysts per 10 liters. Recoveries of oocysts ranged from 2.0 to 61% for viable oocysts and from 3.0 to 59% for modified oocysts. The recoveries between the two seeding procedures were highly correlated (r = 0.802) and were not significantly different. Recoveries by using modified oocysts, therefore, were comparable to recoveries by using conventional seeding procedures.
To improve monitoring for Cryptosporidium oocysts in water, the U.S. Environmental Protection Agency (USEPA) developed methods 1622 (30) and 1623 (31), which consist of filtration, concentration, immunomagnetic separation (IMS), fluorescent antibody and 4′,6′-diamidino-2-phenylindole (DAPI) counter staining, and microscopic detection and enumeration. Method 1622 detects and enumerates only Cryptosporidium in water samples; method 1623 detects and enumerates Cryptosporidium oocysts and Giardia cysts in water samples and incorporates improvements over the previous method. Both are performance-based methods, allowing components to be replaced by other more efficient or effective components, provided the substitutions meet required quality control acceptance criteria.
Recoveries of oocysts from environmental water samples with method 1622 or 1623, however, have been found to be highly variable (15, 26, 32). Because of variable recoveries, matrix spikes are routinely analyzed to determine the effect of the environmental matrix on the method's recovery efficiency for the target organisms. In conventional seeding procedures for these methods, two samples are analyzed, one of which is seeded with known numbers of viable organisms (matrix spike) and the other of which is unseeded. A potential modification for method 1623 is to use an alternative seeding procedure, ColorSeed, which enables the analyst to determine the percent recovery and number of environmental oocysts in one water sample. In this product, organisms are modified to incorporate a fluorochrome that is used to distinguish seeded organisms from unmodified organisms that may be present in the environmental sample. Modified organisms are gamma irradiated to increase stability, whereas conventional seeding procedures use viable organisms. Gamma irradiation also inactivates the oocysts, decreasing the health risk for the laboratory analyst compared to the risk of using viable organisms. If the modified seeding procedure is found to yield equivalent percent recoveries to that of the conventional seeding procedure, sampling and analytical time would be significantly reduced for processing environmental water samples.
In this study, 30 stream water samples were collected from 20 sites throughout the United States to compare the recovery efficiencies of Cryptosporidium oocysts by method 1623 with modified and conventional seeding procedures. For each sample, one unseeded and two seeded subsamples were analyzed. The collection and processing of these samples afforded the opportunity to address other issues regarding the use of method 1623 for monitoring environmental waters. These issues included determining whether water quality factors affected recoveries of oocysts by method 1623 and whether fecal indicators (Escherichia coli, Clostridium perfringens, and somatic and F-specific coliphage) could be used as surrogates for the presence of Cryptosporidium in stream waters.
Several studies have attempted to identify water quality properties and constituents that affect recoveries of oocysts by using method 1622 or 1623. Many investigators found that turbidity was related to recoveries of oocysts (4, 5, 7, 15, 26). The efficiency of the IMS technique was shown to decrease when the adjusted pH deviated from 7.0 (16) or when dissolved iron concentrations were greater than 4 mg/liter (38). In contrast, in the Information Collection Rule Supplemental Survey (ICRSS), wherein 430 samples were collected from 87 source waters, all measured water quality properties, including turbidity, were found to be unrelated or weakly related to recoveries of oocysts (32).
In addition to improving the performance of method 1623, there is continued interest in identifying a microbiological surrogate for the presence of Cryptosporidium in water. Microbiological surrogates investigated in previous studies have included E. coli, C. perfringens, fecal coliforms, total coliforms, and somatic and F-specific coliphage. Previous surface water studies have shown significant relations between detection and enumeration of oocysts and some of these fecal indicators (2, 18, 19, 20, 32). In contrast, other researchers found no significant relations between indicator organisms and Cryptosporidium in wastewater (3), in drainage canals (W. E. McElroy, S. Pillai, E. Cabello, and S. Hernandez, Abstr. 101st Gen. Meet. Am. Soc. Microbiol. Abstr. Q-230, p. 629-630, 2001), or in surface waters (21, 22, 23).
In this study, we tested the use of the modified seeding procedure and found it to be an acceptable alternative seeding procedure for the detection of Cryptosporidium by using method 1623. We demonstrated the importance of including appropriate quality control samples in any monitoring program. The relations between recoveries of Cryptosporidium and water quality properties were examined. Of the water quality properties tested, a negative relation was found between recovery and turbidity or suspended-sediment concentrations. Because Cryptosporidium oocysts were found in a small percentage of the 30 samples collected, we were not able to examine the relation between the detection of Cryptosporidium and concentrations of fecal indicators.
MATERIALS AND METHODS
Site selection and sample collection.
Twenty sites were selected to include broad geographic coverage of the United States (Table 1). If two sites were in the same state, they are labeled as state 1 and state 2 in Table 1. The sites are part of the U.S. Geological Survey (USGS) National Water Quality Assessment Program, a national water quality program that focuses on “study units” that represent major hydrologic systems of the United States (13). A major criterion for site selection was to include sites with high probabilities of detecting target organisms, for example, sites potentially receiving human sewage or livestock inputs.
TABLE 1.
Characteristics of stream water sampling sites, 2002 to 2003
| Stream water sitea | Stream flow conditionb | Land use | Potential sources of fecal contaminationc | Drainage area (square miles) | Population density (people/square mile) | Stream flow at time of sampling (cubic feet/s)f |
|---|---|---|---|---|---|---|
| Alabama 1 | Elevated | Mixed (agricultural and forest) | Feedlots and septic systems | 374 | 33,000 | 163 |
| Alabama 2ad | Elevated | Agricultural | Free-range cattle and septic systems | 29 | 2,100 | 36 |
| Alabama 2bd | Elevated | Agricultural | Free-range cattle and septic systems | 29 | 2,100 | 44 |
| Colorado 1e | Effluent | Agricultural | CAFOs, applied manure, wildlife, and septic systems | 567 | 5.9 | NA |
| Colorado 2 | Elevated | Mixed (forest, rangeland, agricultural, and urban) | WWTPs, CAFOs, applied manure, and wildlife | 9,598 | 300 | 628 |
| Georgia 1 | Base | Mixed (urban, forest, and agricultural) | Large urban WWTP and combined-sewer overflows | 2,060 | 690 | 1,140 |
| Georgia 1 | High | Mixed (urban, forest, and agricultural) | Large urban WWTP and combined-sewer overflows | 2,060 | 690 | 8,400 |
| Georgia 2 | High | Urban residential | Septic systems | 29 | 2,100 | 183 |
| Indiana | Elevated | Agricultural | Cows, septic systems, and WWTPs | 610 | 61 | 142 |
| Iowad | Base | Agricultural | Hog CAFOs and manure | 230 | 16 | 48 |
| Iowad | High | Agricultural | Hog CAFOs and manure | 230 | 16 | 967 |
| Louisianad | Base | Urban residential and commercial | WWTP and combined-sewer overflows | 15 | 3,000 | 2.2 |
| Louisianad | Elevated | Urban residential and commercial | WWTP and combined-sewer overflows | 15 | 3,000 | 20 |
| Nebraska | High | Agricultural | Feedlots and septic systems | 369 | 11 | 153 |
| Nebraska | Base | Agricultural | Feedlots and septic systems | 369 | 11 | 23 |
| Ohio | Elevated | Agricultural | Cows and WWTPs | 310 | 130 | 209 |
| Ohio | High | Agricultural | Cows and WWTPs | 310 | 130 | 668 |
| South Carolina 1 | Base | Agricultural | CAFOs | 23 | 210 | 7.4 |
| South Carolina 1 | Elevated | Agricultural | CAFOs | 23 | 210 | 15 |
| South Carolina 2 | Elevated | Urban | WWTPs, street runoff, and raw sewage | 60 | 1,800 | 65 |
| Texas | Elevated | Mixed (urban and agricultural) | WWTPs, street runoff, and raw sewage | 6,278 | 410 | 4,030 |
| Texas | Base | Mixed (urban and agricultural) | WWTPs, street runoff, and raw sewage | 6,278 | 410 | 340 |
| Virginia 1 | Base | Urban residential and commercial | Wildlife, recreation, and WWTPs | 24 | 110,000 | 2.2 |
| Virginia 2d | Elevated | Agricultural | Feedlots and septic systems | 14 | 110 | 14 |
| Washington 1ad | Base | Agricultural | Dairy CAFOs and applied manure | 62 | 49 | 53 |
| Washington 1bd | Base | Agricultural | Dairy CAFOs and applied manure | 62 | 49 | 38 |
| Washington 2 | High | Urban residential and commercial | WWTPs, street runoff, and wildlife | 12 | 5,600 | 31 |
| Wisconsin 1 | High | Agricultural | Septic systems, dairy farms, and WWTPs | 96 | 85 | 155 |
| Wisconsin 2 | Base | Mixed (urban and agricultural) | Septic systems, feedlots, applied manure, and WWTPs | 696 | 520 | 88 |
| Wisconsin 2 | High | Mixed (urban and agricultural) | Septic systems, feedlots, applied manure, and WWTPs | 696 | 520 | 993 |
If two sites were in the same state, they are labeled as state 1 and state 2. If two samples were collected at the same site and streamflow category, they are labeled as “a” and “b.”
High, streamflow was greater than 90% of all measured streamflows at that site; elevated, streamflow was between 50 and 90% of all measured streamflows at that site; base, streamflow was less than 50% of all measured streamflows at that site.
CAFO, concentrated animal feeding operation; WWTP, wastewater treatment plant.
Ten-year period of record data was not available, so streamflow categories were estimated.
Sample was collected from a drainage ditch on a cattle feedlot.
NA, not applicable.
The site information in Table 1 shows that samples were collected from sites with a range of land use practices, drainage areas, and population densities. Thirty samples were collected between March 2002 and June 2003, 29 of which were collected from streams. The exception was the Colorado 1 effluent sample, which was collected from a drainage ditch on a cattle feedlot. Instantaneous stream flows were estimated at the time of sampling (Table 1) from a stage-stream flow relation developed for each site (14). Samples were placed into three categories based on historical stream flow data: (i) high (stream flow was greater than 90% of all measured stream flows at that site), (ii) elevated (stream flow was between 50 and 90% of all measured stream flows at that site), and (iii) base (stream flow was less than 50% of all measured stream flows at that site). For 10 stream water sites, samples were collected twice; eight of these samples were collected during base flow and elevated or high flow. These sites were expected to receive fecal contamination during different flow conditions. The two Washington 1 samples were collected at base flow during the irrigation season and the two Alabama 2 samples were collected at elevated flow. The “a” and “b” designations for these samples in Table 1 indicate two samples collected at the same site with stream flows in the same category. For the 10 sites in which only one sample was collected, based on the sampler's experience, a base-, elevated-, or high-flow sample was collected to sample when fecal contamination was expected to be greatest.
For Cryptosporidium analyses, three 10-liter sterile, disposable, collapsible cubitainers (Eagle Picher Technologies, Miami, Okla.) were filled with water collected from several vertical sampling intervals in the stream cross section. Samples were similarly collected in a 3-liter sterile sample bottle and analyzed for C. perfringens, coliphage, E. coli, and turbidity. Specific conductance, temperature, dissolved oxygen, and pH were measured in the stream with a four-parameter water quality meter and USGS protocols (37). Alkalinity was determined by field crews using the incremental titration technique (37). Samples for water quality constituents were collected by using USGS protocols (24). Human population densities for each site were obtained from the U.S. Bureau of the Census (27) and were calculated from countywide coverages by applying densities as a percentage of the county in the watershed.
Cryptosporidium parvum oocysts.
Viable Cryptosporidium parvum (Harley Moon strain) oocysts used for the conventional seeding procedure were obtained from C. Sterling, University of Arizona, and were propagated in dexamethasone-immunosuppressed C57BL female mice at the USEPA facility in Cincinnati, Ohio (6). The oocysts were purified with sieving, step sucrose gradients, and cesium chloride purification (1). These highly purified oocysts were suspended in reagent-grade water containing 100 U of penicillin and 100 μg of streptomycin/ml, stored at 4°C, and used within 5 weeks of purification.
A FACS VantageSE (Beckton Dickinson, Palo Alto, Calif.) flow cytometry system equipped with CloneCyt software was used for the preparation of all viable oocyst seeding preparations. Isoton II (Coulter Corp., Hialeah, Fla.) was used as the sheath fluid during cell sorting. A gate was drawn around the unstained oocysts by using forward scatter (FSC) and side scatter (SSC), with both FSC and SSC measured linearly with a gain of 1, an SSC detector set at 399 V, and an FSC threshold set at 32 V. For each seeding suspension, 100 oocysts were sorted into a 1.5-ml microcentrifuge tube containing 1 ml of aqueous 0.01% Tween 20. In addition to the tubes used for seeding, additional tubes were similarly prepared for microscopic verification and trip controls that are further described below. The oocysts were sent on ice by overnight courier to the USGS Ohio District Microbiology Laboratory and were used within 1 week of cell sorting.
The modified oocysts used during this study came from the manufacturer (Biotechnology Frontiers, North Ryde, New South Wales, Australia) in sealed vials containing approximately 100 Cryptosporidium parvum oocysts. These oocysts were sorted by the manufacturer using flow cytometry after having been gamma irradiated and embedded with a fluorochrome by using a proprietary method. Each vial arrived with a certificate of analysis that documented the mean oocyst concentration and standard deviation in each vial.
Filtration, elution, and concentration of Cryptosporidium from water samples.
The three replicate 10-liter water samples to be analyzed for Cryptosporidium oocysts were sent to the USGS Ohio District Microbiology Laboratory within 48 h of sample collection. At the laboratory, the samples were seeded (when applicable), filtered, eluted, and concentrated by use of method 1623 with some modifications to obtain (i) a regular sample (no seed), (ii) a sample seeded with modified oocysts, and (iii) a sample seeded with viable oocysts by the conventional seeding method.
Samples were processed by using a Hemoflow F80A ultrafilter, a polysulfone hollow-fiber single-use unit with an 80,000 molecular weight cutoff (Fresenius USA, Lexington, Mass.) (25). A peristaltic pump (Geotech Environmental Equipment, Inc., Denver, Colo.) was used to recirculate the water within a closed ultrafiltration system at a pressure of 20 to 25 lb/in2. When the volume of the sample was reduced to the hold-up volume of the ultrafilter system, oocysts were eluted from the ultrafilter by recirculating 250 ml of eluting solution (100 mM phosphate-buffered saline with 1% Laureth 12) at low pressure (5 to 10 lb/in2) through the system. The eluate was collected in a 250-ml centrifuge tube along with any additional liquid that was purged from the ultrafilter with air pressure (less than 25 lb/in2). The oocysts were further concentrated by centrifugation at 1,500 × g and 4°C for 15 min in an International Equipment Company (Needham Heights, Mass.) model IEC PR-7000 M centrifuge with a swinging bucket rotor. The supernatant was aspirated from the concentrated sample to the 5-ml mark on the centrifuge tube or to 3 ml above the pellet, whichever volume was greater. The pellet was transferred to a 15-ml centrifuge tube that was pretreated with SigmaCote (Sigma, St. Louis, Mo.), and the 250-ml centrifuge tube was rinsed twice with sterile reagent water. The pellet was sent to the University of North Carolina (UNC) laboratory at Chapel Hill, N.C., on ice by overnight courier for further processing and analysis.
IMS and staining of Cryptosporidium oocysts.
At the UNC laboratory, anti-Cryptosporidium IMS kits (Dynal Inc., Lake Success, N.Y.) were used to separate oocysts within the samples from other extraneous particulate matter by using the protocol described in method 1623 with one major modification: the acid-based dissociation step was replaced with heat dissociation (35). For heat dissociation, the sample was heated at 80°C for 10 min in a heating block. Only one IMS purification was performed for each sample. Following IMS, samples were transferred to well slides (Meridian Diagnostics, Cincinnati, Ohio) and dried in a desiccating chamber for approximately 2 h or overnight. The slides were fixed with absolute methanol and stained with a fluorescein-labeled direct antibody kit (Crypt-a-Glo, Waterborne, Inc., New Orleans, La.) and counterstained with DAPI (0.02 mg/ml; Sigma). After the slides were stained, a glycerol-1,4-diazabicyclo-[2.2.2] octane (DABCO) mounting medium (Sigma) was added to each well and a coverslip was applied. Slides were stored in the desiccator in the dark until they were microscopically examined, which was done within 72 h of staining.
Quality control samples for Cryptosporidium analysis.
Quality control samples to demonstrate method proficiency included initial precision and recovery (IPR) and ongoing precision and recovery (OPR) experiments, method blanks, IMS controls, and stain controls. Four IPRs and three OPRs were performed by processing three 10-liter volumes of reagent-grade water, one unseeded (method blank) and two seeded with 100 Cryptosporidium oocysts (either viable or modified) as described above. Positive and negative IMS and stain controls were included with each set of samples. The negative IMS control was prepared by replacing the sample with reagent water at the beginning of the IMS procedure. The positive IMS control was prepared by replacing the sample with 1 ml of viable flow-counted oocysts brought to volume with reagent water. IMS controls were then processed in the same manner as the regular environmental water samples. The IMS controls were included to demonstrate the recovery efficiency of the IMS part of the method. Stain controls were processed according to the manufacturer's instructions to ensure reliable oocyst staining during each set of experiments.
Microscopic counts analyzed with each set of samples were used to verify concentrations of oocyst seeds (confirmation controls) or to monitor the effects of travel and time on oocysts (trip controls). For all microscopic counts, a test tube containing oocysts prepared for seeding was shaken by hand for 30 s, the sample was transferred by pipette and filtered through a 0.8-μm-porosity, 13-mm-diameter black polycarbonate filter, and the tube was rinsed with 1 ml of reagent water that was then filtered. The oocysts were stained with 100 μl of 1× Crypta-glo stain solution for 30 min. After the filter was transferred to a glass slide and mounted in DAPCO mounting medium, the oocysts were microscopically enumerated. In this manner, the following counts of viable oocysts prepared at USEPA were done: (i) initial counts at USEPA, (ii) confirmation controls shipped from USGS to UNC and counted at UNC, (iii) trip controls shipped from USGS to USEPA and counted at USEPA, and (iv) trip controls shipped from USGS to UNC to USEPA and counted at USEPA. For modified oocysts, confirmation controls were microscopically analyzed at the UNC laboratory.
Analysis of water samples for microbiological indicators and water quality.
Water samples were analyzed for microbiological indicators and turbidity at the USGS Ohio laboratory within 24 h of sample collection. Samples for enumeration of C. perfringens were analyzed by use of the mCP agar method (29) with the following modifications: samples were not heated to remove vegetative cells, holding times exceeded the recommended 8 h, and plates were incubated at 42°C instead of 44.5°C. For C. perfringens, 30-, 10-, and 3-ml sample volumes were processed to obtain a countable dilution for each sample analyzed. Somatic and F-specific coliphage in 100-ml sample volumes were analyzed by using the single-agar layer method (33). Analyses of water samples for E. coli were done by using the mTEC membrane filtration method (28), and sample volumes of 100, 30, 10, 3, 1, and 0.3 ml were chosen to obtain plates within the ideal count range. Quality assurance and quality control procedures for microbiological indicators are described in detail at the USGS Ohio District Microbiology website (http://oh.water.usgs.gov/micro/lab.html#qcm).
Turbidity was measured by means of a Hach (Loveland, Colo.) model 2100P portable turbidimeter, and all turbidity measurements were made in duplicate. Water samples were also analyzed for concentrations of suspended sediment, nitrite plus nitrate, total phosphorus, silica, and iron at the USGS National Water Quality Laboratory in Lakewood, Colo., by USGS methods (8, 9, 10).
Data analysis and statistical methods.
For environmental water samples that yielded pellet volumes greater than 0.5 ml, the entire sample volume was not examined. Percent recoveries (REC) were adjusted for the pellet volume and calculated as follows:
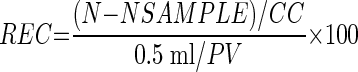 |
(1) |
where N is the number of oocysts found in the seeded environmental sample, NSAMPLE is number of oocysts found in the unseeded environmental sample, CC is the certified oocyst count (100 for viable oocysts or certificate of analysis for modified oocysts, both sorted by flow cytometry), and PV is the pellet volume.
At the 10 sites where two samples were collected, the percent differences in recoveries between the two samples (REC1,2) were calculated as follows:
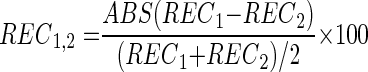 |
(2) |
where ABS is the absolute value, REC1 is the percent recovery for sample 1, and REC2 is percent recovery for sample 2.
Estimated oocysts in environmental samples per 10 liters, adjusted for average recoveries, were calculated as follows:
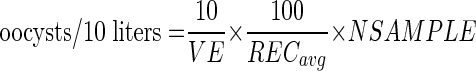 |
(3) |
where VE is volume examined (liters), RECavg is average recovery of modified and viable oocysts, and NSAMPLE is number of oocysts found in the unseeded environmental sample. For samples in which no oocysts were found, the above equation was used with a value of 1 for NSAMPLE, and results were expressed in terms of “less than.”
Because not all of the data were distributed normally, nonparametric statistical methods were used. The Wilcoxon signed-rank test was used to compare two groups of paired data. For comparing more than two groups of unpaired data, the nonparametric analysis of variance, the rank transform test, was used. If the analysis of variance showed significant differences among groups at α = 0.05, the Tukey-Kramer multiple comparison test was used to determine which group's median ranks differed from each other (11).
RESULTS
Quality control samples.
The results of IPRs, OPRs, method blanks, and IMS controls are summarized in Table 2. Recoveries for three out of four IPRs were within acceptable limits for viable and modified oocysts, and the relative standard deviations were within the acceptable percentage, as described in method 1623. All OPRs for Cryptosporidium were within acceptable limits. Although viable oocyst recoveries were higher than modified oocyst recoveries in five out of seven IPR and OPR trials, a paired statistical comparison indicated no significant difference between these two groups. No oocysts were detected in any of the method blanks. Recoveries for the IMS positive controls ranged from 64 to 82% during IPR and OPR experiments.
TABLE 2.
Quality control samples for Cryptosporidium analysis with method 1623
| Sample | Date (mo/day/yr) | % Recovery (RSD) witha:
|
Method blank | % IMS controls (RSD)
|
||
|---|---|---|---|---|---|---|
| Viable oocysts | ColorSeed | Positive | Negative | |||
| IPR samples | ||||||
| 1 | 12/04/2001 | 45 | 34 | 0 | 70 | 0 |
| 2 | 12/11/2001 | 47 | 40 | 0 | 64 | 0 |
| 3 | 01/08/2002 | 41 | 55 | 0 | 69 | 0 |
| 4 | 01/29/2002 | 23 | 31 | 0 | 66 | 0 |
| Avg | 39 (28)b | 40 (27)b | 67 (4) | |||
| OPR samples | ||||||
| 1 | 07/08/2002 | 55 | 51 | 0 | 82 | 0 |
| 2 | 12/09/2002 | 47 | 41 | 0 | 76 | 0 |
| 3 | 06/10/2003 | 57 | 52 | 0 | 50d | 0 |
| Avg | 53 (10)c | 48 (13)c | 0 | 79 (5) | 0 | |
RSD is the relative standard deviation.
Acceptable range for recovery is 24 to 100%; acceptable maximum RSD is 55%.
Acceptable range for recovery is 11 to 100%.
Slide was dropped and cracked, so counts are underestimated; this value was not used in average and statistical analyses.
Microscopic counts for five groups (A through E) of confirmation or trip controls are listed in Table 3. Microscopic counts were always less than certified flow cytometry counts. Certified counts for viable oocysts were 100 for all tubes. Certified counts for modified oocysts were 98 for sample sets 1 and 2 and 99 for the other sample sets. A different analyst counted the oocysts in groups B and E from groups A, C, and D, and different sources of oocysts were counted in groups B and E. Therefore, to determine whether viable oocysts degraded during shipping and handling, the results from microscopic counts of trip controls counted by the same analyst and obtained from the same source were examined by using statistical tests. After removal of outliers that were known to be biased low because of lab error, paired statistical tests between groups A, C, and D were examined. Statistical tests showed no significant difference between groups A and C (P = 0.394) and groups C and D (P = 0.0743). There was, however, a statistically significant difference between groups A and D (P = 0.0007). This result indicates that degradation of viable oocysts occurred as the result of shipping and handling during four-legged trips (from USEPA to USGS to UNC to USEPA [group A to group D]) but not during three-legged trips (from USEPA to USGS to USEPA [group A to group C]).
TABLE 3.
Microscopic counts of viable and modified oocysts in confirmation and trip controls
| Sample set | No. of viable oocysts
|
No. of modified oocysts
|
|||
|---|---|---|---|---|---|
| Initial counts at USEPA (group A)a | Confirmation controls counted at UNC (group B)b | Trip controls from USGS to USEPA counted at USEPA (group C)e | Trip controls from USGS to UNC to USEPA counted at USEPA (group D) | Confirmation counts at UNC (group E) | |
| 1 | 97 | 94 | 94 | 97 | 80d |
| 2 | 98 | 91 | 97 | 94 | 89 |
| 3 | 95 | 88 | 98 | 93 | 97 |
| 4 | 98 | 101 | 99 | 97 | 92 |
| 5 | 93 | 97 | 98 | 92 | 96 |
| 6 | 94 | 88 | 94 | 92 | 93 |
| 7 | 97 | 92 | 92 | 94 | 89 |
| 8 | 96 | 53c | 90 | 88 | 51c |
| 9 | 94 | 90 | ND | 88 | 96 |
| 10 | 93 | 90 | 100 | 93 | 98 |
| 11 | 94 | 94 | 91 | 91 | 84 |
| 12 | 95 | 88 | 92 | 94 | 90 |
| 13 | 96 | 94 | 98 | 94 | 94 |
| 14 | 96 | 88 | 87 | 90 | 90 |
| Avg | 95.4 | 91.9 | 94.6 | 92.6 | 92.3 |
| SD | 1.7 | 4.0 | 4.0 | 2.8 | 4.1 |
Each value is the mean of three counts done by the same analyst.
Each value is the mean of two counts done by the same analyst.
Slide did not stain well, so counts are underestimated; this value was not used in average and statistical analyses.
Slide had air bubbles, so counts are underestimated; this value was not used in average and statistical analyses.
ND, not determined.
Recoveries of oocysts from stream water samples by using modified and conventional seeding procedures.
Cryptosporidium recoveries and associated sample-processing data from stream water samples seeded with viable and modified oocysts are shown in Table 4. The volumes of environmental water samples that were filtered ranged from 5.0 to 10.5 liters. The cubitainers used in this study can hold up to 10.5-liter volumes, but field crews often filled the containers to less than capacity, or water was lost during transport or processing. Because only up to 0.5 ml of concentrated pellet volume was examined by IMS, the volume examined was sometimes less than the volume filtered. For two samples, only about half of the volume was filtered because the filter clogged and filtration was no longer efficient; in these samples, the turbidities were 59 for the Georgia 1 high sample and 2,700 for the Nebraska high sample. Adjusted recoveries of oocysts ranged from 2.0 to 61% for viable oocysts and 3.0 to 59% for modified oocysts. IMS positive control recoveries were higher than adjusted recoveries, ranging from 58 to 87%.
TABLE 4.
Cryptosporidium recoveries in samples seeded with viable and modified oocysts
| Stream water site | Stream flow condition | Viable oocysts
|
Modified oocysts
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Vol filtered (liters) | Pellet vol (ml) | Vol examined (liters) | Adjusted oocyst recovery (%)a | IMS positive controls | Vol filtered (liters) | Pellet vol (ml) | Vol examined (liters) | Adjusted oocyst recovery (%)a | ||
| Alabama 1 | Elevated | 8.3 | 0.3 | 8.3 | 17 | 64 | 9.0 | 0.3 | 9.0 | 22 |
| Alabama 2a | Elevated | 8.3 | 0.1 | 8.3 | 37 | 77 | 8.5 | 0.1 | 8.5 | 59 |
| Alabama 2b | Elevated | 7.0 | 0.5 | 7.0 | 2.0 | 67 | 7.5 | 0.8 | 4.7 | 13 |
| Colorado 1 | Effluent | 9.0 | 1.5 | 3.0 | 24 | 82 | 9.5 | 1.0 | 4.8 | 12 |
| Colorado 2 | Elevated | 8.5 | 0.3 | 8.5 | 20 | 61 | 9.3 | 0.3 | 9.3 | 19 |
| Georgia 1 | Base | 9.5 | 0.3 | 9.5 | 10 | 61 | 8.5 | 0.3 | 8.5 | 9.1 |
| Georgia 1 | High | 5.0 | 0.5 | 5.0 | 10 | 67 | 5.0 | 0.5 | 5.0 | 13 |
| Georgia 2 | High | 8.8 | 1.0 | 4.4 | 14 | 67 | 9.0 | 0.5 | 9.0 | 3.1 |
| Indiana | Elevated | 9.0 | 0.5 | 9.0 | 32 | 50b | 9.3 | 0.5 | 9.3 | 29 |
| Iowa | Base | 8.8 | 0.2 | 8.8 | 29 | 76 | 8.8 | 0.2 | 8.8 | 22 |
| Iowa | High | 9.8 | 1.0 | 4.9 | 2.0 | 74 | 9.8 | 1.0 | 4.9 | 4.0 |
| Louisiana | Base | 8.8 | 0.1 | 8.8 | 61 | 87 | 9.5 | 0.1 | 9.5 | 51 |
| Louisiana | Elevated | 8.0 | 0.5 | 8.0 | 23 | 76 | 9.0 | 0.5 | 9.0 | 20 |
| Nebraska | High | 3.0 | 0.7 | 2.1 | 2.8 | 61 | 5.0 | 1.5 | 1.6 | 3.0 |
| Nebraska | Base | 9.5 | 0.5 | 9.5 | 42 | 77 | 9.5 | 0.5 | 9.5 | 32 |
| Ohio | Elevated | 9.8 | 0.3 | 9.8 | 15 | 61 | 9.8 | 0.3 | 9.8 | 25 |
| Ohio | High | 9.8 | 0.5 | 9.8 | 40 | 82 | 9.3 | 0.5 | 9.3 | 51 |
| South Carolina 1 | Base | 9.8 | 0.1 | 9.8 | 49 | 67 | 9.8 | 0.1 | 9.8 | 30 |
| South Carolina 1 | Elevated | 10 | 0.3 | 10 | 32 | 58 | 10.5 | 0.3 | 10.5 | 28 |
| South Carolina 2 | Elevated | 10 | 0.6 | 8.3 | 17 | 58 | 9.8 | 0.6 | 8.1 | 21 |
| Texas | Elevated | 7.5 | 0.8 | 4.7 | 18 | 87 | 8.2 | 1.0 | 4.1 | 12 |
| Texas | Base | 9.5 | 0.4 | 9.5 | 25 | 79 | 8.5 | 0.4 | 8.5 | 24 |
| Virginia 1 | Base | 8.5 | 0.1 | 8.5 | 21 | 77 | 8.5 | 0.1 | 8.5 | 12 |
| Virginia 2 | Elevated | 9.0 | 0.2 | 9.0 | 34 | 74 | 9.5 | 0.2 | 9.5 | 42 |
| Washington 1a | Base | 10 | 0.2 | 10 | 28 | 79 | 10 | 0.2 | 10 | 25 |
| Washington 1b | Base | 10 | 0.5 | 10 | 21 | 82 | 10 | 0.5 | 10 | 23 |
| Washington 2 | High | 8.5 | 0.5 | 8.5 | 10 | 67 | 7.8 | 0.5 | 7.8 | 15 |
| Wisconsin 1 | High | 8.5 | 2.5 | 1.7 | 15 | 61 | 9.5 | 2.5 | 1.9 | 20 |
| Wisconsin 2 | Base | 9.0 | 0.9 | 5.0 | 27 | 79 | 8.8 | 0.8 | 6.5 | 37 |
| Wisconsin 2 | High | 7.0 | 0.5 | 7.0 | 38 | 74 | 7.0 | 0.6 | 5.8 | 29 |
| Avg | 8.4 | 0.5 | 7.3 | 23.8 | 71.8 | 8.6 | 0.6 | 7.7 | 23.6 | |
If pellet volume was greater than 0.5 ml, the entire sample volume was not examined. Oocyst recovery was adjusted for the pellet volume examined.
Slide was dropped and cracked, so counts are underestimated; this value was not used in average and statistical analyses.
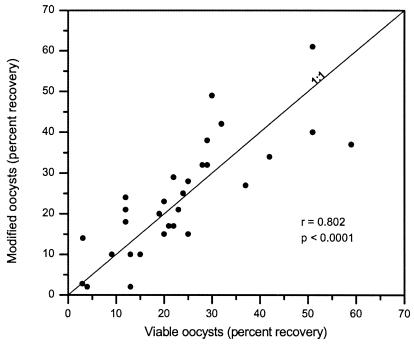
Recoveries of viable oocysts are compared to recoveries of modified oocysts for each sample on a scatter plot (Fig. 1). The recoveries between the two seeding procedures were highly correlated (r = 0.802), and the data were evenly scattered around the equal recovery line. Results from a paired two-way test showed that modified and viable oocyst recoveries were not significantly different (P = 0.8128). Because there was no significant difference between modified and viable oocyst recoveries, average recoveries were used in all subsequent data analyses.
FIG. 1.
Recoveries of viable oocysts (conventional seeding procedure) compared to recoveries of modified oocysts; r, Pearson's correlation coefficient; p, significance of the correlation.
Relation between Cryptosporidium recoveries and water quality characteristics.
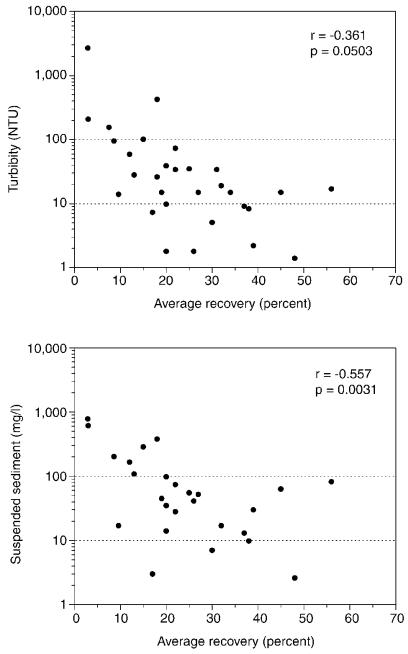
The average recoveries obtained by using viable and modified oocysts were used to examine the relations between average recoveries and water quality properties and constituents (Table 5). Correlation analysis showed no significant relation (α = 0.05) between average recovery and pH, specific conductance, dissolved iron or silica, or alkalinity. Significant negative correlations were found between average oocyst recovery and turbidity or suspended sediment (Fig. 2). For samples with turbidities greater than 100 nephelometric turbidity units (NTU) and suspended-sediment concentrations greater than 100 mg/liter (Fig. 2, top of each graph), average recoveries were less than 20%. For samples with turbidities in the middle range (10 to 100 NTU), a wide range of average recoveries was found (8.6 to 56%); for those in the low range (<10 NTU), a slightly narrower range of average recoveries was found (17 to 48%). A similar pattern was seen in a comparison of average recoveries for suspended-sediment results.
TABLE 5.
Physical and chemical water quality analyses of stream water samplesa
| Stream water site | Stream flow condition | % Average recovery of modified and viable oocysts | % Difference in recoveries for samples from the same site | pH | Specific conductance (μS/cm) | Dissolved iron (mg/liter) | Dissolved silica (mg/liter) | Turbidity (NTU) | Suspended sediment (mg/liter) | Alkalinity (mg/liter) |
|---|---|---|---|---|---|---|---|---|---|---|
| Alabama 1 | Elevated | 20 | 7.8 | 165 | <10 | 6.2 | 39 | 35 | 63 | |
| Alabama 2a | Elevated | 48 | 146 | 8.0 | 86 | 25 | 4.5 | 1.4 | 2.6 | 24 |
| Alabama 2b | Elevated | 7.5 | 7.8 | 96 | 172 | 3.5 | 155 | ND | 39 | |
| Colorado 1 | Effluent | 18 | 6.5 | 3,850 | 384 | 15 | 26 | ND | ND | |
| Colorado 2 | Elevated | 20 | 8.1 | 1,380 | 24 | 10 | 9.8 | 14 | 189 | |
| Georgia 1 | Base | 9.6 | 22 | 7.1 | 186 | 107 | 8.4 | 14 | 17 | 31 |
| Georgia 1 | High | 12 | 7.1 | 190 | 125 | 7.1 | 59 | 165 | 20 | |
| Georgia 2 | High | 8.6 | 7.1 | 80 | 70 | 8.2 | 95 | 202 | 21 | |
| Indiana | Elevated | 31 | 8.0 | 662 | 684 | 7.6 | 34 | ND | 223 | |
| Iowa | Base | 26 | 159 | 7.5 | 906 | 18 | 17 | 1.8 | 41 | 313 |
| Iowa | High | 3.0 | 8.0 | 751 | <10 | 20 | 208 | 610 | 244 | |
| Louisiana | Base | 56 | 87 | 7.8 | 467 | 72 | 17 | 17 | 82 | 149 |
| Louisiana | Elevated | 22 | 6.8 | 199 | 144 | 9.1 | 34 | 74 | 64 | |
| Nebraska | High | 2.9 | 171 | 8.1 | 545 | <10 | 14 | 2,700 | 778 | 232 |
| Nebraska | Base | 37 | 8.4 | 533 | <10 | 15 | 9.1 | 13 | 205 | |
| Ohio | Elevated | 20 | 79 | 7.8 | 736 | ND | ND | 1.8 | 98 | ND |
| Ohio | High | 46 | 8.1 | 632 | 8.0 | 7.3 | 15 | 63 | 234 | |
| South Carolina 1 | Base | 40 | 29 | 7.1 | 132 | 51 | 5.6 | 2.2 | 30 | 30 |
| South Carolina 1 | Elevated | 30 | 6.3 | 108 | 143 | 7.5 | 5.1 | 7.0 | 21 | |
| South Carolina 2 | Elevated | 19 | 6.2 | 48 | 153 | 3.4 | 15 | 45 | 5.0 | |
| Texas | Elevated | 15 | 50 | 7.8 | 447 | 7.0 | 4.0 | 101 | 287 | 126 |
| Texas | Base | 25 | 7.3 | 667 | 18 | 7.8 | 35 | 55 | 100 | |
| Virginia 1 | Base | 17 | 7.1 | 345 | 53 | 7.8 | 7.3 | 3.0 | 60 | |
| Virginia 2 | Elevated | 38 | 8.0 | 443 | 15 | 3.6 | 8.3 | 9.8 | 183 | |
| Washington 1a | Base | 27 | 20 | 8.0 | 327 | 22 | 22 | 15 | 52 | 112 |
| Washington 1b | Base | 22 | 8.0 | 406 | <10 | 30 | 73 | 28 | 136 | |
| Washington 2 | High | 13 | 7.3 | 72 | 85 | 6.4 | 28 | 108 | 24 | |
| Wisconsin 1 | High | 18 | 7.5 | 519 | 36 | 4.9 | 425 | 378 | 190 | |
| Wisconsin 2 | Base | 32 | 6 | 8.3 | 724 | 13 | 7.3 | 19 | 17 | 202 |
| Wisconsin 2 | High | 34 | 7.8 | 714 | ND | ND | 15 | ND | 192 |
ND, not determined.
FIG. 2.
Relations between average recoveries of oocysts (viable and modified) and water quality variables; r, Pearson's correlation coefficient; p, significance of the correlation.
At the 10 sites where two samples were collected, the percent differences in average recoveries between the two samples were calculated and ranged from 6 to 171% (Table 5). The three largest percent differences in recoveries (Alabama 2, Iowa, and Nebraska) were found for sample pairs that also had large differences in turbidities. The lowest percent difference (Wisconsin 2) was a sample pair collected at base and high flows with similar turbidities. Of the remaining six sample pairs in the intermediate group, all but two had higher recoveries in the sample with lower turbidity.
Concentrations of Cryptosporidium oocysts and microbiological and chemical indicators.
Wide ranges of concentrations of E. coli, C. perfringens, somatic and F-specific coliphage, and chemical constituents associated with fecal contamination were found in the stream water samples (Table 6). The only sample in which E. coli was not detected was a high-flow sample from the Wisconsin 2 site, a mixed urban and agricultural area with the potential for contamination from septic systems, feedlots, manure, and wastewater treatment plants. C. perfringens and somatic coliphage were detected in all samples, and F-specific coliphage was detected in 72% of the samples. The highest concentrations of C. perfringens (Nebraska), somatic coliphage (Alabama 2), and F-specific coliphage (Washington 1) were found at three strictly agricultural sites. Two samples from Iowa had concentrations of nitrate above the maximum contaminant level for drinking water (10 mg/liter).
TABLE 6.
Cryptosporidium oocyst detections and concentrations of E. coli, C. perfringens, somatic and F-specific coliphage, and water quality constituents in stream water samplesa
| Stream water site | Stream flow condition |
Cryptosporidium
|
E. coli (CFU/ 100 ml) | C. perfringens (CFU/ 100 ml) | Somatic coliphage (PFU/100 ml) | F-specific coliphage (PFU/100 ml) | Nitrite plus nitrate (mg/liter) | Total phosphorus (mg/liter) | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Oocysts (per 10 liters) | Vol examined (liters) | Estimated oocysts adjusted for avg recovery (per 10 liters) | ||||||||
| Alabama 1 | Elevated | 1 | 9.0 | 6 | 260 | 20 | 120 | 9 | 1.52 | 0.177 |
| Alabama 2a | Elevated | 0 | 8.0 | <2 | 7 | 12 | 18 | 1 | 1.31 | 0.002 |
| Alabama 2b | Elevated | 6 | 5.0 | 80 | 1,800 | 250 | >3,000 | 5 | ND | ND |
| Colorado 1 | Effluent | 3 | 3.0 | 18 | 30 | 240 | 180 | 23 | ND | ND |
| Colorado 2 | Elevated | 0 | 9.2 | <6 | 19 | 150 | 440 | 35 | 7.02 | 0.910 |
| Georgia 1 | Base | 0 | 8.3 | <13 | 23 | 120 | 15 | 3 | 2.72 | 0.070 |
| Georgia 1 | High | 0 | 5.0 | <17 | 730 | 190 | 220 | 50 | 0.83 | 0.136 |
| Georgia 2 | High | 0 | 5.3 | <22 | 2,000 | 470 | 250 | 66 | 0.60 | 0.170 |
| Indiana | Elevated | 0 | 9.2 | <4 | 110 | 67 | 74 | <1 | 0.92 | 0.135 |
| Iowa | Base | 0 | 9.0 | <4 | 11 | 20 | 46 | <1 | 17.8 | 0.015 |
| Iowa | High | 0 | 5.0 | <66 | 510 | 200 | 200 | 4 | 21.1 | 0.490 |
| Louisiana | Base | 0 | 9.5 | <2 | 390 | 200 | 30 | 15 | 0.08 | 0.380 |
| Louisiana | Elevated | 0 | 7.5 | <6 | 2,200 | 270 | 800 | 360 | 0.35 | 0.310 |
| Nebraska | High | 0 | 1.6 | <215 | 18,000 | 900 | 1,400 | 29 | 3.37 | 1.09 |
| Nebraska | Base | 0 | 9.5 | <3 | 7 | 70 | 17 | <1 | 0.96 | 0.163 |
| Ohio | Elevated | 0 | 9.2 | <5 | 200 | 30 | 260 | 10 | 4.10 | 0.080 |
| Ohio | High | 0 | 9.5 | <2 | 210 | 80 | 150 | 3 | 4.57 | 0.061 |
| South Carolina 1 | Base | 0 | 9.5 | <3 | 140 | 45 | 150 | 84 | 2.19 | 0.010 |
| South Carolina 1 | Elevated | 0 | 10.2 | <3 | 180 | 150 | 270 | <1 | 1.98 | 0.049 |
| South Carolina 2 | Elevated | 0 | 8.3 | <6 | 93 | 260 | 11 | 2 | 0.23 | 0.055 |
| Texas | Elevated | 0 | 4.0 | <17 | 160 | 56 | 130 | 4 | 2.05 | 0.490 |
| Texas | Base | 2 | 8.5 | 10 | 2,700 | 100 | 31 | 6 | 8.43 | 1.94 |
| Virginia 1 | Base | 0 | 8.8 | <7 | 41 | 35 | 10 | 1 | 0.65 | 0.033 |
| Virginia 2 | Elevated | 0 | 9.8 | <3 | 12 | 22 | 480 | <1 | 6.62 | 0.069 |
| Washington 1a | Base | 1 | 10 | 4 | 300 | 28 | 1,300 | 620 | 0.87 | 0.188 |
| Washington 1b | Base | 0 | 10 | <4 | 170 | 90 | 24 | <1 | 2.96 | 0.266 |
| Washington 2 | High | 0 | 8.8 | <9 | 770 | 400 | 94 | 5 | 0.43 | 0.156 |
| Wisconsin 1 | High | 0 | 1.7 | <33 | 13,000 | 190 | 2,800 | <1 | 4.97 | 0.620 |
| Wisconsin 2 | Base | 0 | 4.6 | <7 | 100 | 120 | 16 | 5 | <0.05 | 0.180 |
| Wisconsin 2 | High | 0 | 6.2 | <5 | <1 | 200 | 100 | <1 | 0.97 | 0.126 |
| Median | 180 | 120 | 140 | 4 | 1.75 | 0.160 | ||||
ND, not determined.
Five samples were positive for Cryptosporidium, two at elevated flow, two at base flow, and one from effluent (Table 6). Concentrations in the five positive samples, not adjusted for average recoveries, ranged from 1 to 6 oocysts/10 liters. When these concentrations were adjusted for average recoveries, they increased to a range of 4 to 80 oocysts/10 liters. Detection limits, adjusted for recoveries, ranged from <2 to <215 oocysts/10 liters. The five positive samples were in unseeded samples; no endemic oocysts were found in any of the samples seeded with modified oocysts. Of the four positive stream water samples, three were from agricultural areas where animals were present (Alabama 1 and 2b and Washington 1) and one was from a mixed urban and agricultural area (Texas). Both Alabama sites were positive for Cryptosporidium; feedlots are upstream from Alabama 1, and free-range cattle have access to the stream at Alabama 2. The Texas site represents a large watershed, whereas the other positive sites are smaller watersheds (Table 1). The Alabama 1 site has the highest population density and the Washington 1 site has the lowest population density (Table 1) among the four stream water sites positive for Cryptosporidium. Because oocysts were found in a small percentage of samples (16.7%), the relations between detections of Cryptosporidium and concentrations of microbiological and chemical indicators could not be determined.
DISCUSSION
Method 1623 is widely used to monitor source waters and drinking water supplies and is the required method in the proposed Long-Term 2 Enhanced Surface Water Treatment Rule (34). Therefore, modifications to reduce analytical costs and to improve recoveries are needed. In this study, a variety of stream water samples were collected from diverse geographical locations throughout the United States to test a new product, ColorSeed, to determine if it would yield results comparable to those of conventional seeding procedures. This study also afforded the opportunity to collect data on the use of method 1623 for routine monitoring. We demonstrated the importance of including appropriate quality control samples in any monitoring program, examined the effects of water quality variables on method recovery efficiency of oocysts, and determined that the low percentage of environmental samples positive for Cryptosporidium precluded examining the relations between detections of oocysts and concentrations of potential microbiological or chemical indicators of fecally impacted waters.
Quality control samples provide insight into method efficiency and laboratory performance. IPRs, OPRs, and method blanks are required components of method 1623, whereas IMS controls are not required. Confirmation and trip control microscopic counts of oocysts to be used in matrix spikes are also not required components of method 1623. In this study, most IPRs and all OPRs were within acceptable limits and were comparable for viable and modified oocysts. IMS controls were always higher than IPR and OPR recoveries (except when there was a laboratory error), indicating that oocysts were consistently lost during the filtration and concentration steps. The IMS controls indicated that the IMS reagents and process were working to the level expected. In examining confirmation and trip controls, microscopic counts were always less than certified counts of viable or modified oocysts. This observation could be due to oocyst degradation over time and during travel, the inability to transfer all the seed oocysts from their container to the sample, losses during other processing steps, or a failure to identify all oocysts on the slides by the analyst. Nevertheless, the differences showed minimal effects of travel and time on the condition of oocysts except when oocysts traveled through a four-legged trip (groups A and D). During analysis of environmental samples in this study, each set of samples traveled a three-legged trip (from USEPA to USGS to UNC).
As specified by method 1623, matrix spikes are required for every 20th sample or when a new water source is tested. We included a matrix spike for every sample analyzed. We found that, in some cases, recoveries were quite different between two samples collected at the same site, especially when turbidities were different. We also found that when average recoveries were considered in the calculations for some samples, concentrations of ambient oocysts increased considerably over those calculated without considering average recoveries. Determining recovery efficiency also provided data to calculate detection limits; in this study, a wide range of detection limits was found among sites and samples. It would appear prudent to collect and process matrix spikes not only when a new water source is tested but also when a sample is collected at a different stream flow or turbidity than previously collected and tested at a site.
During the course of this study, we found no statistically significant differences in recoveries when modified seeding procedures were used compared to recoveries when conventional seeding procedures were used. This finding is in contrast to a recently published Australian study of 247 raw water and 247 finished water samples in which recoveries were significantly lower for ColorSeed oocysts than for unmodified oocysts (36). The authors maintained, however, that the differences in recoveries were small, with mean value differences ranging from 3 to 5%, and concluded that ColorSeed is suitable for use as a control for routine monitoring (36). Our study supports this conclusion and showed that the use of modified oocysts was comparable to conventional seeding procedures. Unfortunately, we did not have the simultaneous occurrence of modified and environmental oocysts, so we could not evaluate the recognition of modified oocysts in a mixed sample.
In this study, average recoveries of oocysts (modified and viable) ranged from 2.9 to 56%. This wide range is similar to those found in other studies using method 1622 or 1623. In the analysis of 11 stream water samples, percent recoveries ranged from 2 to 63% (26). In the ICRSS, 65% of the 430 samples collected from 87 source waters had recoveries ranging from 20 to 60%, 13% had recoveries less than 20%, and 22% had recoveries greater than 60% (32). Kuhn and Oshima (15) analyzed 19 surface water sites and found that recoveries ranged from 9.3 to 87.7%.
For the environmental waters processed during this study, turbidity and suspended sediment were the only measured water quality characteristics that were significantly related to recoveries. An inverse relation was found for these characteristics and method efficiency, resulting in a decrease in recovery for high turbidities or suspended-sediment concentrations (>100 NTU or >100 mg/liter, respectively). These results are similar to those of other studies in which the effect of turbidity was investigated by adding known numbers of organisms to whole-water samples before the samples were processed by using method 1622 or 1623. Using a high-volume capsule filter, investigators found that recoveries of oocysts from source waters ranged from 36 to 75%; the lowest recovery was in the sample with the highest turbidity (99 NTU), and no relation between turbidity and recovery was found for samples with low to moderate turbidities (7). Kuhn and Oshima (15) found similar results with an analysis of 19 surface water samples. When turbidity was greater than 40 NTU, some decreases in recovery did occur; no relation was observed between oocyst recoveries and low or moderate turbidities (15). In this study, the effect of turbidity on recoveries was shown on a site-by-site basis at the 10 sites where we collected samples twice (Table 5). Of the 10 sample pairs, only 2 pairs behaved contrary to conventional wisdom (Georgia 1 and Ohio); that is, for these two samples, average recovery was higher when turbidity was lower. This result demonstrates that one can generally expect lower recoveries with higher turbidities at the same sampling site.
In this study, initial water pH, specific conductance, dissolved iron, dissolved silica, and alkalinity did not affect recoveries of oocysts. Kuhn et al. (16) found that the IMS kit did not adequately maintain an optimum pH of 7.0 in some water samples. By adjusting the pH of concentrated water samples to optimum pH, recoveries of oocysts increased by 26.4% compared to recoveries from samples where pH was not adjusted (16). We did not evaluate the pH for the concentrated water sample prior to IMS but instead found that the initial sample pH and alkalinity did not affect recovery efficiency. Yakub and Stadterman-Knauer (38) found that dissolved iron concentrations greater than 4 mg/liter affected recoveries of oocysts; however, this concentration was greater than iron concentrations found in the course of our study (the highest dissolved iron concentration was 684 μg/liter) or in typical flowing surface waters (12). Similarly, silica concentrations in this study were typical of those in natural waters (1 to 30 mg/liter) (12) and did not affect recoveries of oocysts. In the ICRSS, total organic carbon concentrations, total suspended solids, temperature, dissolved organic carbon, bromide, ammonia, and UV254 did not affect method performance. Total dissolved solids, pH, specific conductance, and alkalinity weakly affected method performance (32).
Considerable effort was taken to collect samples in streams with potential sources of fecal contamination and under conditions with a greater potential for contamination of water samples. Nevertheless, Cryptosporidium oocysts were detected in only 16.7% of samples collected during this study and at low concentrations. In this study, concentrations ranged from 1 to 6 oocysts per 10 liters before adjustment for recoveries and 4 to 80 oocysts per 10 liters after adjustment for recoveries. Similar detection frequencies and lower concentrations were found in other recent source water studies wherein samples were analyzed by use of method 1622 or 1623. For the ICRSS, 12% of the source water samples were positive for oocysts (32). In a study of six source waters (17), Cryptosporidium oocysts were detected in 60 of 593 (10.1%) samples. Average concentrations, not adjusted for recoveries, ranged from <0.01 to 0.69 oocysts per 10 liters; the highest concentration was 10.17 oocysts per 10 liters (17).
In summary, variable recoveries of oocysts from the studied source waters suggest that water resource managers should consider increasing the frequency of the collection of matrix spikes. This is especially important when source water quality conditions change in response to increased stream flow. The use of a modified seeding procedure provided a reliable estimate of oocyst recoveries compared to the conventional seeding procedure and could be used to reduce the cost of additional matrix spikes. Turbidity and suspended-sediment concentrations affected oocyst recoveries, especially when turbidity was examined on a site-by-site basis.
Acknowledgments
This research was funded by an Interagency Agreement between the U.S. Environmental Protection Agency Office of Research and Development and the U.S. Geological Survey.
Use of trade, product, or firm names is for descriptive purposes only and does not imply endorsement by the U.S. Government.
REFERENCES
- 1.Arrowood, M. J., and K. Donaldson. 1996. Improved purification methods for calf-derived Cryptosporidium parvum using discontinuous sucrose and cesium chloride gradients. J. Eukaryot. Microbiol. 43:89S. [DOI] [PubMed] [Google Scholar]
- 2.Atherholt, T. B., M. W. LeChevallier, W. D. Norton, J. J. Voorhees, and J. S. Rosen. 1998. Effect of rainfall on Giardia and Cryptosporidium. J. Am. Water Works Assoc. 90:66-80. [Google Scholar]
- 3.Bonadonna, L., R. Briancesco, M. Ottaviani, and E. Veschetti. 2002. Occurrence of Cryptosporidium oocysts in sewage effluents and correlation with microbial, chemical, and physical water variables. Environ. Monit. Assess. 75:241-252. [DOI] [PubMed] [Google Scholar]
- 4.Bukhari, Z., R. M. McCuin, C. R. Fricker, and J. L. Clancy. 1998. Immunomagnetic separation of Cryptosporidium parvum from source water samples of various turbidities. Appl. Environ. Microbiol. 64:4495-4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campbell, A., and H. Smith. 1997. Immunomagnetic separation of Cryptosporidium oocysts from water samples: round robin comparison of techniques. Water Sci. Technol. 35:397-401. [Google Scholar]
- 6.Cicmanec, J. L., and D. J. Reasoner. 1997. Enhanced production of Cryptosporidium parvum oocysts in immunosuppressed mice, p. 127-132. In C. R. Fricker, J. L. Clancy, and P. A. Rochelle (ed.), Proceedings of the International Symposium on Waterborne Cryptosporidium. American Water Works Association, Denver, Colo.
- 7.DiGiorgio, C. L., D. A. Gonzalez, and C. C. Huitt. 2002. Cryptosporidium and Giardia recoveries in natural waters by using Environmental Protection Agency method 1623. Appl. Environ. Microbiol. 68:5952-5955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fishman, M. J. 1993. Methods of analysis by the U.S. Geological Survey National Water Quality Laboratory—determination of inorganic and organic constituents in water and fluvial sediments. U.S. Geological Survey open-file report 93-125. U.S. Geological Survey, U.S. Department of the Interior, Denver, Colo.
- 9.Fishman, M. J., and L. C. Friedman. 1989. Methods for determination of inorganic substances in water and fluvial sediments. U.S. Geological Survey TWRI 5A1. U.S. Geological Survey, U.S. Department of the Interior, Washington, D.C.
- 10.Guy, H. P. 1969. Laboratory theory and methods for sediment analysis. U.S. Geological Survey TWRI 5C1. U.S. Geological Survey, U.S. Department of the Interior, Washington, D.C.
- 11.Helsel, D. R., and R. M. Hirsch. 1992. Statistical methods in water resources. Elsevier Science Publishing Company, New York, N.Y.
- 12.Hem, J. D. 1989. Study and interpretation of the chemical characteristics of natural water, 3rd ed. U.S. Geological Survey water supply paper 2254. U. S. Geological Survey, U.S. Department of the Interior, Reston, Va.
- 13.Hirsch, R. M., W. M. Alley, and W. G. Wilber. 1988. Concepts for a national-water quality assessment program. U.S. Geological Survey circular 1021. U. S. Geological Survey, U.S. Department of the Interior, Reston, Va.
- 14.Kennedy, E. J. 1984. Discharge ratings at gaging stations. U.S. Geological Survey TWRI 3A10. U.S. Geological Survey, U.S. Department of the Interior, Washington, D.C.
- 15.Kuhn, R. C., and K. H. Oshima. 2002. Hollow-fiber ultrafiltration of Cryptosporidium parvum oocysts from a wide variety of 10-L surface water samples. Can. J. Microbiol. 48:542-549. [DOI] [PubMed] [Google Scholar]
- 16.Kuhn, R. C., C. M. Rock, and K. H. Oshima. 2002. Effects of pH and magnetic material on immunomagnetic separation of Cryptosporidium oocysts from concentrated water samples. Appl. Environ. Microbiol. 68:2066-2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.LeChevallier, M. W., G. D. Di Giovanni, J. L. Clancy, Z. Bukhari, S. Bukhari, J. S. Rosen, J. Sobrinho, and M. M. Frey. 2003. Comparison of method 1623 and cell culture-PCR for detection of Cryptosporidium spp. in source waters. Appl. Environ. Microbiol. 69:971-979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.LeChevallier, M. W., W. D. Norton, and R. G. Lee. 1991. Occurrence of Giardia and Cryptosporidium spp. in surface water supplies. Appl. Environ. Microbiol. 57:2610-2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Payment, P., A. Berte, M. Prevost, B. Menard, and B. Barbeau. 2000. Occurrence of pathogenic microorganisms in the Saint Lawrence River (Canada) and comparison of health risks for populations using it as their source of drinking water. Can. J. Microbiol. 46:565-576. [PubMed] [Google Scholar]
- 20.Payment, P., and E. Franco. 1993. Clostridium perfringens and somatic coliphages as indicators of the efficiency of drinking water treatment for viruses and protozoan cysts. Appl. Environ. Microbiol. 59:2418-2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rose, J. B., H. Darbin, and C. P. Gerba. 1988. Correlations of the protozoa, Cryptosporidium and Giardia, with water quality variables in a watershed. Water Sci. Technol. 20:271-276. [Google Scholar]
- 22.Rose, J. B., C. P. Gerba, and W. Jakubowski. 1991. Survey of potable water supplies for Cryptosporidium and Giardia. Environ. Sci. Technol. 25:1393-1400. [Google Scholar]
- 23.Rouquet, V., F. Homer, J. M. Brignon, P. Bonne, and J. Cavard. 2000. Source and occurrence of Giardia and Cryptosporidium in Paris rivers. Water Sci. Technol. 41:79-86. [Google Scholar]
- 24.Shelton, L. R. 1994. Field guide for collecting and processing stream-water samples for the National Water-Quality Assessment Program. U.S. Geological Survey open-file report 94-455. U. S. Geological Survey, U.S. Department of the Interior, Sacramento, Calif.
- 25.Simmons, O. D., III, M. D. Sobsey, C. D. Heaney, F. W. Schaefer III, and D. S. Francy. 2001. Concentration and detection of Cryptosporidium oocysts in surface water samples by method 1622 using ultrafiltration and capsule filtration. Appl. Environ. Microbiol. 67:1123-1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Simmons, O. D., III, M. D. Sobsey, F. W. Schaefer III, D. S. Francy, R. A. Nally, and C. D. Heany. 2001. Evaluation of USEPA method 1622 for detection of Cryptosporidium oocysts in stream waters. J. Am. Water Works Assoc. 93:78-87. [Google Scholar]
- 27.U.S. Bureau of the Census. 1991. Census of population and housing, 1990. Data Tech. Doc., Public Law 94-171. Census Bureau, Department of Commerce, Washington, D.C.
- 28.U.S. Environmental Protection Agency. 1985. Test method for Escherichia coli and enterococci in water by the membrane-filter procedure. U.S. Environmental Protection Agency publication no. 600/4-85/076. U.S. Environmental Protection Agency, Cincinnati, Ohio.
- 29.U. S. Environmental Protection Agency. 1996. EPA Information Collection Rule microbial laboratory manual. U.S. Environmental Protection Agency publication no. 600/R-95/178. U.S. Environmental Protection Agency, Washington, D.C.
- 30.U.S. Environmental Protection Agency. 2001. Method 1622—Cryptosporidium and Giardia in water by filtration/IMS/FA. U.S. Environmental Protection Agency publication no. 821/R-01-026. U.S. Environmental Protection Agency, Washington, D.C.
- 31.U.S. Environmental Protection Agency. 2001. Method 1623—Cryptosporidium and Giardia in water by filtration/IMS/FA. U.S. Environmental Protection Agency publication no. 821/R-01-025. U.S. Environmental Protection Agency, Washington, D.C.
- 32.U.S. Environmental Protection Agency. 2001. Implementation and results of the information collection rule supplemental surveys. U.S. Environmental Protection Agency publication no. 815-R-01-003. U.S. Environmental Protection Agency, Washington, D.C.
- 33.U.S. Environmental Protection Agency. 2001. Method 1602—male-specific and somatic coliphage in water by single agar layer (SAL) procedure. U.S. Environmental Protection Agency publication no. 821/R-01-029. U.S. Environmental Protection Agency, Washington, D.C.
- 34.U. S. Environmental Protection Agency. 2003. Source water monitoring guidance manual for public water systems for the Long Term 2 Enhanced Surface Water Treatment Rule (LT2 Rule). U.S. Environmental Protection Agency publication no. 815/D-03-005. U.S. Environmental Protection Agency, Washington, D.C.
- 35.Ware, M. W., L. Wymer, H. Lindquist, and F. W. Schaefer III. 2003. Evaluation of an alternative IMS dissociation procedure for use with method 1622—detection of Cryptosporidium in water. J. Microbiol. Methods 55:575-583. [DOI] [PubMed] [Google Scholar]
- 36.Warnecke, M., C. Weir, and G. Vesey. 2003. Evaluation of an internal positive control for Cryptosporidium and Giardia testing in water samples. Lett. Appl. Microbiol. 37:244-248. [DOI] [PubMed] [Google Scholar]
- 37.Wilde, F. D., and D. B. Radtke. 1998. National field manual for the collection of water-quality data—field measurements. U.S. Geological Survey TWRI 9A6. U.S. Geological Survey, U.S. Department of the Interior, Reston, Va.
- 38.Yakub, G. P., and K. L. Stadterman-Knauer. 2000. Evaluation of immunomagnetic separation for recovery of Cryptosporidium parvum and Giardia duodenalis from high-iron matrices. Appl. Environ. Microbiol. 66:3628-3631. [DOI] [PMC free article] [PubMed] [Google Scholar]