Norovirus is a leading cause of acute gastroenteritis. This nationally representative case-control study found that children's Fucocyltransferase 2 (FUT2) (secretor) genotype strongly influences susceptibility to norovirus infection, particularly with GII.4 strains. Hispanics have the highest frequency of the FUT2 susceptibility genotype.
Keywords: norovirus, FUT2, gastroenteritis, diarrhea, histo-blood group antigens
Abstract
Background. Norovirus is a leading cause of acute gastroenteritis (AGE). Noroviruses bind to gut histo-blood group antigens (HBGAs), but only 70%–80% of individuals have a functional copy of the FUT2 (“secretor”) gene required for gut HBGA expression; these individuals are known as “secretors.” Susceptibility to some noroviruses depends on FUT2 secretor status, but the population impact of this association is not established.
Methods. From December 2011 to November 2012, active AGE surveillance was performed at 6 geographically diverse pediatric sites in the United States. Case patients aged <5 years were recruited from emergency departments and inpatient units; age-matched healthy controls were recruited at well-child visits. Salivary DNA was collected to determine secretor status and genetic ancestry. Stool was tested for norovirus by real-time reverse transcription polymerase chain reaction. Norovirus genotype was then determined by sequencing.
Results. Norovirus was detected in 302 of 1465 (21%) AGE cases and 52 of 826 (6%) healthy controls. Norovirus AGE cases were 2.8-fold more likely than norovirus-negative controls to be secretors (P < .001) in a logistic regression model adjusted for ancestry, age, site, and health insurance. Secretors comprised all 155 cases and 21 asymptomatic infections with the most prevalent norovirus, GII.4. Control children of Meso-American ancestry were more likely than children of European or African ancestry to be secretors (96% vs 74%; P < .001).
Conclusions. FUT2 status is associated with norovirus infection and varies by ancestry. GII.4 norovirus exclusively infected secretors. These findings are important to norovirus vaccine trials and design of agents that may block norovirus-HBGA binding.
(See the Editorial Commentary by Debbink on pages 1639–41.)
Norovirus is associated with nearly one-fifth of acute gastroenteritis (AGE) cases worldwide and is the leading cause of pediatric AGE in countries with universal rotavirus vaccination [1, 2]. Norovirus causes 200 000 pediatric deaths each year in developing countries [3]. Though rarely fatal in developed countries, norovirus AGE causes substantial morbidity and is responsible for 20 million cases annually in the United States alone [4]. The illness is characterized by sudden onset of severe vomiting and dehydrating diarrhea lasting 1–3 days [5]. Its high transmissibility makes norovirus an important public health concern, particularly in closed settings such as long-term care facilities, schools and child care centers, and military barracks [5, 6].
Human susceptibility to norovirus infection is thought to be at least partially dependent upon an individual's fucosyltransferase 2 (FUT2) genotype [7–16]. FUT2 controls the secretion of ABO histo-blood group antigens (HBGAs) at the gut surface. These carbohydrates serve as binding ligands and presumed receptors for caliciviruses, the virus family that includes norovirus [7]. However, in vitro and in silico studies have shown that noroviruses bind to HBGAs in a strain-specific manner [8, 9].
At least 1 functional FUT2 allele is present in 70%–80% of individuals, who are referred to as FUT2 “secretors” [17]. FUT2 is inactivated in the remaining 20%–30% (“nonsecretors”) by homozygous 428G > A nonsense mutations in European and African populations, or less common variants such as the 385A > T missense mutation found in Asian populations [17]. Individuals of Meso-American descent rarely carry these inactivating mutations [10, 17].
In challenge studies, FUT2 nonsecretors have demonstrated resistance to specific noroviruses, developing neither symptoms nor antibody response to norovirus genotypes GI.1 or GII.4 [11, 12, 18]. In epidemiologic studies, FUT2 nonsecretors have demonstrated significantly lower susceptibility to symptomatic infection by some norovirus genotypes (GI.1, GII.3, GII.4) [13–15, 18] but not others (GI.3, GII.3) [15, 16].
Although numerous outbreak studies have shown increased susceptibility of FUT2 secretors to specific noroviruses, the impact of FUT2 on overall risk of sporadic norovirus in racially/ethnically diverse populations has not been examined. Furthermore, secretor genetics in relation to asymptomatic shedding of norovirus has not been studied in populations in which the nonsecretor genotype is common. Finally, although in vitro work suggests that A and B blood group antigens may modify risk of norovirus infection in secretor individuals [8], this question has not been well addressed in epidemiologic studies.
With norovirus vaccines currently in development [2], understanding population patterns of susceptibility to this pathogen is crucial. Our study utilizes a large, prospective, geographically diverse AGE surveillance network to identify sporadic pediatric norovirus infections. We use these surveillance data to determine the relationship of FUT2 secretor status to symptomatic and asymptomatic norovirus infection and modification of that relationship by non-O blood group types, and to describe FUT2 secretor status in the United States by racial/ethnic background.
MATERIALS AND METHODS
Recruitment and Data Collection
The New Vaccine Surveillance Network (NVSN) performs active surveillance of pediatric diseases, coordinated by the US Centers for Disease Control and Prevention (CDC). For this study, enrollment occurred during the 12-month period of December 2011 to November 2012 at the University of Rochester Medical Center (Rochester, New York), Vanderbilt University Medical Center (Nashville, Tennessee), Cincinnati Children's Hospital Medical Center (Cincinnati, Ohio), Texas Children's Hospital (Houston, Texas), Children's Mercy Hospital and Clinics (Kansas City, Missouri), and Seattle Children's Hospital (Seattle, Washington). Approval was obtained from the institutional review boards at each site and the CDC.
Surveillance methods have been described previously [19, 20]. In brief, prospective active surveillance was conducted in the emergency departments and inpatient units of NVSN institutions. Eligible cases were between 14 days and 5 years of age with AGE symptoms of <10 days’ duration. AGE symptoms were defined as diarrhea ≥3 episodes or vomiting ≥1 episode within 24 hours. Children were excluded if they had a noninfectious cause of diarrhea or vomiting, immunodeficiency, or had transferred from another hospital.
Healthy controls between 14 days and 5 years of age were enrolled during scheduled well-child visits at affiliated clinics. They were excluded if they had symptoms of AGE 14 days prior to enrollment, immunodeficiency, or symptoms of acute respiratory infection 3 days prior to enrollment. The enrollment of healthy controls was frequency-matched to the enrollment of AGE cases at each site by age and calendar month.
Written informed consent was obtained in English at all sites, or in Spanish at the Kansas City, Seattle, and Houston sites. Eligible children were not enrolled if the consenting adult did not speak an available language. Demographic and clinical data were systematically collected from all subjects. Whole saliva was collected from subjects at enrollment and stored in DNA stabilization buffer (Oragene Discover for assisted collection, DNA Genotek). Whole stool samples were obtained from both cases and controls, with 98% of cases providing stool within 7 days of enrollment. Stool samples were tested for GI and GII norovirus by real-time reverse transcription polymerase chain reaction (RT-PCR) assay [21] that included an MS2 coliphage extraction control followed by amplification of the positive samples by conventional RT-PCR and sequencing. Noroviruses were genotyped by comparing the sequences to a CaliciNet database of norovirus prototype strains [21].
Human Genotyping and Ancestry Determination
Human DNA from saliva was genotyped using both RT-PCR and the Immunochip (Illumina Infinium), which has been described previously [22]. The Immunochip was selected for its coverage of the FUT2 and ABO locus as well as its inclusion of genetic ancestral informative markers (AIMs).
Samples were prepared for Immunochip analysis using Illumina protocols. Only samples and single-nucleotide polymorphisms (SNPs) with call rates >95% were retained. Samples were checked for agreement with self-identified sex and autosomal heterozygosity <0.4.
O/non-O blood type was determined using SNP rs505922, which acts as a surrogate for the deletion site that creates the typical O phenotype [23].
FUT2 SNPs rs601338 (428G > A nonsense mutation) and rs1800030 (rare 849G > A nonsense mutation) were directly genotyped from the Immunochip. The additional nonsecretor mutations 385T > G and 571C > T were imputed from Immunochip data using the program IMPUTE2. This program calculates the genotype of unobserved SNPs with >0.9 probability by comparing highly associated observed SNPs to a database of haplotypes constructed from the 1000 Genomes Project [24]. For quality control, 10% of all samples as well as samples that failed Immunochip analysis were analyzed at the FUT2 428 position using RT-PCR (TaqMan, Life Technologies). Samples from subjects of self-identified Asian ancestry were further analyzed at position 385 by TaqMan assay.
Genetic ancestry was determined using 1723 AIMs on the Immunochip. Principal components analysis was used to identify and remove genetic outliers of >6 standard deviations. Genetic clusters were then created using the well-described program STRUCTURE [25]. In brief, STRUCTURE uses Markov chain Monte Carlo methods to create genetically similar clusters by maximizing within-group Hardy–Weinberg equilibrium. Program parameters included 3 assumed genetic sources with admixture among groups. The resulting clusters were labeled according to the self-identified race/ethnicity most prevalent in each group.
Statistical Analysis
Demographic and clinical data for both cases and controls were examined in relation to secretor genotype by χ2 or Fisher exact test. The Cochran–Mantel–Haenszel test was used to control for race, ethnicity, and genetic ancestry as appropriate. Logistic regression was used to generate odds ratios (ORs) adjusted for covariates. The regression model was stratified by genetic ancestry.
RESULTS
Enrolled Norovirus Cases and Controls
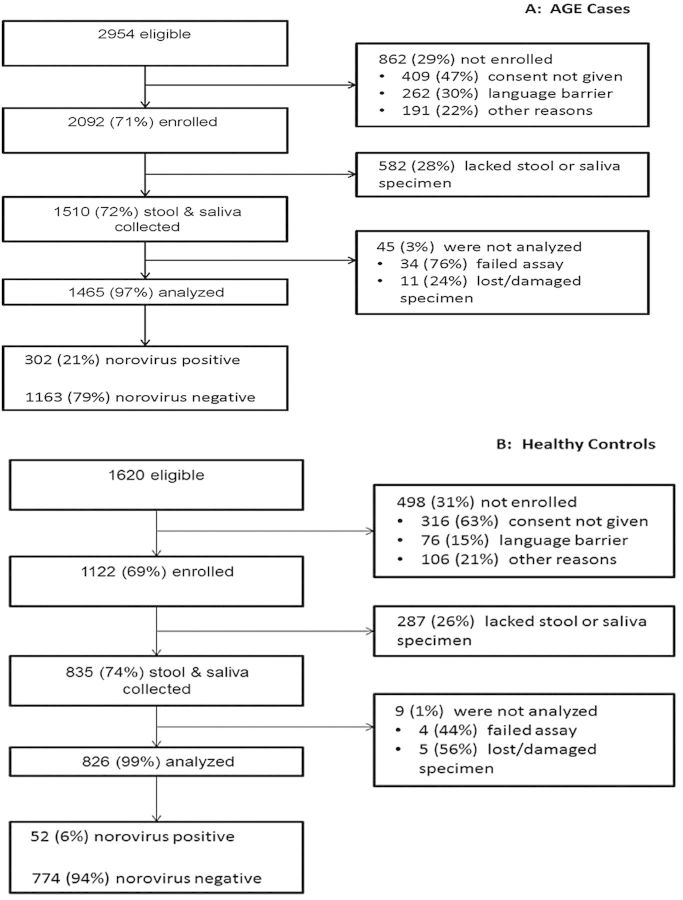
During the year of prospective surveillance, 2954 children with AGE were identified, of whom 2092 (71%) were enrolled in the study (Figure 1). There were 1122 children enrolled as healthy controls. Among enrolled children, testing was completed on stool and saliva specimens from 1465 (70%) children with AGE and 826 (74%) healthy controls. Sequence-confirmed GI and GII norovirus RNA was detected in stool specimens from 302 (21%) AGE cases and 52 (6%) controls (Figure 1). Norovirus AGE cases were comparable to controls for demographic variables such as sex and insurance status but differed modestly from controls by age, study site, and self-identified race/ethnicity (Table 1).
Figure 1.
Flow diagram of subject participation in the study, including eligibility assessment, enrollment, specimen collection, testing for norovirus, and final determination of norovirus status. A, Process of acute gastroenteritis (AGE) case inclusion. B, Process of control inclusion. In each panel, the left-hand column of boxes indicates subjects included at each study step; the right-hand column of boxes details the subjects excluded from each study step and the primary reasons for exclusion.
Table 1.
Demographics of Norovirus Acute Gastroenteritis Cases and All Healthy Control Study Completers
| Characteristic | Norovirus AGE Cases, No. (%) | Healthy Controls, No. (%) |
|---|---|---|
| Total | 302 (100) | 826 (100) |
| Sex | ||
| Male | 171 (57) | 428 (52) |
| Female | 131 (43) | 398 (48) |
| Study site | ** | |
| Nashville | 66 (22) | 157 (19) |
| Rochester | 8 (3) | 58 (7) |
| Cincinnati | 47 (16) | 208 (25) |
| Seattle | 23 (8) | 54 (7) |
| Houston | 43 (14) | 107 (13) |
| Kansas City | 115 (38) | 242 (29) |
| Health insurance | ||
| Public insurance | 231 (76) | 627 (76) |
| Private insurance | 59 (20) | 180 (22) |
| None/unknown | 12 (4) | 19 (2) |
| Age | ** | |
| 0–5 mo | 34 (11) | 218 (26) |
| 6–11 mo | 81 (27) | 165 (20) |
| 12–23 mo | 94 (31) | 225 (27) |
| 24–35 mo | 51 (17) | 95 (12) |
| 36–59 mo | 42 (14) | 123 (15) |
| Self-identified race | ** | |
| White | 145 (48) | 324 (39) |
| Black | 99 (33) | 376 (46) |
| Multiple/other | 58 (19) | 126 (15) |
| Self-identified ethnicity | ** | |
| Non-Hispanic | 210 (70) | 681 (82) |
| Hispanic | 92 (30) | 145 (18) |
| Genetic ancestry | ** | |
| White | 135 (45) | 335 (41) |
| Black | 101 (33) | 392 (47) |
| Hispanic | 53 (18) | 74 (9) |
| Othera | 13 (4) | 25 (3) |
Abbreviation: AGE, acute gastroenteritis.
a Outliers and those without ancestry data.
** P < .001, norovirus AGE cases vs controls.
Genetic Ancestry Grouping
A total of 2241 (98%) subjects had Immunochip data for detailed analysis of genetic ancestry. Principal components analysis using AIMs identified 21 genetic outliers (all self-identified Asian/Pacific Islander). These outlier subjects did not form a separate cluster, and none was homozygous for either the FUT2 428 or 385 inactivating mutations; they could thus not be analyzed as a distinct subgroup. The remaining 2220 subjects were classified by STRUCTURE into 3 genetically similar clusters (white, black, and Hispanic [Meso-American]), which had 95% agreement with self-identified race/ethnicity. Genetic ancestry was considered to agree with self-identified racial/ethnic background if any self-identified category was matched. Of the 582 (26%) subjects who self-identified multiple race/ethnicities, 62% reported backgrounds that were Hispanic and white.
Individuals in the genetically determined Meso-American subgroup were significantly more likely than white or black subgroups to be secretors (in controls, 96% vs 74%; P < .001). This association was also significant using self-identified Hispanic ethnicity (Table 2).
Table 2.
Relationship of Secretor Status to Norovirus Infection (Norovirus Acute Gastroenteritis Cases), Norovirus-Asymptomatic Carriers (Norovirus-Positive Controls), and Norovirus-Negative Controls
| Characteristic | Secretors in Norovirus AGE Cases, No. (%) | Secretors in Asymptomatic Carriers, No. (%) | Secretors in Norovirus-Negative Controls, No. (%) |
|---|---|---|---|
| Total | 276/302 (91) | 47/52 (90) | 591/774 (76) |
| Self-identified race | ** | * | |
| White | 136/145 (94) | 17/18 (94) | 240/306 (78) |
| Black | 85/99 (86) | 23/25 (92) | 253/351 (72) |
| Multiple/other | 55/58 (95) | 7/9 (78) | 98/117 (84) |
| Self-identified ethnicity | ** | * | |
| Non-Hispanic | 185/210 (88) | 37/42 (88) | 475/639 (74) |
| Hispanic | 91/92 (99) | 10/10 (100) | 116/135 (86) |
| Genetic ancestrya | ** | * | |
| White | 123/135 (91) | 17/19 (89) | 235/316 (74) |
| Black | 87/101 (86) | 24/27 (89) | 267/365 (73) |
| Hispanic | 53/53 (100) | 5/5 (100) | 66/69 (96) |
| Otherb | 13/13 (100) | 1/1 (100) | 23/24 (96) |
Abbreviation: AGE, acute gastroenteritis.
a Genetic ancestry: 92% of individuals in white genetic ancestry group self-identified as white; 98% of individuals in black genetic ancestry group self-identified as black; 97% of individuals in Hispanic genetic ancestry group self-identified as Hispanic.
b Outliers and those without ancestry data.
* P < .05 vs norovirus-negative controls.
** P < .001 vs norovirus-negative controls.
Secretor Genotype and Norovirus Infection
The only FUT2 nonsecretor polymorphism observed in our study population was at position 428, and this SNP was therefore used to determine secretor status. Of the demographic variables shown in Table 1, race, ethnicity, genetic ancestry, and study site were significantly associated with FUT2 secretor status. After controlling for AGE case status, study site was not significant. The varying secretor status distribution among controls by race, ethnicity, and genetic ancestry reflected the expected population distribution.
Cases with norovirus AGE were significantly more likely to be secretors than norovirus-negative healthy controls (OR, 2.78; 95% confidence interval [CI], 1.77–4.37; P < .001) in a logistic regression model that included genetic ancestry, age, study site, and health insurance. Results were similar regardless of self-identified or genetically determined ancestral background (Table 2), when norovirus-positive (asymptomatic carrier) children were included or excluded from the controls, or within a model of children aged <12 months that included breastfeeding status.
Among the children studied, 14 different norovirus genotypes were detected. Only 1 coinfection with multiple noroviruses (GI.6 and GII.1) was detected. The most commonly detected genotype, norovirus GII.4, was detected in 155 (51%) norovirus AGE cases including 28 (9%) GII.4 Den Haag 2006b, 71 (24%) GII.4 New Orleans 2009, and 56 (19%) GII.4 Sydney 2012. All (100%) GII.4-positive samples from AGE cases were collected from children who were FUT2 secretors (Figure 2). The second most common genotype, GII.6, was present in 43 (14%) children with norovirus AGE and also exhibited significant secretor specificity (P = .01). The remaining norovirus genotypes did not display secretor specificity (Figure 2).
Figure 2.
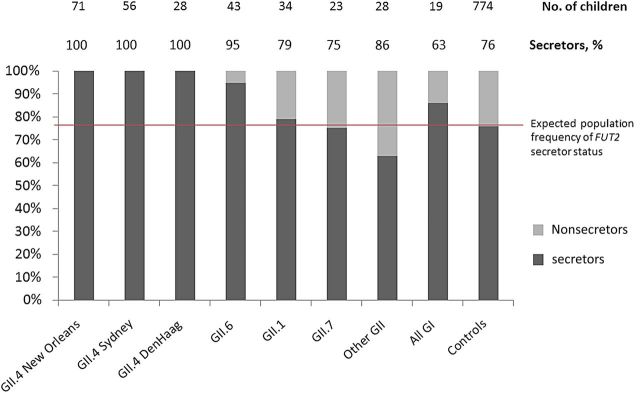
Stacked bar chart representing the percentage of FUT2 secretors and nonsecretors in norovirus acute gastroenteritis (AGE) cases compared with norovirus-free healthy controls. The first 8 individual bars from the left represent the secretor status distribution of all 302 norovirus AGE cases infected with a specific norovirus genogroup, genotype, or strain. The most commonly detected norovirus genotype, GII.4, was detected in 155 AGE cases. The GII.4 strains identified—New Orleans, Sydney, and Den Haag—were only detected in FUT2 secretors (P < .001, compared with the frequency of secretors in controls). The second most common norovirus genotype, GII.6, also exhibited a secretor specificity (P = .01). Noroviruses GII.1, GII.7, GI, and others, occurred less frequently and did not display secretor specificity. The “Other GII” category represents 4 less commonly detected norovirus GII genotypes (GII.2, GII.3, GII.8, and GII.13) and 1 case coinfected with GII.1 and GI.6. FUT2 secretors represented 76% of the norovirus-free healthy controls (bar, far right), consistent with the expected population frequency. Not depicted are the 52 asymptomatic norovirus carriers who were enrolled as healthy controls. The distribution of strains, and secretor specificity of strains carried asymptomatically, was consistent with the pattern found in norovirus AGE (see text).
Asymptomatic norovirus carriers (norovirus-positive healthy controls) were also more likely to be secretors compared with norovirus-negative healthy controls (OR, 5.64; 95% CI, 1.16–7.69; P = .02) after adjusting for genetic ancestry (Table 2). All of the 21 GII.4 and 3 GII.6 carriers (46% of asymptomatic carriers) were secretors.
Modification of O/Non-O Blood Type
Among norovirus-negative controls, 44% were blood type O, consistent with the prevalence of blood type O in norovirus AGE cases (45%), secretor norovirus-negative controls (45%), and secretor norovirus AGE cases (45%). Analysis of norovirus genotype-specific O/non-O distribution including all AGE subjects and restricted to secretor-only AGE subjects showed no significant associations.
DISCUSSION
This large, multisite study found that a point nonsense mutation that is homozygous in nearly one-quarter of the global population dramatically impacts risk of norovirus infection. FUT2 secretors were at significantly greater risk for both symptomatic and asymptomatic norovirus infections. Similar associations of norovirus AGE and FUT2 status were recently found in single-hospital surveillance studies in West Africa, China, and Vietnam [15, 26, 27]. This finding should be considered in designing phase 3 clinical trials, as innate susceptibility to norovirus infections varies by this genetic factor. Challenge studies using noroviruses known to have a secretor preference are already incorporating the secretor status of test subjects into the study design [28].
We confirmed that secretor genotype varies by genetic ancestry and self-identified ethnicity. Norovirus-free control children of Hispanic ethnicity, whether identified by self-report or genetic analysis, were significantly more likely to be FUT2 secretors. This ancestral difference in a norovirus susceptibility factor should be taken into consideration in epidemiologic surveillance, disease modeling, and norovirus vaccine effectiveness studies. This principle may apply to other AGE pathogens as well. Some rotaviruses has been shown to have HBGA preferences by both Lewis and secretor blood types [29, 30]. Indeed, ethnic variation in Lewis binding to a component of the rotavirus vaccine was recently proposed as an explanation for reduced vaccine efficacy in an African population [29]. The significant association between FUT2 secretor status and risk of norovirus infection reported in this study is driven by the specificity of norovirus genotypes GII.4 and GII.6. These noroviruses comprised 67% of infections in symptomatic children and 46% of infections in asymptomatic children. Our finding that all GII.4 norovirus symptomatic and asymptomatic children were genetic secretors suggests that the nonsecretor genetic characteristic offers near-total protection from this predominant viral genotype. These findings are consistent with in vitro work with 6 different GII.4 strains showing that, while each strain had differential HBGA binding patterns, none bound the nonsecretor phenotype [9].
The results of this study could potentially influence decisions to prioritize vaccination of individuals who lack innate protection. However, we note that nonsecretors are susceptible to many noroviruses and thus remain candidates for vaccination. Furthermore, an individual's secretor status is rarely known, and targeted immunization is challenging to enact.
Noroviruses are known to bind to various HBGAs with strain-dependent binding patterns [8, 9]. It has been proposed that compounds that block HBGA binding sites could be used as novel therapeutics [31]. This strategy is already at play in nature. Human milk from FUT2 secretors has been shown to have norovirus-binding elements, and secretor infants fed with human milk higher in these elements have lower risk of AGE [31–33].
We examined the relationship of FUT2 to norovirus susceptibility because FUT2 secretor status alters gut HBGA expression. FUT2 enzymes cause secretion of H-antigen at the gut surface. Further glycosylation of the H-antigen is possible, such as the addition of A or B antigens by ABO [23]. We saw an association between norovirus and FUT2 secretor status, but within the secretor group did not see an association with O vs non-O blood type.
Our study has several limitations. We studied the circulating noroviruses only for 2011–2012 within the United States, and noroviruses are known to shift over time and geography. However, our 2011–2012 surveillance shows that 21% of children <5 years old with AGE tested positive for norovirus, similar to 2008–2010 surveillance [19]. Norovirus continues to be the leading cause of epidemic and sporadic AGE across all age groups, and our surveillance of sporadic AGE found a distribution of norovirus genotypes similar to that found during outbreak surveillance over the same period [6]. Our asymptomatic norovirus carriage rate of 6% is similar to recently published estimates of global norovirus asymptomatic carriage of 7% [1]. A recent longitudinal cohort study of children in Ecuador with an 18% asymptomatic carriage rate also found GII.4 norovirus only in secretor children. Interestingly, nonsecretor children in this cohort were significantly more likely to be infected with a non-GII.4 norovirus than secretor children, a finding not replicated in this study [34].
Furthermore, in our study, we found only the 428G > A inactivating polymorphism, although we examined 4 known FUT2 inactivating polymorphisms. The lack of other polymorphisms may be explained by the limited representation of children of Asian ancestry in our study population. In Asians, FUT2 polymorphisms other than 428G > A often define nonsecretors or low secretors. Though we cannot fully extrapolate our results to Asia or other regions where other FUT2 inactivating mutations are found, we expect results to be similar for any inactivating mutation [17]. In a recent study of 34 Chinese children hospitalized for AGE, homozygosity for the 385A > T “low secretor” mutation provided incomplete protection from norovirus GII.3 and GII.4 [27]. Additionally, genotype may not fully capture expression of gut HBGAs. Some genetic secretors have low expression of H-antigen and a phenotype similar to nonsecretors [35]. Future studies should examine whether low H-antigen expression in genetic secretors might also confer protection against norovirus.
In conclusion, our study indicates that FUT2 genotype is a major factor driving pediatric norovirus infection risk, particularly with the predominant GII.4 viruses. Our findings of strain- and ancestry-dependent susceptibility are relevant to understanding variation in norovirus risk within and between populations and may help guide the design and interpretation of norovirus vaccine trials.
Notes
Acknowledgments. We thank the numerous dedicated staff members at each New Vaccine Surveillance Network site, including Cincinnati team members Monica McNeal, Courtney Rhorer, Jayme Alexander, Sahle Amsalu, Nancy Back, Kyle Connolly, Vanessa Florian, Michelle Roth, Elizabeth Stahl, Michael Whalen, and Marilyn Rice; Houston team members Carol Baker, Marcie Rench, and Leila Sahni; Kansas City team members Karisa Deculus, Tiffany Hefner, Usha Kallemuchikkal, Mary Moffatt, Ashley Smith, Kirsten Weltmer, Ashley Willingham, and Cecilia Trombino; Nashville team members Jim Chappell, Rendie McHenry, Natalee Rathert, Yesenia Romero-Herazo, Angela Mendoze, and Becca Smith; Rochester team members Gerry Lofthus, Kenneth Schnabel, and Lynne Shelley; and Seattle team members Cathy Bull, Diane Kinnunen, Kirsten Lacombe, Jennifer Lenahan, and Bonnie Strelitz. We thank Morrow laboratory staff Myra Johnson Willwerth and Diana Taft for laboratory support, and Barbara Davidson and Donna Wuest for administrative support. We thank John Harley and Sara Lazaros-Palacios for advising and for use of equipment, and Leah Kottyan for consulting on Immunochip analysis.
Disclaimer. The findings and conclusions in this report are those of the author(s) and do not necessarily represent the views of the Centers for Disease Control and Prevention (CDC) or the Agency for Toxic Substances and Disease Registry.
Financial support. This work was supported by the CDC and the National Institutes of Health (grant numbers P01 HD013021 to A. L. M., F30 AI109893 to R. L. C., T32 GM063483, and T32 ES010957).
Potential conflicts of interest. All authors: No potential conflicts of interest.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Ahmed SM, Hall AJ, Robinson AE, et al. Global prevalence of norovirus in cases of gastroenteritis: a systematic review and meta-analysis. Lancet Infect Dis 2014; 14:725–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Debbink K, Lindesmith LC, Baric RS. The state of norovirus vaccines. Clin Infect Dis 2014; 58:1746–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Patel MM, Widdowson MA, Glass RI, Akazawa K, Vinje J, Parashar UD. Systematic literature review of role of noroviruses in sporadic gastroenteritis. Emerg Infect Dis 2008; 14:1224–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hall AJ, Lopman BA, Payne DC, et al. Norovirus disease in the United States. Emerg Infect Dis 2013; 19:1198–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Glass RI, Parashar UD, Estes MK. Norovirus gastroenteritis. N Engl J Med 2009; 361:1776–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vega E, Barclay L, Gregoricus N, Shirley SH, Lee D, Vinje J. Genotypic and epidemiologic trends of norovirus outbreaks in the United States, 2009 to 2013. J Clin Microbiol 2014; 52:147–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ruvoen-Clouet N, Ganiere JP, Andre-Fontaine G, Blanchard D, Le Pendu J. Binding of rabbit hemorrhagic disease virus to antigens of the ABH histo-blood group family. J Virol 2000; 74:11950–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Han L, Kitov PI, Kitova EN, et al. Affinities of recombinant norovirus P dimers for human blood group antigens. Glycobiology 2013; 23:276–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tian P, Yang D, Jiang X, et al. Specificity and kinetics of norovirus binding to magnetic bead-conjugated histo-blood group antigens. J Appl Microbiol 2010; 109:1753–62. [DOI] [PubMed] [Google Scholar]
- 10.Bucardo F, Kindberg E, Paniagua M, et al. Genetic susceptibility to symptomatic norovirus infection in Nicaragua. J Med Virol 2009; 81:728–35. [DOI] [PubMed] [Google Scholar]
- 11.Lindesmith L, Moe C, Marionneau S, et al. Human susceptibility and resistance to Norwalk virus infection. Nat Med 2003; 9:548–53. [DOI] [PubMed] [Google Scholar]
- 12.Frenck R, Bernstein DI, Xia M, et al. Predicting susceptibility to norovirus GII.4 by use of a challenge model involving humans. J Infect Dis 2012; 206:1386–93. [DOI] [PubMed] [Google Scholar]
- 13.Carlsson B, Kindberg E, Buesa J, et al. The G428A nonsense mutation in FUT2 provides strong but not absolute protection against symptomatic GII.4 norovirus infection. PLoS One 2009; 4:e5593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tan M, Jin M, Xie H, Duan Z, Jiang X, Fang Z. Outbreak studies of a GII-3 and a GII-4 norovirus revealed an association between HBGA phenotypes and viral infection. J Med Virol 2008; 80:1296–301. [DOI] [PubMed] [Google Scholar]
- 15.Van Trang N, Vu HT, Le NT, Huang P, Jiang X, Anh DD. Association between norovirus and rotavirus infection and histo-blood group antigen types in Vietnamese children. J Clin Microbiol 2014; 52:1366–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nordgren J, Kindberg E, Lindgren PE, Matussek A, Svensson L. Norovirus gastroenteritis outbreak with a secretor-independent susceptibility pattern, Sweden. Emerg Infect Dis 2010; 16:81–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ferrer-Admetlla A, Sikora M, Laayouni H, et al. A natural history of FUT2 polymorphism in humans. Mol Biol Evol 2009; 26:1993–2003. [DOI] [PubMed] [Google Scholar]
- 18.Parrino TA, Schreiber DS, Trier JS, Kapikian AZ, Blacklow NR. Clinical immunity in acute gastroenteritis caused by Norwalk agent. N Engl J Med 1977; 297:86–9. [DOI] [PubMed] [Google Scholar]
- 19.Payne DC, Vinje J, Szilagyi PG, et al. Norovirus and medically attended gastroenteritis in U.S. children. N Engl J Med 2013; 368:1121–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Payne DC, Boom JA, Staat MA, et al. Effectiveness of pentavalent and monovalent rotavirus vaccines in concurrent use among US children <5 years of age, 2009–2011. Clin Infect Dis 2013; 57:13–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vega E, Barclay L, Gregoricus N, Williams K, Lee D, Vinje J. Novel surveillance network for norovirus gastroenteritis outbreaks, United States. Emerg Infect Dis 2011; 17:1389–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cortes A, Brown MA. Promise and pitfalls of the Immunochip. Arthritis Res Ther 2011; 13:101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rummel S, Shriver CD, Ellsworth RE. Relationships between the ABO blood group SNP rs505922 and breast cancer phenotypes: a genotype-phenotype correlation study. BMC Med Genet 2012; 13:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet 2009; 5:e1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics 2000; 155:945–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nordgren J, Nitiema LW, Ouermi D, Simpore J, Svensson L. Host genetic factors affect susceptibility to norovirus infections in Burkina Faso. PLoS One 2013; 8:e69557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu P, Wang X, Lee JC, et al. Genetic susceptibility to norovirus GII.3 and GII.4 infections in Chinese pediatric diarrheal disease. Pediatr Infect Dis J 2014; 33:e305–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Atmar RL, Opekun AR, Gilger MA, et al. Determination of the 50% human infectious dose for Norwalk virus. J Infect Dis 2014; 209:1016–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nordgren J, Sharma S, Bucardo F, et al. Both Lewis and secretor status mediate susceptibility to rotavirus infections in a rotavirus genotype–dependent manner. Clin Infect Dis 2014; 59:1567–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Imbert-Marcille BM, Barbe L, Dupe M, et al. A FUT2 gene common polymorphism determines resistance to rotavirus A of the P[8] genotype. J Infect Dis 2014; 209:1227–30. [DOI] [PubMed] [Google Scholar]
- 31.Morrow AL, Ruiz-Palacios GM, Jiang X, Newburg DS. Human-milk glycans that inhibit pathogen binding protect breast-feeding infants against infectious diarrhea. J Nutr 2005; 135:1304–7. [DOI] [PubMed] [Google Scholar]
- 32.Jiang X, Huang P, Zhong W, Morrow AL, Ruiz-Palacios GM, Pickering LK. Human milk contains elements that block binding of noroviruses to histo-blood group antigens in saliva. Adv Exp Med Biol 2004; 554:447–50. [DOI] [PubMed] [Google Scholar]
- 33.Newburg DS, Ruiz-Palacios GM, Altaye M, et al. Innate protection conferred by fucosylated oligosaccharides of human milk against diarrhea in breastfed infants. Glycobiology 2004; 14:253–63. [DOI] [PubMed] [Google Scholar]
- 34.Lopman BA, Trivedi T, Vicuna Y, et al. Norovirus infection and disease in an Ecuadorian birth cohort: association of certain norovirus genotypes with host FUT2 secretor status. J Infect Dis 2014; doi:10.1093/infdis/jiu672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morrow AL, Meinzen-Derr J, Huang P, et al. Fucosyltransferase 2 non-secretor and low secretor status predicts severe outcomes in premature infants. J Pediatr 2011; 158:745–51. [DOI] [PMC free article] [PubMed] [Google Scholar]