Abstract
Although a large number of key odorants of Swiss-type cheese result from amino acid catabolism, the amino acid catabolic pathways in the bacteria present in these cheeses are not well known. In this study, we compared the in vitro abilities of Lactobacillus delbrueckii subsp. lactis, Lactobacillus helveticus, and Streptococcus thermophilus to produce aroma compounds from three amino acids, leucine, phenylalanine, and methionine, under mid-pH conditions of cheese ripening (pH 5.5), and we investigated the catabolic pathways used by these bacteria. In the three lactic acid bacterial species, amino acid catabolism was initiated by a transamination step, which requires the presence of an α-keto acid such as α-ketoglutarate (α-KG) as the amino group acceptor, and produced α-keto acids. Only S. thermophilus exhibited glutamate dehydrogenase activity, which produces α-KG from glutamate, and consequently only S. thermophilus was capable of catabolizing amino acids in the reaction medium without α-KG addition. In the presence of α-KG, lactobacilli produced much more varied aroma compounds such as acids, aldehydes, and alcohols than S. thermophilus, which mainly produced α-keto acids and a small amount of hydroxy acids and acids. L. helveticus mainly produced acids from phenylalanine and leucine, while L. delbrueckii subsp. lactis produced larger amounts of alcohols and/or aldehydes. Formation of aldehydes, alcohols, and acids from α-keto acids by L. delbrueckii subsp. lactis mainly results from the action of an α-keto acid decarboxylase, which produces aldehydes that are then oxidized or reduced to acids or alcohols. In contrast, the enzyme involved in the α-keto acid conversion to acids in L. helveticus and S. thermophilus is an α-keto acid dehydrogenase that produces acyl coenzymes A.
Amino acid catabolism by the microflora is a major process for the formation of a large number of key aroma compounds in Swiss-type cheeses such as gruyere and emmental (7, 14, 17). However, the amino acid catabolic pathways in the different bacteria present in these cheeses are not well known. This knowledge could lead to the development of cultures with optimized aromatic properties.
Amino acid catabolism by Lactococcus lactis and mesophilic lactobacilli has been extensively studied recently (2, 8, 11, 19, 26; S. Gummalla and J. R. Broadbent, Abstract, J. Dairy Sci. 79(Suppl. 1):101, 1996). In these lactic acid bacteria (LAB), amino acid catabolism is mainly initiated by a transamination reaction, which requires the presence of an α-keto acid as the amino group acceptor. The production of an α-keto acid acceptor by LAB often limits amino acid catabolism, but strains exhibiting glutamate dehydrogenase (GDH) activity are capable of producing α-ketoglutarate (α-KG) from glutamate (Glu) and therefore are capable of degrading amino acids in a reaction medium containing Glu (20).
The key aroma compounds identified in Swiss-type cheese result mainly from the catabolism of branched-chain amino acids (3-methylbutanal, isobutyric acid, isovaleric acid, and their derived esters), methionine (methional, methanethiol, and dimethyl trisulfide), and phenylalanine (phenylacetic acid and phenylacetaldehyde). Most of these compounds are considered beneficial to the cheese flavor. Isobutyric acid and isovaleric acid participate in the sweaty and strong cheese notes, respectively, while 3-methylbutanal has a malty aroma. All the compounds derived from methionine have sulfur notes that are appreciated in cheese. Finally, phenylacetic acid and phenylacetaldehyde have a floral or fruity note. However, a few of them, such as methional and compounds derived from tryptophan such as indole, have been identified as being responsible for off flavors when they are present in too high amounts (13, 17).
The microflora of Swiss-type cheese consists mainly of propionibacteria and thermophilic LAB, especially Lactobacillus helveticus, Lactobacillus delbrueckii subsp. lactis, and Streptococcus thermophilus. Recently, Thierry et al. have shown that propionibacteria are capable of producing isovaleric acid from leucine in vitro (21) and are mainly responsible for its production in Swiss cheese (22). However, propionibacteria do not seem to be capable of producing aldehydes from amino acids, while these compounds are very important for Swiss cheese aroma. Among the thermophilic LAB, only amino acid conversion by L. helveticus was partially studied. In vitro studies have shown that L. helveticus can produce acetaldehyde from threonine (10), volatile sulfur compounds from methionine (5, 18), and, in the presence of α-KG, benzaldehyde from phenylalanine (12). Under simulated cheese-ripening conditions, L. helveticus cells mainly produces carboxylic acids and hydroxyacids from phenylalanine and tyrosine (9).
Investigations of amino acid catabolism by S. thermophilus and L. delbrueckii subsp. lactis have been limited. Cystathionine-β-lyase activity, which is involved in the production of volatile sulfur compounds from methionine in Lactococcus lactis (6), has been detected in only one strain of L. delbrueckii subsp. lactis of five strains tested (3).
The aim of the present study was to compare the in vitro abilities of L. delbrueckii subsp. lactis, L. helveticus, and S. thermophilus to produce aroma compounds from three amino acids, leucine, methionine, and phenylalanine, and to elucidate the catabolic pathways used by these bacteria. Here we report the results obtained with one laboratory strain of each species, but otherwise we used two other industrial strains for each species to determine if the amino acid catabolism was strain dependent.
MATERIALS AND METHODS
Bacterial strains and preparation of cells for amino acid degradation.
The three strains used in this study were obtained from the Centre National de la Recherche Zootechnique culture collection (CNRZ, INRA, Jouy-en-Josas, France): L. helveticus CNRZ 32, L. delbrueckii subsp. lactis CNRZ 207, and S. thermophilus CNRZ 302. Six other strains of S. thermophilus were used for the determination of GDH activity: CNRZ 390, CNRZ 1446, CNRZ 1447, CNRZ 1591, CNRZ 385, and LMG 18311 (LMG collection, Laboratory of Microbiology, University of Gest, Gest, Belgium). All strains were stored at −80°C in MRS broth (Difco) containing 20% (vol/vol) glycerol. Strains were cultivated (1% inoculum) in 10% Nilac low-heat skim milk powder (Netherlands Institute for Dairy Research, Ede, The Netherlands) at 42°C for streptococci and 45°C for lactobacilli. Cells in the late exponential growth phase were harvested by centrifugation (5,000 × g, 10 min, 4°C), washed twice with 50 mM sodium glycerophosphate buffer (pH 7.5), and suspended in the same buffer to an optical density at 480 nm of 200. Cell dry weights were determined by drying 100 to 300 μl of cell suspensions under vacuum (3 h) with a SpeedVac concentrator. Aliquots of cell suspensions (200 μl) were stored at −80°C until used.
Determination of GDH activity in cell extracts.
Cell extracts were prepared by disrupting cells with a mini-bead beater 8TM cell disrupter, as previously described (11). NAD- and NADP-dependent GDH activities were determined in fresh cell extracts with the test based on the colorimetric glutamic acid assay of Boehringer, as previously described (11). Briefly, the reduced cofactor (NADH or NADPH) produced by oxidative deamination of glutamate reacts with iodonitrotetrazolium in the presence of diaphorase to produce a colored product that absorbs at a wavelength of 492 nm. Quantification was made with calibration curves established with pure NADH and NADPH. The test was performed with enzyme-linked immunoassay plates. One unit of GDH activity corresponds to the production of 1 nmol of NAD(P)H per min of reaction. The results are the means of three determinations (plus or minus the standard deviation).
Analysis of amino acid catabolism by HPLC.
The catabolism of Phe, Leu, and Met by the different strains was investigated in different reaction mixtures. The basic reaction mixture consisted of 70 mM potassium phosphate buffer (pH 5.5), 3 mM unlabeled amino acid (Phe, Leu, or Met), and 0.05 μM radiolabeled amino acid (l-[2,3,4,5,6-3H]phenylalanine, 100 to 140 Ci/mmol; l-[4,5-3H]leucine, 120 to 190 Ci/mmol; or l-[methyl-3H]methionine, 70 to 85 Ci/mmol) (Amersham Pharmacia Biotech). In some experiments, 0.05 mM pyridoxal phosphate (PLP) and 10 mM α-ketoglutarate (α-KG) or 10 mM glutamate (Glu) were added to the reaction medium. When specified, 10 mM sodium m-arsenite (NaAsO2; Sigma Chemical Co., St. Louis, Mo.) was added as an inhibitor of keto acid dehydrogenases. Resting cell suspension (0.05 ml) was added to 0.45 ml of the different reaction mixtures, and the mixtures were incubated for 40 h at 37°C. Aliquots (100 μl) of the reaction mixture were taken after 0, 10, 20, and 40 h of incubation, and cells were removed by centrifugation (5,000 × g, 5 min).
Cell lysis was evaluated during incubation by measuring cell protein release in the supernatant of the reaction medium. The protein concentration was determined in the supernatant and in cells (after cell disruption with a cell disrupter as described above) by the method of Bradford (4).
The soluble metabolites were then analyzed by reverse-phase high-pressure liquid chromatography (HPLC) and ion exclusion HPLC with both UV and radioactivity detection as previously described (24). Identification was made by comparison of the retention times with those of appropriate standard compounds, obtained from Sigma Chemical Co. Table 1 shows the common names of metabolites resulting from Phe, Leu, and Met catabolism.
TABLE 1.
Common names of compounds resulting from Phe, Leu, and Met degradation
| Type | Formula | Compound formed
|
||
|---|---|---|---|---|
| Phe | Leu | Met | ||
| α-Keto acid | R-CH2-CO-COOH | Phenylpyruvate | α-Ketoisocaproate | α-Ketomethylthiobutyrate |
| Hydroxy acid | R-CH2-CHO-COOH | Phenyllactate | Hydroxyisocaproate | Hydroxymethylthiobutyrate |
| Carboxylic acid | R-CH2-COOH | Phenylacetic acid | Isovaleric acid | 3-Methylthiopropionic acid |
| Aldehyde | R-CH2-CHO | Phenylacetaldehyde | 3-Methylbutanal or isovaleraldehyde | Methional |
| Alcohol | R-CH2-CHOH | 3-Methylbutanol | Methionol | |
| Other volatile sulfur compounds | Methanethiol, dimethyl disulfide, dimethyl trisulfide | |||
Analysis of amino acid catabolism by headspace gas chromatography-mass spectrometry.
The catabolism of Leu and Met by the different strains was investigated in the reaction mixture containing PLP and α-KG but without radiolabeled amino acid. A volume of 0.15 ml of the cell suspension was added to 1.35 ml of the reaction mixture, and incubations were performed for 40 h at 37°C. Aliquots (500 μl) of the reaction mixture were taken after 0, 20, and 40 h of incubation, and cells were removed by centrifugation (5,000 × g, 5 min). The supernatants were stored at −20°C until analysis.
The neutral volatile compounds were analyzed with a system composed of a 3100 Purge and Trap concentrator (Tekmar and Dohrmann, Cincinnati, Ohio) fitted with a sorbent trap (Tenax, 60 to 80 mesh, 0.25 g, 30 by 0.32 cm; Tekmar Inc.) and a cryofocusing module. The concentrator was coupled to a gas chromatograph (GC 6890+, Agilent Technologies Inc., Palo Alto, Calif.) connected to a mass spectrometer detector (MS 5973N; Agilent Technologies). The reaction mixture diluted with pure water (0.5 ml of the supernatant plus 1.5 ml of boiled MilliQ water) was heated at 60°C and then purged for 15 min with high-purity helium at 2.88 ml min−1 (5 lb/in2). The volatile compounds were concentrated by adsorption to the Tenax trap maintained at 30°C for analysis of Leu metabolites and at 10°C for analysis of Met metabolites. Water was then removed by flushing the trap with helium for 5 min (dry purge). The trap was heated at 225°C for 2 min to desorb the volatile compounds, which were directly transferred at 150°C to the head of a capillary column and condensed by cryofocusing at −150°C.
The condensed volatile compounds were then injected by heating (225°C) on a nonpolar capillary column (HP-5MS, 30.0 m by 0.25 mm, 0.25-μm film thickness) flushed with high-purity helium at a constant pressure of 30 lb/in2. The oven temperature was held at 5°C for 8 min. Then the metabolites were eluted with a first linear gradient of 3°C min−1 from 5 to 20°C, followed by a second linear gradient of 10°C min−1 from 20 to 150°C. The temperature was then held at 150°C for 10 min. The compounds were identified and quantified by mass spectrometry. The column was directly connected to the mass sensitive detector by an interface heated at 280°C. The electron impact energy was set at 70 eV, and data were collected in the range of 20 to 200 atomic mass units. Components were identified by their retention times and by comparison of their mass spectra with those of the NIST98 library (National Institute of Standards and Technology). The concentration of components was calculated by external standard calibration.
Control of experimental conditions used for the study of amino acid catabolism.
The method used to study amino acid catabolism by the different strains consisted of incubating resting cells stored at −80°C at 37°C for 40 h in a phosphate buffer at pH 5.5 (average pH of cheese) in the presence of one amino acid and in the presence or the absence of cofactors (PLP and α-KG). Before beginning the study, we verified three points of the experimental protocol. First, we verified that similar quantities of cells from the different strains were added when we used optical density for measuring the cell concentration of cell suspensions. Indeed, the dry weights of the cells per ml of suspension (optical density at 480 nm = 200) were similar whatever the strain used (27.0 ± 4.7 mg [dry weight] ml−1). Second, we checked that storage of cells at −80°C did not affect the amino acid catabolism of the different strains. The comparison between fresh cells and cells stored at −80°C was made for each strain by incubating the cells in reaction medium containing α-KG and PLP. The amino acid catabolism observed was quantitatively and qualitatively similar for both types of cells (data not shown). Finally, we verified that cells were not lysed after incubation by measuring intracellular protein release in the supernatants of the reaction medium after incubation with cells of each strain. Less than 0.2% of the total cell protein was detected in the supernatants of all reaction mixtures after 40 h of incubation, indicating that the strains did not lyse significantly under our incubation conditions.
RESULTS
Amino acid catabolism in the absence of α-KG in the reaction medium and GDH activity.
Amino acid catabolism by L. helveticus CNRZ 32, L. delbrueckii subsp. lactis CNRZ 207, and S. thermophilus CNRZ 302 was studied in vitro with radiolabeled Phe, Leu, and Met as tracers. In reaction medium without α-KG, only little or no amino acid degradation was observed with the two lactobacilli, while S. thermophilus significantly degraded Phe, Leu, and Met (Table 2). Figure 1 shows that S. thermophilus degraded the three amino acids to various metabolites, which were identified as α-keto acids, products of amino acid transamination or deamination, and their further degradation products, hydroxy acids and carboxylic acids. The major metabolites produced from Phe and Met were carboxylic acids (phenylacetic acid and methylthiopropionic acid). Carboxylic acid was also the main degradation product of the α-keto acid derived from Leu, but a large proportion of the α-keto acid was not degraded.
TABLE 2.
Amino acid degradation by Lactobacillus and Streptococcus strains after incubation for 40 h in reaction mixture without α-KGa
| Strain | % Amino acid degradation ± SD
|
||
|---|---|---|---|
| Phe | Leu | Met | |
| L. helveticus CNRZ 32 | 2.3 ± 0.9 | 0.6 ± 1.1 | 1.3 ± 2.1 |
| L. delbrueckii subsp. lactis CNRZ 207 | 1.3 ± 1.2 | 0.5 ± 0.6 | 0 |
| S. thermophilus CNRZ 302 | 17.0 ± 7.1 | 21.1 ± 4.3 | 8.0 ± 3.6 |
Results are expressed as a percentage of the initial amino acid concentration and are the means of three independent experiments ± standard deviation.
FIG. 1.
Metabolites produced from phenylalanine, leucine, and methionine by S. thermophilus CNRZ 302 after incubation for 40 h in reaction medium without α-KG addition but containing Glu. Metabolites were analyzed by HPLC with radioactivity detection. Results are expressed as percentages of the initial amino acid concentration and are the means of three independent experiments. Error bars indicate the variation in the total of metabolites. The names of metabolites resulting from amino acid catabolism are given in Table 1.
Since it has been shown previously that the ability of several LAB to catabolize amino acids in reaction medium without α-KG addition is closely related to their GDH activity, we searched for both NAD- and NADP-GDH activities in the three thermophilic LAB strains. S. thermophilus CNRZ 302 exhibited high NADP and NAD activities (221 and 16 U mg of protein −1, respectively) while no activity was detected for the lactobacilli. Therefore, six other Streptococcus strains were further screened. NADP-dependent activity was detected in all the strains tested except S. thermophilus LMG 18311, but the level of activity was strain dependent, while NAD-dependent activity was found only in S. thermophilus CNRZ 302. Since strains possessing GDH activity produce α-KG from Glu, we examined the ability of S. thermophilus CNRZ 302 to degrade amino acids in a reaction medium containing Glu. The levels of amino acid degradation in the presence (Fig. 1) and in the absence (data not shown) of Glu were similar, suggesting that the intracellular pool of Glu was sufficient to produce the α-KG required for amino acid transamination.
Amino acid catabolism in the presence of α-KG in the medium.
Amino acid catabolism by L. helveticus CNRZ 32, L. delbrueckii subsp. lactis CNRZ 207, and S. thermophilus CNRZ 302 was also studied in the reaction mixture containing α-KG. Figure 2 shows the metabolites produced from Phe, Leu, and Met by the three LAB species after incubation for 40 h. Amino acid catabolism by the three strains in the presence of α-KG was much higher than in its absence, as previously observed with L. lactis (15, 25). Indeed, previously it was shown that α-KG addition stimulated the amino acid catabolism initiated by a transamination step because α-KG is the cosubstrate of aminotransferases. Therefore, we can conclude that the catabolism of Phe, Leu, and Met by the three LAB species studied is also initiated by a transamination reaction. α-Keto acids resulting from transamination (phenylpyruvate, ketoisocaproate, and ketomethylthiobutyrate) were further differently transformed by the three species of LAB.
FIG. 2.
Metabolites produced from phenylalanine, leucine, and methionine in reaction medium containing α-KG and PLP by L. helveticus CNRZ 32 (A), L. delbrueckii subsp. lactis CNRZ 207 (B), and S. thermophilus CNRZ 302 (C). Metabolites were analyzed after 40 h of incubation by HPLC with radioactivity detection. Results are expressed as percentages of the initial amino acid concentration and are the means of three independent experiments. Error bars indicate the variation in the total of metabolites. The names of metabolites resulting from amino acid catabolism are given in Table 1.
From Phe and Leu, lactobacilli produced larger quantities of key aroma compounds such as carboxylic acids, aldehydes, and alcohols (Fig. 2A and B) than S. thermophilus CNRZ 302, which mainly produced α-keto acids and, to a lesser extent, hydroxy acids (phenyllactate), which are not aroma compounds (Fig. 2C). S. thermophilus CNRZ 302 also produced 3 to 13% carboxylic acids from the three amino acids, but the production of phenylacetic acid from Phe was fourfold lower than in the reaction medium without α-KG addition (Fig. 1). A similar observation was previously reported for GDH-positive lactobacilli (11) and for a genetically modified L. lactis strain overproducing a GDH (16). The mechanism of such behavior is not yet understood, but it may be related to a change in the intracellular oxidoreduction potential caused by the oxidative deamination of Glu to α-KG.
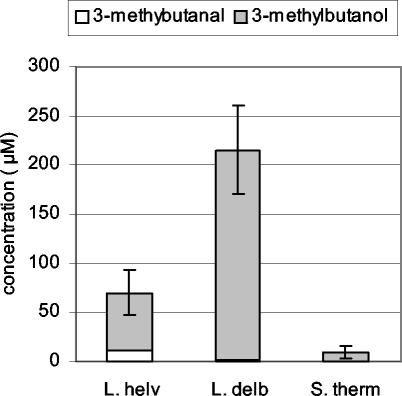
L. helveticus CNRZ 32 (Fig. 2A) mainly transformed phenylpyruvate into phenylacetic acid and, to a lesser extent, to phenylethanol and α-ketoisocaproate into isovaleric acid. From 50 to 100 μmol of 3-methylbutanol and 3-methylbutanal per liter was also detected by headspace gas chromatography-mass spectrometry (Fig. 3). The quantity of aldehydes produced from Phe and Leu after 20 h of incubation was higher than the quantity produced after 40 h of incubation, while the quantity of alcohols was the highest after 40 h (data not shown), suggesting that aldehydes were further transformed to alcohols.
FIG. 3.
Production of volatile compounds from leucine in reaction medium containing α-KG and PLP by L. helveticus CNRZ 32 (L. helv), L. delbrueckii subsp. lactis CNRZ 207 (L. delb), and S. thermophilus CNRZ 302 (S. therm). Metabolites were analyzed by headspace gas chromatography-mass spectrometry after 40 h of incubation.
L. delbrueckii subsp. lactis CNRZ 207 (Fig. 2B) produced much less carboxylic acids but larger amounts of alcohol from Phe and Leu than L. helveticus. About 200 μmol of 3-methylbutanol per liter was also found by headspace gas chromatography-mass spectrometry (Fig. 3), but aldehydes were not detected even after 20 h of incubation.
From Met, L. helveticus and S. thermophilus mainly produced α-keto acid (ketomethylthiobutyrate) and 5 to 10% carboxylic acid. L. delbrueckii subsp. lactis mainly converted ketomethylthiobutyrate to hydroxy acid and carboxylic acid. Negligible quantities of volatile sulfur compounds were detected by headspace gas chromatography-mass spectrometry in the reaction medium containing Met and resting cells of the three strains (data not shown).
Amino acid catabolism pathways.
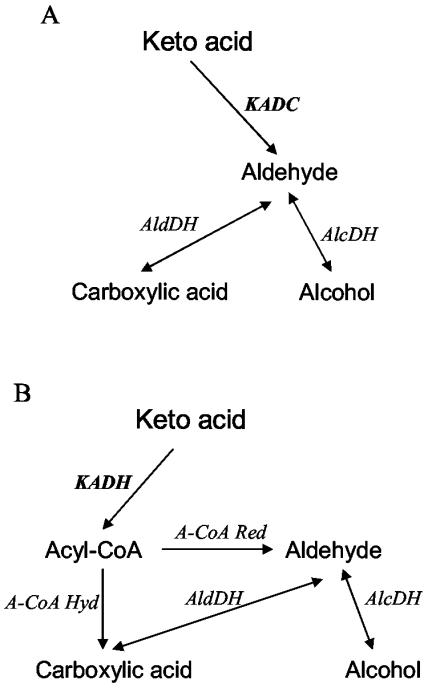
To identify the catabolic pathways used by the three strains for the production of acids, alcohols, and aldehydes, resting cells were incubated in reaction medium in the presence of arsenite, which is a specific inhibitor of α-keto acid dehydrogenases (23). In fact, acids, alcohols, and aldehydes could be produced from α-keto acids via two different pathways (Fig. 4). The first pathway (Fig. 4A) consists of α-keto acid decarboxylation to aldehydes by an α-keto acid decarboxylase (KADC), followed by an aldehyde reduction or oxidation to alcohol or acid, respectively. The second pathway (Fig. 4B) consists of oxidative decarboxylation of α-keto acid to acyl coenzyme A (acyl-CoA) by an α-keto acid dehydrogenase (KADH). Acyl-CoA can then be directly converted into carboxylic acid by an acyl-CoA hydrolase or reduced to aldehyde by an acyl-CoA reductase. Aldehyde can be further oxidized or reduced to carboxylic acid or alcohol, respectively.
FIG. 4.
Two possible pathways for the formation of acid, aldehyde, and alcohol from α-keto acid: the KADC pathway (A) and the KADH pathway (B). The names of metabolites resulting from amino acid catabolism are given in Table 1.
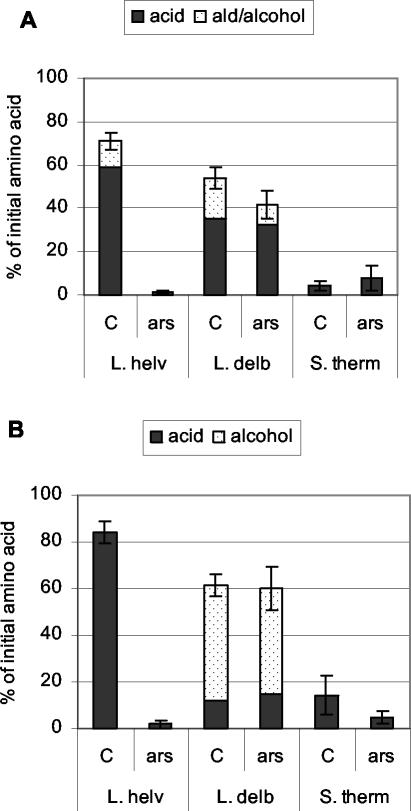
Sodium arsenite addition did not affect the total percentage of Phe and Leu degradation (data not shown), indicating that amino acid conversion to α-keto acids was not affected, and did not affect hydroxy acid formation. But Fig. 5 shows that sodium arsenite totally inhibited the conversion of α-keto acids derived from Phe and Leu into aldehydes, alcohols, and acids by the L. helveticus strain and slightly reduced the formation of carboxylic acids from the α-keto acid of Leu by the S. thermophilus strain. In the presence of sodium arsenite, α-keto acids accumulated in the reaction medium. On the contrary, sodium arsenite did not affect the formation of these compounds by L. delbrueckii subsp. lactis. These results indicate that L. helveticus mainly used the KADH pathway, while L. delbrueckii subsp. lactis used the KADC pathway.
FIG. 5.
Metabolites produced from Phe (A) and Leu (B) by L. helveticus CNRZ 32 (L. helv), L. delbrueckii subsp. lactis CNRZ 207 (L. delb), and S. thermophilus CNRZ 302 (S. therm) in reaction medium containing α-KG and PLP in the presence of sodium arsenite (ars) or in its absence (C). Metabolites were analyzed after 40 h of incubation by HPLC with radioactivity detection. Results are expressed as percentages of the initial amino acid concentration and are the means of three independent experiments. Error bars indicate the variation in the total of metabolites. The names of metabolites resulting from amino acid catabolism are given in Table 1.
We also verified the presence of aldehyde and alcohol dehydrogenases in lactobacilli by incubating the cells with phenylacetaldehyde (derived from Phe) and isovaleraldehyde (derived from Leu). Indeed, both lactobacilli produced alcohols and acids from these aldehydes (results not shown).
DISCUSSION
As previously demonstrated for many LAB, including L. lactis, mesophilic lactobacilli, and propionibacteria (2, 8, 11, 19, 26; Gummall and Broadbent, abstract), our results showed that catabolism of Phe, Leu, and Met in L. helveticus, L. delbrueckii subsp. lactis, and S. thermophilus is initiated by an amino acid transamination step, which requires the presence of an α-keto acid such α-KG as an amino group acceptor for the aminotransferases. This finding was confirmed with two other strains of each species. Among the three LAB species tested in this study, only S. thermophilus generally exhibited GDH activity that produced α-KG from glutamate. Consequently, only S. thermophilus was capable of catabolizing amino acids in the reaction medium without α-KG addition.
GDH activity appeared to be widespread among S. thermophilus species, but the intensity of activity was strain dependent. Although S. thermophilus LMG 18311 did not exhibit significant GDH activity, a gene sharing high homology with gdh genes from other bacteria has been found in its genome (UCL Life Sciences Institute website, www.biol.ucl.ac.be/gene/genome). However, it is not possible to know whether the gene is functional or not because the open reading frame has not been totally sequenced yet. Surprisingly, although the GDH activity of S. thermophilus CNRZ 302 was much higher (50-fold) than the activity previously determined in some L. plantarum and L. paracasei strains, the level of amino acid degradation by S. thermophilus CNRZ 302 was not higher than the degradation observed with the L. plantarum or L. paracasei strains under similar conditions (20). Amino acid degradation by S. thermophilus is not limited by accumulation of α-keto acids (without further degradation of these compounds) because, in the presence of α-KG, amino acid degradation was threefold higher than without α-KG addition. Therefore, our results suggest that the GDH of S. thermophilus is less efficient than that of other GDH-positive LAB for producing α-KG. However, the reason for such poor efficiency remains to be explained. GDH activity was not detected in the three L. helveticus strains or in the three L. delbrueckii subsp. lactis strains, but more strains should be tested before generalizing because this activity appeared to be quite rare in certain LAB species, such as L. lactis (20).
In the presence of α-KG, thermophilic lactobacilli produced much more varied aroma compounds than S. thermophilus. Indeed, L. helveticus and L. delbrueckii subsp. lactis produced acids, alcohols, and aldehydes from Phe and Leu, while S. thermophilus mainly produced α-keto acids, which are not by themselves aroma compounds. This finding was verified with two other strains of each species. L. helveticus mainly produced phenylacetic acid from Phe and isovaleric acid from Leu, as previously observed for propionibacteria (21), while L. delbrueckii subsp. lactis produced large amounts of alcohols (and/or aldehydes). Among the LAB studied previously, only a few strains of L. lactis and Lactobacillus maltaromicus also produced large amounts of aldehyde (26). The catabolism of Leu in S. thermophilus looks like that previously described for L. lactis (11), since both produced mainly α-ketoisocaproate from Leu. L. lactis produced more carboxylic acids from Phe and Met than S. thermophilus in medium containing α-KG, but in medium without α-KG addition, S. thermophilus also produced large amounts of carboxylic acids.
The formation of alcohols and acids by L. delbrueckii subsp. lactis mainly results from the action of a KADC (Fig. 6B). KADC produces aldehydes that are then either oxidized to acids or reduced to alcohols, probably depending on the redox potential inside the cell or in the medium, since lactobacilli exhibited both aldehyde and alcohol dehydrogenase activities. The formation of aldehyde by certain L. lactis strains is also due to a KADC that was recently partially purified (1). On the contrary, L. helveticus uses the KADH pathway (Fig. 6A). KADHs produce acyl-CoAs from α-keto acids, but the pathway for further conversion of acyl-CoAs to acids and aldehydes has not been identified yet. Other experiments are necessary to discriminate between the acyl-CoA hydrolase and acyl-CoA reductase pathways. The pathway used by S. thermophilus has not been clearly identified because acid production was very low and not really significant.
FIG. 6.
Hypothetical pathways for amino acid catabolism by L. helveticus CNRZ 32 (A) and L. delbrueckii subsp. lactis CNRZ 207 (B). The major metabolites produced are in bold, and the most abundant is underlined. AT, aminotransferase; GDH, glutamate dehydrogenase; HADH, hydroxy acid dehydrogenase; KADH, keto acid dehydrogenase; KADC, keto acid decarboxylase; AlcDH, alcohol dehydrogenase; AldDH, aldehyde dehydrogenase. The names of metabolites resulting from amino acid catabolism are given in Table 1.
Our results suggest a major role of lactobacilli, especially L. delbrueckii subsp. lactis, in flavor development in cheese because they have the enzymatic potential to produce potent and varied aroma compounds from amino acids. However, these lactobacilli do not seem to be capable of producing the α-KG required for amino acid transamination, which initiates the amino acid conversion. S. thermophilus generates α-KG from Glu due to its GDH activity and therefore could initiate amino acid catabolism by producing α-keto acids. Cooperation in amino acid catabolism may occur between S. thermophilus and thermophilic lactobacilli, as has been shown in vitro for L. lactis and L. paracasei (11). Indeed, the GDH-positive lactobacilli degraded amino acids to α-keto acids that were further transformed to carboxylic acids by L. lactis. Further studies are necessary to verify that such cooperation between S. thermophilus and thermophilic lactobacilli occurs in cheese.
Acknowledgments
This work was supported by the Entremont Group, Annecy, France.
We thank Pascal Courtin for technical assistance, Véronique Monnet, Catherine Tanous, and Agnieszka Kieronczyk for critical reading of the manuscript, and Annick Lacombe from INRA's translation unit for revising the English version of the manuscript.
REFERENCES
- 1.Amarita, F., D. Fernandez-Espla, T. Requena, and C. Pelaez. 2001. Conversion of methionine to methional by Lactococcus lactis. FEMS Microbiol. Lett. 204:189-195. [DOI] [PubMed] [Google Scholar]
- 2.Amarita, F., T. Requena, G. Taborda, L. Amigo, and C. Pelaez. 2001. Lactobacillus casei and Lactobacillus plantarum initiate catabolism of methionine by transamination. J. Appl. Microbiol. 90:971-978. [DOI] [PubMed] [Google Scholar]
- 3.Aubel, D., J. E. Germond, C. Gilbert, and D. Atlan. 2002. L'activité β-cystathionase au sein de l'espèce Lactobacillus delbrueckii. Sci. Aliments 22:161-166. [Google Scholar]
- 4.Bradford, M. M. 1976. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 5.Dias, B., and B. Weimer. 1998. Conversion of methionine to thiols by lactococci, lactobacilli, and brevibacteria. Appl. Environ. Microbiol. 64:3320-3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fernandez, M., W. van Doesburg, G. A. M. Rutten, J. D. Marugg, A. C. Alting, R. van Kranenburg, and O. P. Kuipers. 2000. Molecular and functional analyses of the metC gene of Lactococcus lactis, encoding cystathionine β-lyase. Appl. Environ. Microbiol. 66:42-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Friedrich, J. E., and T. E. Acree. 1998. Gas chromatography olfactometry (GC/O) of dairy products. Int. Dairy J. 8:235-241. [Google Scholar]
- 8.Gummalla, S., and J. R. Broadbent. 1999. Tryptophan catabolism by Lactobacillus casei and Lactobacillus helveticus cheese flavor adjuncts. J. Dairy Sci. 82:2070-2077. [DOI] [PubMed] [Google Scholar]
- 9.Gummalla, S., and J. R. Broadbent. 2001. Tyrosine and phenylalanine catabolism by Lactobacillus cheese flavor adjuncts. J. Dairy Sci. 84:1011-1019. [DOI] [PubMed] [Google Scholar]
- 10.Hickey, M. W., A. J. Hillier, and G. R. Jago. 1983. Enzymic activities associated with lactobacilli in dairy products. Aust. J. Dairy Technol. 38:154-157. [Google Scholar]
- 11.Kieronczyk, A., S. Skeie, T. Langsrud, and M. Yvon. 2003. Cooperation between Lactococcus lactis and nonstarter lactobacilli in the formation of cheese aroma from amino acids. Appl. Environ. Microbiol. 69:734-739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klein, N., M. B. Maillard, A. Thierry, and S. Lortal. 2001. Conversion of amino acids into aroma compounds by cell-free extracts of Lactobacillus helveticus. J. Appl. Microbiol. 91:404-411. [DOI] [PubMed] [Google Scholar]
- 13.Parliment, T. H., M. G. Kolor, and D. J. Rizzo. 1982. Volatile components of Limburger cheese. J. Agric. Food Chem. 30:1006-1008. [Google Scholar]
- 14.Preininger, M., and W. Grosch. 1994. Evaluation of key odorants of the neutral volatiles of Emmentaler cheese by the calculation of odour activity values. Lebensm.-Wiss. Technol. 27:237-244. [Google Scholar]
- 15.Rijnen, L., S. Bonneau, and M. Yvon. 1999. Genetic characterization of the major lactococcal aromatic aminotransferase and its involvement in conversion of amino acids to aroma compounds. Appl. Environ. Microbiol. 65:4873-4880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rijnen, L., P. Courtin, J.-C. Gripon, and M. Yvon. 2000. Expression of a heterologous glutamate dehydrogenase gene in Lactococcus lactis highly improves the conversion of amino acids to aroma compounds. Appl. Environ. Microbiol. 66:1354-1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rychlik, M., and J. O. Bosset. 2001. Flavour and off-flavours compounds of Swiss Gruyère cheese. Identification of key odorants by quantitative instrumental and sensory studies. Int. Dairy J. 11:903-910. [Google Scholar]
- 18.Seefeldt, K. E., and B. C. Weimer. 2000. Diversity of sulfur compound production in lactic acid bacteria. J. Dairy Sci. 83:2740-2746. [DOI] [PubMed] [Google Scholar]
- 19.Tammam, J. D., A. G. Williams, J. Noble, and D. Lloyd. 2000. Amino acid fermentation in non-starter Lactobacillus spp. isolated from Cheddar cheese. Lett. Appl. Microbiol. 30:370-374. [DOI] [PubMed] [Google Scholar]
- 20.Tanous, C., A. Kieronczyk, S. Helinck, E. Chambellon, and M. Yvon. 2002. Glutamate dehydrogenase activity: a major criterion for the selection of flavour-producing lactic acid bacteria strains. Antonie van Leeuwenhoek 82:271-278. [PubMed] [Google Scholar]
- 21.Thierry, A., M. B. Maillard, and M. Yvon. 2002. Conversion of l-leucine to isovaleric acid by Propionibacterium freudenreichii TL 34 and ITGP23. Appl. Environ. Microbiol. 68:608-615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thierry, A., R. Richoux, and J.-R. Kerjean. Isovaleric acid is mainly produced by Propionibacterium freudenreichii in Swiss cheese. Int. Dairy J., in press.
- 23.Webb, J. L. 1966. Enzyme and metabolic inhibitors. Academic Press, New York, N.Y.
- 24.Yvon, M., S. Berthelot, and J. C. Gripon. 1998. Adding α-KG to semi-hard cheese curd highly enhances the conversion of amino acids to aroma compounds. Int. Dairy J. 8:889-898. [Google Scholar]
- 25.Yvon, M., E. Chambellon, A. Sorokine, and F. Roudot-Algaron. 2000. Characterization and role of the branched-chain aminotransferase (BcaT) isolated from Lactococcus lactis subsp. cremoris NCDO 763. Appl. Environ. Microbiol. 66:571-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yvon, M., and L. Rijnen. 2001. Cheese flavour formation by amino acid catabolism. Int. Dairy J. 11:185-201. [Google Scholar]