Abstract
Assessing and predicting bloom dynamics and toxin production by Microcystis requires analysis of toxic and nontoxic Microcystis genotypes in natural communities. We show that genetic differentiation of Microcystis colonies based on rRNA internal transcribed spacer (ITS) sequences provides an adequate basis for recognition of microcystin producers. Consequently, ecological studies of toxic and nontoxic cyanobacteria are now possible through studies of rRNA ITS genotypic diversity in isolated cultures or colonies and in natural communities. A total of 107 Microcystis colonies were isolated from 15 lakes in Europe and Morocco, the presence of microcystins in each colony was examined by matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS), and they were grouped by rRNA ITS denaturing gradient gel electrophoresis (DGGE) typing. Based on DGGE analysis of amplified ITSa and ITSc fragments, yielding supplementary resolution (I. Janse et al., Appl. Environ. Microbiol. 69:6634-6643, 2003), the colonies could be differentiated into 59 classes. Microcystin-producing and non-microcystin-producing colonies ended up in different classes. Sequences from the rRNA ITS of representative strains were congruent with the classification based on DGGE and confirmed the recognition of microcystin producers on the basis of rRNA ITS. The rRNA ITS sequences also confirmed inconsistencies reported for Microcystis identification based on morphology. There was no indication for geographical restriction of strains, since identical sequences originated from geographically distant lakes. About 28% of the analyzed colonies gave rise to multiple bands in DGGE profiles, indicating either aggregation of different colonies, or the occurrence of sequence differences between multiple operons. Cyanobacterial community profiles from two Dutch lakes from which colonies had been isolated showed different relative abundances of genotypes between bloom stages and between the water column and surface scum. Although not all bands in the community profiles could be matched with isolated colonies, the profiles suggest a dominance of nontoxic colonies, mainly later in the season and in scums.
Mass occurrences (blooms) of toxic cyanobacteria from the genus Microcystis constitute a threat to the safety and ecological quality of surface waters worldwide. The most prominent toxin produced by Microcystis is the hepatotoxin microcystin, a cyclic heptapeptide which is formed nonribosomally by peptide and polyketide synthetases (3). Proper assessment of the hazards of Microcystis blooms necessitates rapid and reliable methods for microcystin detection. For predictions of the development of microcystin concentrations, tools and insights for understanding the dynamics of microcystin production are required. Environmental factors may affect microcystin production in Microcystis cultures by a factor of 3 to 4 (24). However, the capability for microcystin production as such is genetically determined. Strains isolated from the same bloom sample are constitutively microcystin producing or nonproducing (15, 19, 28), and the types and cellular content of microcystins may differ considerably between strains (2, 16, 28). The decisive factors determining the toxicity of a bloom are therefore the ratio of microcystin-producing and non-microcystin-producing genotypes and the amounts and variants of microcystins produced by individual cells (4, 23).
Understanding of the community composition and dynamics of microcystin-producing and non-microcystin-producing Microcystis strains in the field is very limited, due to a lack of suitable identification methods. Traditional characterization of Microcystis based on morphological features is very difficult, and the differentiation that can be attained below genus level is limited. Moreover, transition between morphological characteristics has been observed (20). Identification difficulties may partly explain why correlations between morphology and toxicity have proven to be unreliable (6, 13, 21), emphasizing the need for identification independent of morphological characteristics. Molecular biological methods provide more reliable tools for recognition of Microcystis strains and their properties.
A number of studies have targeted the microcystin synthetase (mcy) gene cluster (5, 25) for the identification of toxic Microcystis strains. This approach has the obvious advantage of a direct relationship between gene detection and toxin production provided that nontoxic strains do not possess the gene. Detection by PCR of the mcyB gene, the mcyA gene, or the adenylation domain within the microcystin synthetase gene cluster showed good correlation with microcystin production, but some anomalies were found. A small number of Microcystis strains tested positive for the mcy gene cluster but lacked detectable microcystins (10, 16-18, 26). Homology of the mcyB region or adenylation domains with other loci (5, 18, 26) or even the presence of the mcy gene cluster in some nontoxic Microcystis (18) strains may explain these discrepancies. The toxic potential of a bloom cannot be determined solely by the isolation and cultivation of strains, due to the biases and limitations inherent to studies of cultured isolates. Toxic strains can be isolated from field samples with undetectable toxin concentrations, and conversely, cultivation of toxic strains from toxin-containing samples can be unsuccessful (28). Therefore, genetic analyses have also been done on colonies collected directly from the water. Pooled size classes or individual colonies were used to investigate the relation between toxic genotypes, based on the detection of mcyB genes, and colony size (12) or morphospecies (13).
In the study presented here, we used an alternative approach to characterize toxic and nontoxic Microcystis colonies in natural populations independent of mcy gene detection. Colonies were isolated directly from water samples, the presence of microcystins was examined by means of matrix-assisted laser desorption ionization-time of flight mass spectrometry(MALDI-TOF MS), and they were differentiated at high resolution based on sequences of the rRNA internal transcribed spacer (ITS). This gene fragment is known to differ considerably between Microcystis strains (9, 21). We show that differentiation of the rRNA ITS by denaturing gradient gel electrophoresis (DGGE) or sequencing is sufficient for resolving the identification of toxic and nontoxic Microcystis genotypes in single isolated colonies and in natural samples.
The benefits of this approach for analysis of Microcystis community composition and dynamics in relation to the toxicity of a water body are discussed.
MATERIALS AND METHODS
Colony isolation and characterization.
Sampling for colony isolation was carried in the summer of 2001 in 15 water bodies from 9 European countries and Morocco by net hauls with a plankton net (40-μm mesh size). The samples were kept cool until they were processed, in each case within 14 days after collection. Colony isolation and morphological description were standardized in the course of a workshop in July 2001 (L. Via-Ordorika, J. Fastner, M. Hisbergues, E. Dittmann, M. Erhard, J. Komarck, R. Kurmayer, and I. Chorus, unpublished data). For colony isolation, samples were diluted and single colonies were picked out by using thin Pasteur pipettes or forceps under binocular microscopes. Colonies were washed in BG11 medium (22) to dispose of any visible secondary colonies or other cyanobacteria and algae. Morphological classification was done by using the criteria proposed by Komárek and Anagnostidis (11). Eight morphotypes were identified, mostly Microcystis aeruginosa and Microcystis ichthyoblabe and less frequently Microcystis botrys, Microcystis flos-aquae, Microcystis panniformis,Microcystis viridis, Microcystis wesenbergii, and Microcystis novacekii (Via-Ordorika et al., unpublished). Colonies were transferred into a reaction tube (0.5 ml) containing culture medium (final sample volume, 4 to 30 μl), and the presence of each colony in the tube was verified microscopically. The tubes were frozen (liquid nitrogen) and thawed several times to disintegrate the colonies and stored at −20°C. Aliquots from each tube were used for gene amplification and for MALDI-TOF MS. In 123 of 151 colony samples we obtained, the presence or absence of toxins had been determined by MALDI-TOF MS, and in 111 of these colonies, the toxin production capacity had been confirmed by PCR detection of the mcy gene. The 28 colonies that had not yielded any MALDI-TOF MS signal were typified as colonies of unknown toxin production. Eight such colonies had been characterized by mcy PCR only.
Cell counts and DNA isolation from natural samples.
Lakes ‘t Joppe and Zeegerplas are eutrophic lakes (maximum depth, approximately 25 m) which are stratified during summer.
Samples were taken from the water column in both Lake ‘t Joppe and Lake Zeegerplas in June and August and in Lake ‘t Joppe only in September 2001. On the latter two sampling dates, surface scums were collected in Lake ‘t Joppe. Water samples were collected 0.5 m below the surface or from surface scum in sterile bottles in the middle of the lake. For counting, water and scum samples were preserved with Lugol's iodine directly after sampling and stored at 4°C. Water samples were concentrated by sedimentation and counted by inverted light microscopy by using a Sedgewick-Rafter counting chamber (Pyser-SGI Limited, Edenbridge, Kent, United Kingdom). In water samples containing high Microcystis colony densities and in scum samples, Microcystis colonies were first disintegrated. For that purpose, 20 ml of fixed sample was collected on a 45-mm-diameter, 0.45-μm-pore-size HA membrane filter (Millipore, Bedford, Mass.), and the filter was transferred to an Erlenmeyer flask containing 20 ml of 0.01 M KOH and incubated for 30 min at 80°C. The disintegrated sample was transferred to a test tube and vortexed for 60 s, and single Microcystis cells were counted as described above. For DNA isolation, water or scum samples of 250 ml, or less if the filter clogged, were filtered over a 25-mm-diameter, 0.2-μm-pore-size mixed esters filter (ME 24; Schleicher & Schuell, Dassel, Germany), and the filters were processed as described before (9).
Microcystin analysis.
MALDI-TOF MS measurements were performed as described previously (6; Via-Ordorika et al, unpublished). Mass signals indicative for known microcystins were in most cases further analyzed by recording fragment ions by using postsource decay (PSD). For microcystin analysis by high-performance liquid chromatography, a 100- to 500-ml water column or scum sample was filtered over glass microfiber filters (25-mm-diameter GF/C; Whatman, Maidstone, United Kingdom). From dense scum samples, 2 ml was transferred to microcentrifuge tubes without filtration. Filters and tubes were stored at −20°C until further processing. After lyophilization, filters and scum samples were extracted with aqueous methanol (7). Microcystins were analyzed by reverse-phase high-performance liquid chromatography with diode array detection (14).
PCR amplification.
DNA from isolated Microcystis colonies was amplified by a nested PCR protocol. First, cyanobacterium-specific 16S rRNA primer CYA 371F and universal 23S rRNA primer ULR were used for amplification of a major part of the 16S, the ITS, and a short section of the 23S from the rRNA gene. The resulting PCR product was diluted and used as a template for a second amplification with cyanobacterium-specific 16S rRNA primer CSIF in combination with ITS primer 373R (amplifying part of the rRNA ITS, ITSa) or in combination with 23S rRNA primer ULR (amplifying the entire rRNA ITS, ITSc) (9). For amplification of DNA isolated from natural samples, only the latter PCR protocol was used. Primer sequences and PCR conditions were according to those described in reference 9. Primer CYA 371F had the sequence 5′-CCT ACG GGA GGC AGC AGT GGG GAA TTT TCC AC-3′. Primer CSIF had the sequence 5′-GYC ACG CCC GAA GTC RTT AC-3′ plus a 40-nucleotide GC clamp added to the 5′ site when PCR products were used for DGGE analysis. Primer 373R had the sequence 5′-CTA ACC ACC TGA GCT AAT-3′, and primer ULR had the sequence 5′-CCT CTG TGT GCC TAG GTA TC-3′.
DGGE profiling.
DGGE was performed as described earlier (9). To enable comparison between DGGE gels of rRNA ITS bands from colonies, each new batch of colonies was analyzed together with a mixture of colonies assigned to different groups and thus representing different gel positions. A marker for band positions in ITSa DGGE gels from natural samples was composed of ITSa amplicons from the following strains of cyanobacteria and a prochlorophyte: IMS101 (Trichodesmium erythraeum), CYA146 (Pseudanabaena catenata), CYA126 (Planktothrix aghardi), PCC9006 (Prochlorothrix hollandica), ATCC 29413 (Anabaena variabilis), CYA 135 (Anabaenopsis arnoldii), S2, K29, PCC7820, SAG17.85 (Microcystis sp.), and isolated Microcystis colonies K16 and K55 (Table 1). A marker for gel positions in ITSc DGGE was composed of DNA amplified with ITSc primers from the following cyanobacterial strains: IMS101 (T. erythraeum), CYA99 (Lyngbya sp.), PCC73110 (Leptolyngbya sp.), PCC6803 (Synechocystis sp.), 1401/7CCAP, S2, V91, Z11, CYA140, PCC7806, PCC7820 (Microcystis sp.), and two excised and reamplified bands from Aphanizomenon isolate T33. For more details on the cyanobacterial cultures, refer to reference 9.
TABLE 1.
Characterization and classification based on rRNA ITS DGGE of isolated Microcystis coloniesa
| Colony type | ITSa group | ITSc group | Colony K: | Microcystis morphospeciesb | Lakec |
|---|---|---|---|---|---|
| Toxin producing | 1 | 5 | 21* | W | J |
| 69* | B | Z | |||
| 107 | B | K | |||
| 146, 147-u | A | W | |||
| 1 | 18 | 147-l | A | W | |
| 2 | 4 | 63-u | B | Z | |
| 4 | 1 | 79-u | I/P | Br | |
| 4 | 2 | 79-l | I/P | Br | |
| 4 | 15 | 112-ue | I | M | |
| 6 | 4 | 15-u, 19-m | B | J | |
| 6 | 5 | 63-l | B | Z | |
| 8 | 5 | 12, 15-l,19-l | B | J | |
| 11 | 5 | 3-u | N | J | |
| 10-u | A | J | |||
| 42-u, 43-u, 59 | B | Z | |||
| 41-u, 46, 47-u, 49,50, 52, 55-u, 57, 58 | A | Z | |||
| 83 | A | Br | |||
| 102 | A | MI | |||
| 111 | A | M | |||
| 139-u | P | A | |||
| 143 | B | W | |||
| 144 | A | W | |||
| 11 | 8 | 51, 72 | B | Z | |
| 11 | 10 | 3-l | N | J | |
| 41-l,47-l,55-l | A | Z | |||
| 42-l, 43-l | B | Z | |||
| 139-l | P | A | |||
| 15 | 15 | 112-le | I | M | |
| 19 | 12 | 60-u | A | Z | |
| 23 | 3 | 114-u | F | M | |
| 23 | 14 | 10-l | A | J | |
| 114-l | F | M | |||
| 26 | 8 | 9, 14, 20 | A | J | |
| 26 | 12 | 13e | A | J | |
| 60-l | A | Z | |||
| 27 | 19 | 39 | A | J | |
| Unknown toxin production | 1 | 4 | 65* | A | Z |
| 7 | 13 | 29* | A | J | |
| 10 | 10 | 61-u*, 70-m*, 66* | A | Z | |
| 13 | 17 | 32* | I | J | |
| 17 | 5 | 67-u* | I | Z | |
| 17 | 11 | 61-l* | A | Z | |
| 17 | 17 | 67-l* | I | Z | |
| 17 | 70-l* | A | Z | ||
| 21 | 18 | 31-l* | A | J | |
| Non-toxin producing | 2 | 11 | 75-u | A | Ba |
| 145 | I | W | |||
| 3 | 16 | 75-l | A | Ba | |
| 142 | I | W | |||
| 5 | ND | 119-u, 120-u | I/P | Pw | |
| 6 | 14 | 128, 129 | A | R | |
| 9 | 16 | 106-u | I | H | |
| 9 | ND | 119-m, 120-l | I/P | Pw | |
| 10 | 6 | 5-ud | N | J | |
| 11 | 14 | 17-m | I | J | |
| 11 | 15 | 99-u, 150 | I | MI | |
| 11 | 16 | 17-l | I | J | |
| 97 | F | MI | |||
| 99-l, 101, 149 | I | MI | |||
| 140 | P | A | |||
| 12 | 1 | 130 | I | R | |
| 151* | A | R | |||
| 12 | 9 | 73, 76 | I | Ba | |
| 132, 134 | I | R | |||
| 12 | 11 | 18 | A | J | |
| 14 | 11 | 16d | W | J | |
| 14 | 16 | 106-l | I | H | |
| 16 | 9 | 108, 110 | I | T | |
| 117 | I | Pc | |||
| 121 | P | Pw | |||
| 123 | I/P | Pw | |||
| 16 | ND | 119-l | P | Pw | |
| 17 | 6 | 74 | A | Ba | |
| 131 | W | R | |||
| 133 | A | R | |||
| 17 | 9 | 5-ld | N | J | |
| 127 | I | R | |||
| 18 | 1 | 78-u | A | Bo | |
| 18 | 2 | 78-l | A | Bo | |
| 18 | 7 | 148 | I | T | |
| 18 | 17 | 105-u | I | Pe | |
| 18 | 18 | 87, 88, 89, 90, 91, 92, 95 | A | F | |
| 93, 94 | I | F | |||
| 100e | I | MI | |||
| 20 | 7 | 33-u | I | J | |
| 21 | 5 | 135, 136 | A | A | |
| 137, 138, 141 | P | A | |||
| 22 | 13 | 34, 35, 36 | I | J | |
| 38* | W | J | |||
| 24 | 17 | 37 | I | J | |
| 68 | I | Z | |||
| 105-l | I | Pe | |||
| 31-u* | A | J | |||
| 25 | 3 | 104-u | I | H | |
| 25 | 11 | 33-l | I | J | |
| 25 | 14 | 104-l | I | H |
Morphospecies assignment and lake of origin are given for each colony. Based on the positions of their bands in the ITSa and ITSe DGGE profiles, colonies K1 to K151 were assigned group numbers. Classes of colonies were distinguished on the basis of combined ITSa and ITSc grouping. Colonies in boldface type contained microcystins as determined by MALDI-TOF MS or, in a few cases, by amplification of the mcy gene. Toxin production data were lacking for colonies indicated by asterisks. Colony classes were organized in three groups: one with classes containing toxin-producing colonies, one with nontoxic colony classes, and one with classes of colonies of unknown toxin production. The few colonies of unknown toxin production that classified with toxic or nontoxic colonies ended up in the group of either. Bands of underlined colonies were sequenced. The suffix “-u” refers to the upper band in the ITSc DGGE profile from a colony, the suffix “-m” refers to the middle band, and the suffix “-l” refers to the lower band. If the upper and lower bands from a particular colony have the same group assignment for, e.g., ITSa, this means that there is one band in the ITSa profile and two bands in the ITSc profile. Note that if a mixture of 2 sequences gave rise to 2 ITSa and 2 ITSc bands, their relative band positions (meaning upper or lower) may be reversed. ND, not determined.
A, M. aeruginosa; B, M. botrys; I, M. ichthyoblabe; F, M. flos-aquae; N, M. novacekii; P, M. panniformis; W, M. wesenbergii.
Lakes: A, Lake Arancio, Sicily, Italy; Ba, Loch Balgavies, Dundee, Scotland; Bo, Lac du Jardin Public, Bordeaux, France; Br, Brno reservoir, Brno, Czech Republic, F, lake Frederiksborg Sløtssø, Hillerød, Denmark; H, Holzöstersee, Salzburg, Austria; J, ’t Joppe, Leiden, The Netherlands; K, Krossinsee, Berlin, Germany; M, Mügelsee, Berlin, Germany; MI, Monikie Island Pond, Dundee, Scotland; T, Lake Takerkoust, Marrakesh, Morocco; Pe, Parque de Cicade pond east, Porto, Portugal; Pw, Parque de Cicade pond west, Porto, Portugal; R, Loch Rescoby, Dundee, Scotland; W, Wannsee, Berlin, Germany; Z, Zeegerplas, Alphen aan de Rijn, The Netherlands.
Toxin production was examined by mcy gene detection only (no MALDI-TOF MS).
The MALDI-TOF MS profile showed only 1 peak at a mass-to-charge ratio (m/z) of 995, which was not analyzed by PSD measurements and could therefore be derived from either microcystin LR or cyanopeptolin.
Sequencing and sequence analysis.
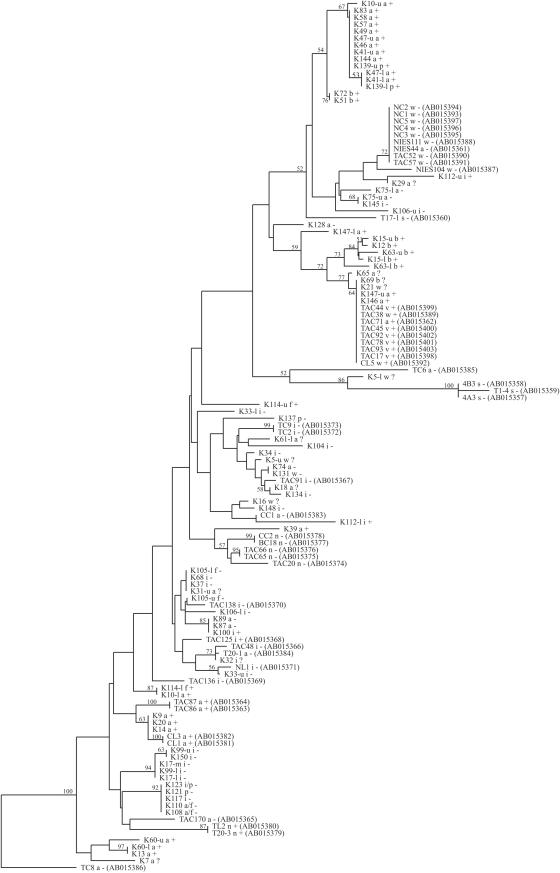
Isolated colonies that produced DGGE profiles with single bands were used directly for sequencing. DNAs from colonies that produced multiple bands in DGGE profiles and from bands in DGGE profiles from natural lake water samples were obtained as follows. A small piece of gel from the middle of the target band was excised from the DGGE gel and incubated in 50 μl of sterile milli-Q purified water for 24 h at 4°C. The eluent was reamplified with the original primers and run on DGGE to confirm its identity. DNA for sequencing was amplified with primers without a GC clamp, and the PCR products were purified and sequenced by Baseclear Labservices (Leiden, The Netherlands). If possible (colonies K60, K105, and K106) (Table 1; see Fig. 2), sequencing of two ITSa bands resolved ambiguous base calls resulting from a mixture of sequences that were not separated by using ITSc DGGE. This was not possible for colony K112, which had many ambiguous base calls in the 3′ site of the rRNA ITS. Sequences were deposited at EMBL (see below). Similarity with sequences deposited in GenBank, EMBL, and DDBJ was checked by using the program BLAST (1) (via http://www.ncbi.nlm.nih.gov/BLAST/). Sequences from the rRNA ITS region were obtained from GenBank, EMBL, and DDBJ and were aligned by using the programs ClustalW, the BioEdit Sequence Alignment Editor (8), and the program package ARB (www.ARB-home.de). Treecon software (27) was used for the construction of distance trees from aligned sequences. The neighbor-joining method was used with multiple substitutions corrected by the method of Jukes and Canter. One thousand bootstrap trials were performed.
FIG. 2.
Distance tree based on the alignment of rRNA ITS sequences from isolated colonies (indicated by K plus a number, as shown in Table 1) and cultured isolates (retrieved from the EMBL database and indicated by strain code and accession number). Bootstrap values higher than 50% are indicated. Sequence TC8 was used as an out-group. Sequences from the entire ITSc amplicons (which includes 100 bp in the 16S rRNA) were available from isolated colonies and from the rRNA ITS only for the cultured Asian strain isolates. From a few colonies (K60, K105, and K106) with 2 ITSa DGGE bands and 1 ITSc DGGE band, sequences were compiled from the sequences of excised ITSa bands plus ITSc sequences. This was not possible for colony K112, and for this colony, only sequences spanning the ITSa amplicon were used. The letters following colony numbers refer to morphospecies assignation (a, M. aeruginosa; b, M. botrys; i, M. ichthyoblabe; f, M. flos-aquae; n, M. novacekii; p, M. panniformis; v, M. viridis; w, M. wesenbergii; s, Microcystis sp.). Symbols: +, microcystin producer; −, non-microcystin producer; ?, unknown microcystin production.
Nucleotide sequence accession number.
Sequences were deposited at EMBL and were assigned accession numbers AJ605140 to AJ605221 for sequences from isolated colonies and AJ619633 to AJ619663 for sequences from bands excised from DGGE profiles of natural samples.
RESULTS
Isolation and characterization of Microcystis colonies.
The Microcystis colonies we used for our investigations originated from 15 lakes from 9 European countries and Morocco. Most of the water samples were collected at the end of June 2001 and were isolated and characterized by experts during a European workshop. Additional colonies from two Dutch lakes (‘t Joppe and Zeegerplas) were isolated and characterized in August and September 2001. With these colonies, a study of the relative abundances and distribution over the sampled lakes of different morphotypes, the occurrence of microcystins measured by MALDI-TOF MS, and diagnostic PCR targeted at the mcyA and mcyB genes has been made by Via-Ordorika et al. (unpublished)
Differentiation of colonies based on rRNA ITS DGGE.
To enable discrimination of Microcystis colonies based on the rRNA ITS, DNA fragments ITSa (spanning part of the 16S rRNA and part of the rRNA ITS) and ITSc (spanning part of the 16S rRNA and the entire rRNA ITS) were amplified and analyzed by DGGE (9). A nested PCR protocol was used to maximize DNA retrieval from the small quantities of cells in colony aliquots that remained after the analyses described above. Of a total of 151 sample aliquots, 107 yielded PCR products. Successful amplification was only slightly biased for lake of origin (between 70 and 100% of the colonies originating from a lake yielded PCR products, with the exception of 25% from the Brno reservoir), morphospecies assignation (between 60 and 85% of the colonies assigned to a morphospecies yielded PCR products, with the exception of the two M. viridis colonies), and presence of toxins (75% of both toxic and nontoxic colonies and 48% of the colonies of unknown toxin production yielded PCR products). Consequently, the collection of colonies that was subjected to DGGE analysis contained Microcystis colonies derived from all 15 water bodies and covering all morphospecies except M. viridis. Toxin production data were available for 96 of the 107 colonies that yielded PCR products.
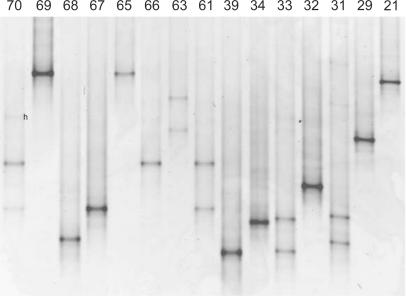
Colonies were differentiated on the basis of the position of their rRNA ITS amplicons on DGGE gels (Fig. 1). Based on ITSa DGGE, colonies could be differentiated into 27 different groups (Table 1). The longer ITSc amplicons showed smaller differences in migration on DGGE, and colonies could be differentiated into 19 different groups (Table 1). Most colonies (76 of 107) yielded ITSa as well as ITSc DGGE profiles containing one sequence. However, several colonies gave rise to profiles with two or more bands (Fig. 1, colonies K61, K63, and K70). Eleven colonies yielded two bands both in ITSa and ITSc profiles (e.g., K63), 14 colonies yielded profiles with one ITSa band and two ITSc bands (e.g., K147), and 5 colonies had profiles with two ITSa bands and one ITSc band (e.g., K112) (Table 1). In the eight profiles with three or four bands (e.g., colony K70) (Fig. 1), the upper one or two bands were heteroduplexes, as shown through excision of these bands, followed by reamplification and DGGE analysis. Only one colony (K119) appeared to contain three different (nonheteroduplex) bands in ITSa DGGE profiles.
FIG. 1.
Example of an ITSa DGGE gel from 15 isolated Microcystis colonies. Each unique gel position was assigned a number (Table 1), e.g., numbers 10 and 17 for colony K70, number 1 for colonies K69, K65, and K21, and number 24 for colony K68. h, heteroduplex band.
Classification of colonies based on the combined ITSa and ITSc DGGE profiles clearly separated toxic and nontoxic colonies (Table 1). From the 59 classes that could be distinguished by ITSa plus ITSc DGGE, 19 classes contained only toxic colonies (plus two colonies of unknown toxin production in the class formed by ITSa group 1 and ITSc group 5, designated class 1a/5c). The other classes were derived from nontoxic colonies (31 classes) or from colonies of unknown toxin production (9 colonies).
Only one colony (K100, class 18a/18c) which was identified as toxic could not be differentiated from nontoxic colonies on the basis of ITS DGGE. However, toxin identification in this colony was ambiguous, since a mass signal that is indicative for microcystin-LR and also for another cyanopeptide could not be further analyzed by PSD analysis due to lack of colony material (Table 1). Consequently, reliable identification of toxic and nontoxic Microcystis colonies could be achieved on the basis of ITSa plus ITSc DGGE profiles. DGGE of ITSa or ITSc alone was not a sufficient determinant for differentiation of toxic and nontoxic colonies. In ITSa group 11 for instance, nontoxic colonies grouped with a majority of toxic ones (Table 1).
There was no apparent relation between morphospecies and classification based on rRNA ITS DGGE profiles, since colonies from each morphospecies were assigned to different ITS DGGE classes. Also, with the exception of Lake Frederiksborg Sløotssøo (Denmark), each lake yielded colonies from multiple ITS DGGE classes. Hence, multiple colonies in an ITS DGGE class were in most cases isolated from different lakes and assigned to different morphospecies (Table 1).
rRNA ITS sequences from colonies.
To substantiate the DGGE-based division of colonies and to allow in-depth analysis of the Microcystis classes that were identified, DNA fragments covering ITSc were sequenced from colonies representative of most of the classes listed in Table 1. Of 59 ITS DGGE classes, we obtained multiple sequences from 15 classes, one sequence from 29 classes, and no sequences from 14 classes. Attempts to obtain sequences failed for some bands (e.g., from colony K78, class 18a/1c and 18a/2c), due to the occurrence of many ambiguous base calls. The sizes of the Microcystis ITS ranged between 354 and 362 bp, and variation between strains was restricted to approximately 11% of the sequence positions. Sequences differed between DGGE classes and were typically identical within a class. Nevertheless, a 1-bp substitution in the 5′ region of the rRNA ITS distinguished sequence 12 from 15-l in class 8a/5c, and a substitution in the 16S rRNA distinguished 10-u from the other sequences in class 11a/5c. This sporadic sequence variability in colonies within a DGGE classes could be explained by the position of the variation in the 5′ high-melting domain of the amplicons, where base differences do not result in altered migration on DGGE. In contrast, if positioned in the 3′ lower-melting domain, differences of only 1-bp could be detected clearly, as illustrated by the upper and lower bands from, e.g., colony K41 (11a/5c and 11a/10c).
A phylogenetic tree was constructed (Fig. 2) based on an alignment of the 78 rRNA ITS sequences from our Microcystis colonies and of 47 Asian Microcystis strains of known microcystin production sequenced by Otsuka et al. (21). Colonies from different DGGE classes ended up in separate branches. Every unique sequence, occupying a distinct branch, was derived from either a microcystin-producing colony or a non-microcystin-producing colony. Therefore, rRNA ITS sequences were well suited for recognition of toxic and nontoxic genotypes. Similar to what was found for differentiation based on DGGE, the only incongruity was formed by toxic colony K100, which had an ITS sequence identical to that of nontoxic colonies K87 and K89 (Table 1, class 18a/18c and above). Clustering of sequences from microcystin producers and from nonproducers was observed (Fig. 2).
Community profiles from original field sample.
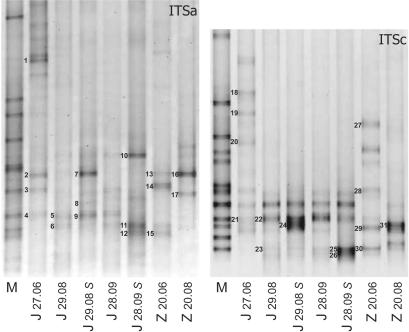
To investigate to what extent the isolated colonies represented the cyanobacterial community in their lakes of origin, we analyzed ITSa and ITSc DGGE profiles from the water samples from which most colonies had been harvested. Cell counts of cyanobacteria and microcystin concentrations in these samples are given in Table 2, and the cyanobacterial ITSa and ITSc DGGE profiles are shown in Fig. 3. In the ITSa community profile from Lake ‘t Joppe, we detected 12 bands (4 of which were less distinct) in the lower part of the gel, where Microcystis sequences were expected, and 6 bands in the upper part. In Lake Zeegerplas ITSa profiles, 8 different Microcystis bands (2 less distinct) were discernible. ITSc DGGE yielded, for both lakes, profiles with 8 bands (1 less distinct) in the lower part of the gels. For identification of the Microcystis strains present in natural samples, bands at different positions in the ITS profiles were excised and reamplified, and their position and purity were verified on DGGE. Sequences were retrieved from the bands indicated in Fig. 3 and Table 3. Compared to ITSa profiles, retrieval of pure bands for sequencing proved more difficult from ITSc profiles.
TABLE 2.
Cell counts of cyanobacteria and microcystin concentrations in natural samples from lakes which were used for isolation of Microcystis colonies
| Lake | Sampling date | Source | No. of cells (cells/liter) ofa:
|
Toxin concn
|
|||
|---|---|---|---|---|---|---|---|
| Microcystis sp. | Anabaena sp. | Aphanizomenon sp. | Microcystin concn (μg/liter) | Amt of microcystin per Microcystis cell (fg/cell) | |||
| ‘t Joppe | 27 June | Water column | 8.1 × 103 | 2.4 × 104 | 4.5 × 102 | 0.22 (0.18) | 27 |
| 29 August | Water column | 5.5 × 104 | 5.3 × 102 | 0 | 2.64 (0.03) | 48 | |
| Scum | 5.6 × 106 | ND | ND | 420 (22.9) | 75 | ||
| 28 September | Water column | 7.8 × 104 | 1.4 × 103 | 88 | 0 | 0 | |
| Scum | 6.1 × 107 | ND | ND | 3,308 (244) | 55 | ||
| Zeegerplas | 20 June | Water column | 5.4 × 104 | 1.8 × 105 | 0 | 0 | 0 |
| 20 August | Water column | 2.8 × 105 | 1.0 × 105 | 0 | 0.60 (0.09) | 2 | |
ND, not detected.
FIG. 3.
Cyanobacterial community composition of lake water samples analyzed by ITSa and ITSc DGGE. Samples were taken from Lake ‘t Joppe (J) in June, August, and September 2001 and from Lake Zeegerplas (Z) in June and August. S following the sampling date (day.month [06, June; 08, August; 09, September]) signifies scum samples. M, marker lane. Bands that yielded useable sequences after excision, reamplification, and sequencing are numbered. Table 3 contains more information about these sequences.
TABLE 3.
Comparison of sequences derived from DGGE gels from natural samples with sequences derived from isolated coloniesa
| ITSa band sequence(s) | ITSc band sequence(s) | Matching colony(ies) |
|---|---|---|
| e3 | e28 | K46, K49, K51b (+) |
| e4, e5, e9 | e21, e22 | K74, K131 (−) |
| e6, e11 | K148, K105-u (−) | |
| e8 | K16 (−) | |
| e12, e15 | K34 (−) | |
| e17 | K17, K99, K150 (−) | |
| e25, e30 | K37, K68b (−) | |
| e2, e7, e13, e16 | e24, e31 | None |
| e10 | e26 | None |
| e14 | e29 | None |
| e23 | None |
DGGE gels from natural samples are shown in Fig. 3. Each row contains identical sequences (ITSa sequences were compared with the 5′ region of the longer ITSc fragments). In the last column, colonies with sequences identical to those of excised bands are presented. Toxin production in these colonies is indicated as follows: (+), toxic; (−), nontoxic.
Not an exhaustive list; there were more colonies for which the sequences matched equally well.
A BLAST search revealed that bands e1, e18, e19, e20, and e27, excised from the upper part of the gels, were derived from Anabaena, Aphanizomenon, and Synechocystis, whereas all other bands contained Microcystis sequences. Sequences from most of the bands excised from ITSa profiles matched with sequences from isolated colonies (Table 3). From the nine different Microcystis sequences we retrieved, only one (derived from band e3) aligned with sequences corresponding to toxic colonies, five aligned with nontoxic sequences (bands e4 = e5 = e9, e6 = e11, e12 = e15, e8, and e17), and three sequences (e2 = e7 = e13 = e16, e10, and e14) did not correspond to any isolated colony (Table 3). The most prominent bands that were amplified contained sequences corresponding to nontoxic colonies (e4 = e5 = e9, e6 = e11, and e12 = e15) or sequences of unknown toxicity (e10 and e7 = e16). From the seven different sequences retrieved from ITSc profiles, one (derived from band e28) corresponded to a toxic strain, two corresponded to nontoxic strains (e21 = e22 and e25 = e30), and four (e23, e24 = e31, e29, and e26) yielded new sequences that did not match any isolated colony (Table 3). The sequences of two strains that became relatively dominant in scum samples were detected in ITSa profiles (bands e7 and e10) as well as in ITSc profiles (bands e24 and e26).
DISCUSSION
Discrimination of toxic and nontoxic Microcystis based on rRNA ITS gene diversity.
Our data show that Microcystis rRNA ITS sequences are sufficiently heterogeneous for differentiation of strains that differ physiologically with respect to toxin production (microcystin producers versus nonproducers). Thus, by analyzing rRNA gene ITS diversity, toxic and nontoxic Microcystis strains can be identified and their ecology can be studied. The correlation between rRNA ITS sequences and microcystin production existed in a diverse array of European and Asian genotypes, which suggests it may be extended to other geographic regions and may possibly be valid worldwide. Genotype analysis by DGGE enabled processing of many samples (without the need to sequence them all), assessment of the purity of isolated cultures or colonies, and analysis of different genotypes in complex natural communities. Genotype analysis by sequencing confirmed the colony classification based on DGGE and enabled the identification of genotypes (characterized as toxic or nontoxic) obtained from isolated colonies or natural samples.
The use of a universal taxonomic marker gene not related to toxin production for recognition of Microcystis strains has several important advantages. First, this approach provides ecologically relevant insights into the dynamics of all cyanobacterial strains that are present in a natural community. Instead of focusing on the limited subgroup of mcy gene-containing Microcystis strains, different toxic and nontoxic strains can be distinguished. Second, even information on the relative toxicity of strains could be implied in the rRNA ITS sequences (provided a correlation with rRNA ITS sequences exists). Besides the ratio of toxic to nontoxic strains, the variants (3) and amount per cell (23) of microcystins that are produced are important factors in determining bloom toxicity. Recently, the production of microcystin variants was found to have a genetic basis (16). Third, a broader range of strain properties could be characterized through rRNA ITS as colony properties other than microcystin production (such as the production of other, potentially toxic, peptides) may also correlate with rRNA ITS classification. Finally, the use of a universal marker gene did not suffer from problems of diagnostic PCR that can be encountered in mcy gene detection. The identification of toxic strains was based on sequence information which was analyzed only after amplification and therefore independent of the detection of a specific gene. In contrast, in a diagnostic PCR, the amplification itself is a crucial step which is sensitive to variations in initial DNA concentration and quality and PCR conditions (notably the number of temperature cycles). The occurrence of contradictory mcyB gene detection, possibly due to PCR inconsistencies, was reported for 5 of 27 parallel samples of microcystin-producing colonies that had been split in half (13).
A prerequisite for the use of rRNA ITS sequences for ecological studies of Microcystis is that all genotypes that can be encountered should be characterized with regard to the strain properties of interest (such as toxin production).
Other colony properties related to rRNA gene ITS diversity.
We did not find a relation between rRNA ITS genotypes and the origin of the isolated Microcystis colonies (Table 1). Although most DGGE classes (or sequences) were derived from strains originating from the same lake (Table 1), this could be explained by the small number of colonies representing most sequences. If genotypes were represented by several colonies, these originated from different locations (e.g., class 5a/11c). Elucidation of the occurrence of local genotypes would require a more comprehensive sampling at each location for extensive colony isolations and generation of community DGGE profiles. There was also no relation between genotypic classification based on rRNA ITS and morphospecies assignation. Every unique DGGE class (Table 1) or sequence (Fig. 2) with a considerable number of colonies (Table 1) contained representatives of different morphospecies (Table 1). This is in agreement with the difficulties inherent to morphological identification (see the introduction) and the finding that different Microcystis morphotypes contain toxic and nontoxic strains (21; Via-Ordorika et al., unpublished).
Multiple bands in DGGE profiles.
We detected more than one Microcystis sequence in 28% of the colonies we analyzed. An explanation for multiple sequences is contamination by aggregated colonies which were not separated despite thorough washings during isolation. We considered it unlikely that additional bands originated from only a few contaminating cells, since the intensities of the multiple bands in most profiles were similar. Highly similar rRNA ITS sequences and identical primer sites in Microcystis strains result in similar amplification efficiencies, as was confirmed by the correlation between initial cell numbers and DGGE band intensities after coamplification of DNA from two Microcystis strains (data not shown). An alternative explanation for multiple sequences is the presence of multiple different rRNA operons in a portion of Microcystis strains. An occurrence of different operons in only some Microcystis strains implies considerable intragenus dissimilarity. Such dissimilarity could be most parsimoniously explained by the presence in the genus Microcystis of at least two rRNA operons, identical in sequence in most strains but with mutations in one operon in some strains. The equal band intensities that were encountered in most colonies with multiple sequences are in agreement with the occurrence of two different operons in one strain. On the other hand, the unambiguous sequences retrieved from all 47 Asian strains (21) argue against the presence of two different operons in a proportion of strains.
The alternative explanations for multiple bands per colony may be true for different colonies. Some colonies may have consisted of aggregated strains, as was supported by the presence of even three different sequences in colony K119, the presence of additional sequences of low intensity (e.g., colony K70) (Fig. 1), or the occurrence of sequences as the sole sequence in one colony and as one of two sequences in another colony (e.g., K46 and K47-u). Conversely, the multiple bands in several colonies could be well explained by a single base pair substitution (e.g., colonies K41, K47, K99, and K139) or an insertion/deletion at one location (K17) in one operon (of two present).
The possible presence of aggregated colonies in a considerable portion of the carefully isolated and washed colonies, highlights that restriction fragment length polymorphism patterns or sequences from any gene obtained from such colonies (13) need to be interpreted with caution. A few isolated colonies (e.g., K78) probably contained a mixture of sequences even if they generated one band on ITSa and ITSc DGGE, as judged from the many ambiguous base calls obtained from sequencing. Toxin data from such colonies are impossible to assign to sequences unless the excised fragments are cloned. Unequivocal correlation of toxin production to a particular sequence was not possible for toxic colonies containing multiple sequences. While two sequences derived from a nontoxic colony could both be linked to nontoxic Microcystis, one of the sequences from a toxic colony could be derived from a nontoxic contaminant.
Natural samples.
We analyzed ITSa DGGE profiles in addition to ITSc profiles because they offered useful supplementary data for identification of toxic and nontoxic genotypes. Greater differences in melting behavior of the smaller ITSa amplicons made the detection and isolation of bands easier. Moreover, the partial rRNA ITS sequences covered by ITSa were sufficiently resolving for the recognition of toxic and nontoxic genotypes from all of our colonies (data not shown) and could thus be confidently applied to our studied lakes (Fig. 3; Table 3).
Microcystin concentrations in the water column of lakes ‘t Joppe and Zeegerplas were undetectable or well below the 10 μg/liter guideline for safe recreational waters (3), but in scums, this concentration was far exceeded. In both lakes, the number of Microcystis cells increased during the summer. Cell counts for the genera Anabaena and Aphanizomenon decreased after June, as was confirmed by the disappearance of their corresponding bands from DGGE profiles (Fig. 3). Although care is needed in quantitative interpretations of DGGE bands, the similar DNA isolation and amplification efficiencies (see above) likely result in a good correlation between the proportion of Microcystis genotypes and DGGE band intensities. Prominent bands in DGGE profiles from natural populations will therefore represent the most abundant Microcystis genotypes. A nontoxic strain (bands e4, e5, e9, e21, and e22) appeared to be abundant in all samples from ‘t Joppe, and a strain of unknown toxin production (bands e2, e7, e13, e16, and e24) was present in June and August but had disappeared in September (Fig. 3 and Table 3). Bands derived from toxic strains were detected only in the samples from June (bands e3 and e28) and not in the samples from August and September, when Microcystis reached the highest densities (Table 2). In contrast, many nontoxic strains were identified in the profiles from August and September (e6, e8, e11, e12, e15, e17, e22, e25, and e30). Most of the toxic colonies isolated from these lakes (90%, 29 of 32) originated from the June samples, and most of the nontoxic colonies (70%, 9 of 13) originated from the August and September samples. Together, these data could suggest that while in the beginning of a bloom toxic colonies were relatively dominant, they were out-competed when the bloom progressed. This is in agreement with the negative correlation between microcystin content per cell and Microcystis abundance during a bloom that was found by Welker et al. (30). However, the microcystin concentrations and cell numbers that were measured at our sampling dates did not fully substantiate this correlation, since the increasing Microcystis populations in Lake ‘t Joppe had the highest microcystin content per cell in August (Table 2).
Some colonies had accumulated in scums, as evidenced by their dominance in DGGE profiles of scum samples compared to water column samples (Fig. 3). Accumulation in scums could be due to a greater capability for gas vesicle formation of these strains or to an increased sinking rate of other strains. Cell density changes resulting from the accumulation of polyglucose ballast are dependent on irradiance and turbulence and may take place in the course of a day (29). It would be interesting to study the distribution of strains over the scum and water column throughout a day, but this requires more intensive sampling. The dominant bands in scum samples appeared to correspond to nontoxic colonies (bands e12 and e25) or colonies of unknown toxin production (bands e7, e10, e24, and e26). The latter strains may be expected to be toxin producers, since the microcystin concentration per cell was higher in scums compared to the water column (Table 2) while no higher abundance of known toxin producers was observed. Alternatively, low numbers of high-toxin-producing genotypes may be present that do not give rise to major bands in DGGE profiles yet have a large impact on overall toxin concentration.
The number of different Microcystis sequences that could be distinguished in ITS DGGE community profiles of water samples was lower than the number that had been identified based on isolated colonies. For instance, in ITSa profiles from ‘t Joppe, we detected 12 Microcystis genotypes (Fig. 3) while the colonies isolated from this lake were differentiated into 18 groups based on ITSa (Table 1). This indicates that the colony isolation procedure also yielded less-abundant genotypes. Other discrepancies also signified a bias in the isolation or processing of colonies. While most isolated Zeegerplas colonies were identified as microcystin producers, two dominant sequences from the Zeegerplas profiles (bands e15 = e30 and e17) corresponded to nontoxic colonies and two other dominant sequences (bands e13 = e16 and e14) corresponded to colonies of unknown toxicity (Fig. 3 and Table 3). Possibly, selectivity against small colonies of the isolation procedure (Via-Ordorika et al., unpublished) resulted in preferential isolation of larger colonies, which are more often toxic (12). Also, the abundance in many community profiles of (identical) bands e2, e7, e13, and e16 should have resulted in the isolation of the corresponding colonies. An explanation for the resistance of these Microcystis strains to isolation could be their formation of small or no colonies that are missed by the colony isolation procedure yet are detected in community profiles which are generated from filtered water samples. A more intensive sampling and colony isolation study is required to elucidate the sequence identity of all bands in the community profiles and thereby substantiate insights in the dynamics of Microcystis genotypes.
Acknowledgments
This work was supported by the Dutch Technology Foundation (STW) project no. ACH.4874. P.M.V. was sponsored by The Netherlands Organization for Scientific Research (NWO) Meervoud no. 836.01.030.
All participants of the European Union workshop CYANOTOX and TOPIC (project no. ENV4-CT98-0802 and FMRX CT98-0246, respectively) are acknowledged for their efforts in isolating and identifying Microcystis colonies. L. Via-Ordorika is gratefully acknowledged for supplying mcy gene PCR data.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. H. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bolch, C. J. S., S. I. Blackburn, G. J. Jones, P. T. Orr, and P. M. Grewe. 1997. Plasmid content and distribution in the cyanobacterial genus Microcystis Kützing ex Lemmermann (cyanobacteria: Chroococcales). Phycologia 36:6-11. [Google Scholar]
- 3.Chorus, I., and J. Bartram. 1999. Toxic cyanobacteria in water. E. & F. N. Spon, London, United Kingdom.
- 4.Chorus, I., V. Niesel, J. Fastner, C. Wiedner, B. Nixdorf, and K.-E. Linden-Schmidt. 2001. Environmental factors and microcystin levels in water bodies, p. 159-177. In I. Chorus (ed.), Cyanotoxins: occurrences, causes, consequences. Springer-Verlag KG, Berlin, Germany.
- 5.Dittmann, E., B. A. Neilan, M. Erhard, H. von Döhren, and T. Börner. 1997. Insertional mutagenesis of a peptide synthetase gene that is responsible for hepatotoxin production in the cyanobacterium Microcystis aeruginosa PCC 7806. Mol. Microbiol. 26:779-787. [DOI] [PubMed] [Google Scholar]
- 6.Fastner, J., M. Erhard, and H. von Döhren. 2001. Determination of oligopeptide diversity within a natural population of Microcystis spp. (Cyanobacteria) by typing single colonies by matrix-assisted laser desorption ionization-time of flight mass spectrometry. Appl. Environ. Microbiol. 67:5069-5076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fastner, J., I. Flieger, and U. Neumann. 1998. Optimised extraction of microcystins from field samples-a comparison of different solvents and procedures. Water Res. 32:3177-3181. [Google Scholar]
- 8.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 9.Janse, I., M. Meima, W. E. A. Kardinaal, and G. Zwart. 2003. High-resolution differentiation of cyanobacteria by using rRNA-internal transcribed spacer denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 69:6634-6643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaebernick, M., T. Rohrlack, K. Christoffersen, and B. A. Neilan. 2001. A spontaneous mutant of microcystin biosynthesis: genetic characterization and effect on Daphnia. Environ. Microbiol. 3:669-679. [DOI] [PubMed] [Google Scholar]
- 11.Komárek, J., and K. Anagnostidis. 1999. Cyanoprokaryota, vol. 1. Teil Chroococcales, p. 225-236. Gustav Fischer Verlag, Jena, Germany.
- 12.Kurmayer, R., G. Christiansen, and I. Chorus. 2003. The abundance of microcystin-producing genotypes correlates positively with colony size in Microcystis sp. and determines its microcystin net production in Lake Wannsee. Appl. Environ. Microbiol. 69:787-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kurmayer, R., E. Dittmann, J. Fastner, and I. Chorus. 2002. Diversity of microcystin genes within a population of the toxic cyanobacterium Microcystis spp. in Lake Wannsee (Berlin, Germany). Microb. Ecol. 43:107-118. [DOI] [PubMed] [Google Scholar]
- 14.Lawton, L. A., C. Edwards, and G. A. Codd. 1994. Extraction and high-performance liquid-chromatographic method for the determination of microcystins in raw and treated waters. Analyst 119:1525-1530. [DOI] [PubMed] [Google Scholar]
- 15.Long, B. M., G. J. Jones, and P. T. Orr. 2001. Cellular microcystin content in N-limited Microcystis aeruginosa can be predicted from growth rate. Appl. Environ. Microbiol. 67:278-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mikalsen, B., G. Boison, O. M. Skulberg, J. Fastner, W. Davies, T. M. Gabrielsen, K. Rudi, and K. S. Jakobsen. 2003. Natural variation in the microcystin synthetase operon mcyABC and impact on microcystin production in Microcystis strains. J. Bacteriol. 185:2774-2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neilan, B. A., E. Dittmann, L. Rouhiainen, R. A. Bass, V. Schaub, K. Sivonen, and T. Börner. 1999. Nonribosomal peptide synthesis and toxigenicity of cyanobacteria. J. Bacteriol. 181:4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nishizawa, T., M. Asayama, K. Fujii, K. Harada, and M. Shirai. 1999. Genetic analysis of the peptide synthetase genes for a cyclic heptapeptide microcystin in Microcystis spp. J. Biochem. 126:520. [DOI] [PubMed] [Google Scholar]
- 19.Orr, P. T., and G. J. Jones. 1998. Relationship between microcystin production and cell division rates in nitrogen-limited Microcystis aeruginosa cultures. Limnol. Oceanogr. 43:1604-1614. [Google Scholar]
- 20.Otsuka, S., S. Suda, R. H. Li, S. Matsumoto, and M. M. Watanabe. 2000. Morphological variability of colonies of Microcystis morphospecies in culture. J. Gen. Appl. Microbiol. 46:39-50. [DOI] [PubMed] [Google Scholar]
- 21.Otsuka, S., S. Suda, R. H. Li, M. Watanabe, H. Oyaizu, S. Matsumoto, and M. M. Watanabe. 1999. Phylogenetic relationships between toxic and non-toxic strains of the genus Microcystis based on 16S to 23S internal transcribed spacer sequence. FEMS Microbiol. Lett. 172:15-21. [DOI] [PubMed] [Google Scholar]
- 22.Rippka, R. 1988. Isolation and purification of cyanobacteria. Methods Enzymol. 167:3-27. [DOI] [PubMed] [Google Scholar]
- 23.Rohrlack, T., M. Henning, and J.-G. Kohl. 2001. Isolation and characterisation of colony-forming Microcystis aeruginosa strains, p. 152-158. In I. Chorus (ed.), Cyanotoxins-occurrence, causes, consequences. Springer-Verlag, Berlin, Germany.
- 24.Sivonen, K., and G. J. Jones. 1999. Cyanobacterial toxins, p. 41-111. In I. Chorus and J. Bartram (ed.), Toxic cyanobacteria in water. E. & F. N. Spon, London, United Kingdom.
- 25.Tillett, D., E. Dittmann, M. Erhard, H. von Döhren, T. Borner, and B. A. Neilan. 2000. Structural organization of microcystin biosynthesis in Microcystis aeruginosa PCC7806: an integrated peptide-polyketide synthetase system. Chem. Biol. 7:753-764. [DOI] [PubMed] [Google Scholar]
- 26.Tillett, D., D. L. Parker, and B. A. Neilan. 2001. Detection of toxigenicity by a probe for the microcystin synthetase A gene (mcyA) of the cyanobacterial genus Microcystis, comparison of toxicities with 16S rRNA and phycocyanin operon (phycocyanin intergenic spacer) phylogenies. Appl. Environ. Microbiol. 67:2810-2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van de Peer, Y., and R. De Wachter. 1994. TREECON for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput. Appl. Biosci. 10:569-570. [DOI] [PubMed] [Google Scholar]
- 28.Vezie, C., L. Brient, K. Sivonen, G. Bertru, J. C. Lefeuvre, and M. Salkinoja-Salonen. 1998. Variation of microcystin content of cyanobacterial blooms and isolated strains in Lake Grand-Lieu (France). Microb. Ecol. 35:126-135. [DOI] [PubMed] [Google Scholar]
- 29.Wallace, B. B., and D. P. Hamilton. 1999. The effect of variations in irradiance on buoyancy regulation in Microcystis aeruginosa. Limnol. Oceanogr. 44:273-281. [Google Scholar]
- 30.Welker, M., H. von Döhren, H. Tauscher, C. E. W. Steinberg, and M. Erhard. 2003. Toxic Microcystis in shallow lake Müggelsee (Germany)-dynamics, distribution, diversity. Arch. Hydrobiol. 157:227-248. [Google Scholar]