Abstract
BACKGROUD
We used single-marker and novel gene-based methods to examine the associations of endothelial system genes with blood pressure (BP) changes and hypertension in a longitudinal family study.
METHODS
The Genetic Epidemiology Network of Salt Sensitivity follow-up study was conducted among 1,768 Chinese participants from 633 families. Nine BP measurements were obtained at baseline and at 2 follow-up visits using a random-zero sphygmomanometer. Mixed-effect models were used to assess the additive associations of 206 single-nucleotide polymorphisms (SNPs) in 15 endothelial system genes with longitudinal BP changes and hypertension incidence. Gene-based analyses were conducted using the truncated product method. The Bonferroni method was used to adjust for multiple testing in all analyses.
RESULTS
Among those free from hypertension at baseline, 512 (32.1%) developed hypertension during the average 7.2 years of follow-up. In single-marker analyses, each copy of the minor alleles of correlated SELE markers rs4656704, rs6427212, and rs5368 were associated with increased risk of developing hypertension (P for trend = 1.48×10−4, 6.69×10−5, and 7.64×10−5, respectively). In addition, the minor allele of SELE marker rs3917436 was associated with smaller diastolic BP (DBP) increases over time. Results of gene-based analyses confirmed associations of the SELE gene with the longitudinal BP phenotypes (P values < 1.00×10−6 for DBP change and hypertension incidence). Furthermore, the DDAH1 and COL18A1 genes were associated with systolic BP change (P < 1.00×10−6 and P = 4.00×10−6, respectively), while EDNRA was associated with hypertension incidence (P = 2.39×10−4).
CONCLUSIONS
The current study provides strong evidence of a role of endothelial system genes in BP progression and hypertension incidence.
Keywords: blood pressure changes, common variants, endothelial, hypertension.
Hypertension is a major public health challenge worldwide due to its high prevalence and associated increases in risks of cardiovascular diseases.1,2 As the leading risk factor for mortality globally, approximately 13.5% of premature deaths were attributable to high blood pressure (BP) in 2001.2 High BP can be described as a complex trait, influenced by multiple environmental and genetic factors. Heritability studies indicate that about 30%–60% of interindividual variation in BP is genetically determined.3,4 Still, the genomic mechanisms underlying BP regulation remain largely unknown.
Several reports have highlighted the importance of the endothelium on relaxing vascular smooth muscle and regulating vasomotor tone.5,6 Such findings suggest a potential role for endothelial dysfunction in the pathogenesis of hypertension. For example, studies have illustrated that endothelial-mediated vasodilatation is reduced not only in animal models of hypertension but also in humans with essential hypertension.7,8 These findings make endothelial system genes attractive candidates for genomic study of BP phenotypes. Several studies have already identified associations between genes encoding components of the endothelial system and prevalent hypertension.9–12 These studies have reported contributions of the endothelial nitric oxide synthase (NOS3), selectin E (SELE), endothelin receptor type A (EDNRA), and endothelin 1 (EDN1) genes to this complex phenotype.9–12 However, genomic research on longitudinal BP phenotypes, such as BP change over time and hypertension incidence, is limited. Only 4 studies of longitudinal BP traits have been conducted,10,13–15 including just 1 study conducted in East Asian participants.15 Furthermore, the previous studies examined only the NOS3 gene with inconsistent results.10,13–15 The paucity of research in this area highlights a need for additional study of the relation between endothelial system genes and longitudinal BP phenotypes.
The present study was undertaken to determine whether common variants in endothelial system genes can predict BP changes or hypertension incidence among 1,768 Han Chinese participants of the Genetic Epidemiology Network of Salt Sensitivity (GenSalt) follow-up study.
METHODS
Study population
The GenSalt study was a unique family-based dietary feeding study designed to examine the interaction of genes and sodium intake on BP. GenSalt participants were recruited from 6 northern, rural Chinese populations with high habitual sodium intake. Detailed information on the study design and methods of the GenSalt study has been published elsewhere.16 Briefly, community-based BP screenings were carried out among persons aged 18–60 years in the 6 study villages to identify potential probands. Those with mean systolic BP (SBP) of 130–160mm Hg and/or diastolic BP (DBP) of 85–100mm Hg and no use of antihypertensive medication were recruited, as well as with their parents, siblings, spouses, and offspring. Individuals who had stage 2 hypertension, secondary hypertension, and a history of clinical cardiovascular disease or diabetes or were pregnant, heavy alcohol drinkers, or currently on a low-sodium diet were excluded from the study.
The baseline examination and subsequent dietary intervention was carried out in 2003–2005. As part of the GenSalt follow-up study, GenSalt participants took part in 2 follow-up examinations that were conducted in 2008–2009 and again in 2011–2012. Among 1,906 eligible individuals from 633 families who participated in the GenSalt baseline examination, 117 individuals were missing BP data at both of the follow-up visits and another 21 individuals were missing genotype data. In total, 1,768 participants (92.28%) were included in the current analysis.
Written informed consents were obtained from all GenSalt participants after detailed explanation of the study. The study has been approved by the Institutional Review Boards at all the participating institutions.
Data collection on phenotype and other covariates
Prior to the dietary intervention, GenSalt participants underwent a 3-day baseline examination that was repeated at each of the 2 follow-up visits. Information on family structure, demographic characteristics, and personal and family medical history was collected by trained staff using a standardized questionnaire. Body weight and height were measured twice with the participants in light indoor clothing without shoes. Body mass index (BMI) was calculated as kilograms per square meter (kg/m2). BP was measured 3 times on each day of the 3-day baseline period and on each day of the two 3-day follow-up visits. Three random-zero BP measurements were obtained with participants in the sitting position after 5 minutes of rest by the same trained and certified technician using a Hawksley random-zero sphygmomanometer (Hawksley & Sons Ltd, Lancing, UK; zero range 0–20mm Hg) according to the protocols recommended by the American Heart Association.17 The appropriate cuff size was selected based on each study participant’s arm circumference.17 Additionally, participants were advised to avoid drinking, cigarette smoking, having coffee/tea, or doing exercise for at least 30 minutes before their BP measurements. BP levels at baseline and during each follow-up visit were calculated as the average of the 9 BP measurements. Hypertension was defined as SBP ≥ 140mm Hg and/or DBP ≥ 90mm Hg or use of antihypertensive medications.
Genotype data and quality control
We conducted a MEDLINE literature search to identify 16 candidate genes in the endothelial system, including EDN2, DDAH1, VCAM1, SELP, SELE, EDNRA, MEF2C, EDN1, SERPINE1, NOS3, VWF, EDNRB, CYBA, ICAM1, TGFB1, and COL18A1 genes. The detailed literature search strategy has been published elsewhere.18 Within the 16 candidate genes (±5,000-bp flanking regions), 347 single-nucleotide polymorphisms (SNPs) were genotyped on the Affymetrix 6.0 platform (Affymetrix, Santa Clara, CA). SNPs with minor allele frequency less than 1%, genotyping call rate less than 95%, and deviation from Hardy–Weinberg equilibrium after correction for multiple testing were excluded. After quality control, we selected functional SNPs and additional tag-SNPs from these genes with pairwise r 2 thresholds of less than 0.9. A total of 206 SNPs in 15 genes were included in the current analysis. Characteristics of genes are shown in Table 1. Characteristics of the SNPs, including information on location, function, and quality, are presented in Supplementary Table 1. Haploview software (version 4.2, http://www.broad.mit.edu/mpg/haploview) was used to conduct quality control and SNP selection.
Table 1.
Endothelial system genes selected for candidate gene study
| Locus | Gene symbol | Gene name | Physical position (±5,000bp) | No. of SNPs | Function |
|---|---|---|---|---|---|
| 1p34.2 | EDN2 | Endothelin 2 | (41939446, 41955344) | 4 | Encodes a potent endothelium-derived vasoconstrictor and mediates endothelial dysfunction through EDNRA |
| 1p22.3 | DDAH1 | Dimethylarginine dimethylaminohydrolase 1 | (85779164, 86048933) | 60 | Inactivate asymmetric dimethylarginine, which is the inhibitor of NO synthase |
| 1p21.2 | VCAM1 | Vascular cell adhesion module 1 | (101180298, 101209601) | 5 | Implicates in the process of leukocyte rolling, firm adhesion, and transmigration |
| 1p13.2 | SELP | Selectin P | (10376511, 10402291) | 10 | Encodes a major adhesion molecule that mediates inflammation |
| 1q24.2 | SELE | Selectin E | (169686781, 169708220) | 13 | Encodes a major adhesion molecule on vascular endothelial surfaces |
| 4q31.23 | EDNRA | Endothelin receptor type A | (148397069, 148471106) | 10 | Leads to long-lasting vasoconstriction after binding to endothelin 1 |
| 5q14.3 | MEF2C | Myocyte enhancer factor 2C | (88008975, 88204922) | 21 | Contribute to the development of endothelial cells |
| 6p24.1 | EDN1 | Endothelin 1 | (12285596, 12302427) | 9 | Encodes a potent endothelium-derived vasoconstrictor and mediates endothelial dysfunction through EDNRA and EDNRB |
| 7q22.1 | SERPINE1 | Serpin peptidase inhibitor, clade E, member 1 | (100765370, 100787547) | 4 | Modulate angiogenesis and contribute to proangiogenic effects |
| 7q36.1 | NOS3 | Nitric oxide synthase 3 (endothelial cell) | (150683083, 150716676) | 2 | Generates NO from L-arginin |
| 12p13.31 | VWF | Von Willebrand factor | (6053040, 6238936) | 40 | Encodes a multifunctional vascular protein and mediates regulation of angiogenesis |
| 13q22.3 | EDNRB | Endothelin receptor type B | (78464616, 78498903) | 7 | Stimulates the release of NO |
| 16q24.3 | CYBA | Cytochrome b-245, alpha polypeptide | (88704691, 88722560) | 1 | Relates to NADH/NADPH oxidase activity |
| 19q13.2 | TGFB1 | Transforming growth factor, beta 1 | (41802492, 41864816) | 1 | Involves in endothelial cells migration, proliferation |
| 21q22.3 | COL18A1 | Collagen, type XVIII, alpha 1 | (46820052, 46938634) | 19 | Endostatin, a fragment of collagen XVIII is an endogenous angiogenesis inhibitor |
Abbreviations: NO, nitric oxide; NADH/NADPH: nicotinamide adenine dinucleotide/nicotinamide adenine dinucleotide phosphate; SNP, single-nucleotide polymorphism.
Statistical analysis
The characteristics of GenSalt follow-up study participants were presented as mean ± SD for continuous variables and as percentages for categorical variables.
To accommodate the longitudinal, family-based GenSalt study design, we used mixed-effect regression models to examine the associations between each SNP and longitudinal BP phenotypes.19,20 The additive associations of each SNP with longitudinal SBP and DBP changes were examined using a mixed-effect linear regression model. Autoregressive (1) and compound symmetry covariance matrices were used to account for the correlations of repeated measurements within individuals and of individuals within families, respectively. SBP and DBP changes for the ith individual from the jth family at the kth visit were modeled using the PROC MIXED procedure in SAS (version 9.2; SAS Institute, Cary, NC) as:
In the formula, is the mean after accounting for covariates, genetic effects, and the interaction term. The terms , , and represent baseline age, gender, and BMI of the ith individual in the jth family, respectively. models the genetic main effect, where the genotype is coded under an additive model. is the follow-up years since baseline for the ith individual in the jth family at the kth visit. The seventh term is the linear interaction between follow-up time and genetic effects. The random effects terms and account for the correlation among individuals in the same family as well as the correlation of repeated measures among individuals nested within families. The last term stands for residual. P values for were used to evaluate the significance of the association of each SNP with longitudinal BP change. For participants taking antihypertensive medication (N = 7 at baseline, 86 at the first follow-up examination, and 166 at the second follow-up examination), BP was imputed by adding 10 and 5mm Hg to SBP and DBP values, respectively.21 To examine the influence of BP imputation among participants taking antihypertension medication, we conducted a sensitivity analysis excluding those participants taking antihypertensive treatment in the month prior to the visit.
After excluding 173 participants with hypertension at baseline, the additive association of SNPs with hypertension incidence was assessed using a multilevel logistic regression model.22 Autoregressive (1) and compound symmetry covariance matrices were again used to account for the correlations of repeated measurements within individuals and of individuals within families, respectively. After accommodating the longitudinal family design, the fixed effects of age, gender, BMI, and follow-up time were adjusted in multivariable analyses using the PROC GLIMMIX procedure in SAS.
The truncated product method, which combines P values from single-marker association analyses, was used to evaluate the overall association of each candidate gene with longitudinal BP changes and hypertension incidence.23,24 For longitudinal BP changes, the P value for the genotype by follow-up time interaction term was used, while for hypertension incidence, the P value of the genotype term was used. The truncation point was set as τ = 0.10, and the P value for truncated product method was estimated by 10,000,000 simulations. Sensitivity analyses were conducted using the truncated product method after excluding significant SNPs within a gene (identified by single-marker analyses) to examine their influence on the gene-based analysis. Gene-based analysis was performed using R software (version 3.0.1; http://www.r-project.org).
Bonferroni correction was used to adjust multiple testing. The α-thresholds for single-marker analysis and gene-based analysis were 2.43×10−4 (0.05/206) and 3.85×10−3 (0.05/13), respectively.
RESULTS
Table 2 presents the characteristics of 1,768 GenSalt participants. On average, participants were 39.0 years of age, had a mean BMI of 23.4kg/m2, and mean baseline SBP and DBP of 116.9mm Hg and 73.8mm Hg, respectively. A total of 924 (52.3%) participants were male and 173 (9.8%) participants had hypertension at baseline. During the average 7.2 years of follow-up, SBP and DBP increased 1.8 and 1.2mm Hg per year, respectively. In addition, among those free from hypertension at baseline, 512 (32.1%) developed hypertension.
Table 2.
Characteristics of 1,768 GenSalt follow-up participants
| Mean (SD) or N (%) | |
|---|---|
| Age | 39.0 (9.3) |
| Male | 924 (52.3%) |
| Body mass index (kg/m2) | 23.4 (3.2) |
| Baseline BP, mm Hg | |
| SBP | 116.9 (14.1) |
| DBP | 73.8 (10.2) |
| Average change in BP, mm Hg (per year) | |
| SBP | 1.8 (1.9) |
| DBP | 1.2 (1.3) |
| Hypertension at baseline | 173 (9.8%) |
| Hypertension incidencea | 512 (32.1%) |
| Follow-up time | 7.2 (0.9) |
Abbreviations: BP, blood pressure; DBP, diastolic blood pressure; SBP, systolic blood pressure.
aExcludes 173 participants who had hypertension at baseline.
Analyses of 206 SNPs in 15 endothelial system genes revealed that 3 SNPs in the SELE gene (rs4656704, rs6427212, and rs5368) were significantly associated with hypertension incidence after Bonferroni correction for multiple testing (Figure 1 and Supplementary Table 1). Each copy of the minor allele of correlated SELE markers rs4656704, rs6427212, and rs5368 were associated with an increased risk of hypertension (P for trend = 1.48×10−4, 6.69×10−5, and 7.64×10−5, respectively). For example, compared to participants with the rs5368 C/C genotype, the relative risks (95% confidence intervals) of hypertension for participants with genotypes C/T and T/T were 1.38 (1.09, 1.77) and 2.33 (1.46, 3.73), respectively. Although the associations of these 3 SNPs with longitudinal BP changes were not significant, a similar trend was observed for average BP change per year (Table 3). In addition, SELE marker rs3917436 was significantly associated with longitudinal DBP change after Bonferroni correction, with the minor allele predicting smaller DBP increases over follow-up. A similar trend identified for SBP change and hypertension incidence (Table 3). Results of sensitivity analyses excluding participants with hypertension from the analyses of BP changes were similar to the overall findings, with only SELE marker rs3917436 associated with DBP change after adjustment for multiple testing (Supplementary Table 2).
Figure 1.
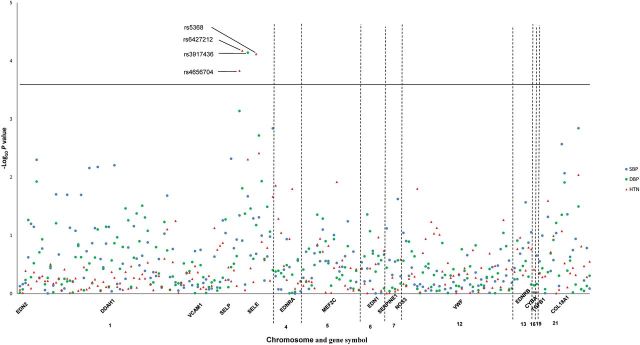
Log10 P values for the 206 SNPs in 15 candidate genes of endothelial system with longitudinal changes in SBP, DBP, and hypertension incidence. The blue solid circle and green solid circle indicate P values for the interactions of follow-up time and genotype of each SNP for SBP and DBP, respectively. The red triangles indicate P values for the association of each SNP with hypertension incidence. All labeled SNPs are statistical significant after Bonferroni correction. Abbreviations: DBP, diastolic blood pressure; HTN, hypertension incidence; SBP, systolic blood pressure; SNP, single-nucleotide polymorphism.
Table 3.
Associations of SELE gene variants with BP changes and hypertension according to genotype
| Gene | SNP | Chra | Physical positionb | Function | MAF | Genotype | SBPc | DBPc | Hypertension incidenced |
|---|---|---|---|---|---|---|---|---|---|
| SELE | rs4656704 | 1 | 169688228 | Unknown | 0.376 | AA | 1.71 (0.08) | 1.11 (0.05) | 1.00 |
| AG | 1.67 (0.07) | 1.23 (0.05) | 1.23 (0.95, 1.59) | ||||||
| GG | 2.13 (0.13) | 1.43 (0.09) | 2.20 (1.50, 3.22) | ||||||
| P e | 0.045 | 7.28×10−4 | 1.48×10−4* | ||||||
| rs6427212 | 1 | 169689041 | Unknown | 0.314 | CC | 1.73 (0.07) | 1.17 (0.05) | 1.00 | |
| CT | 1.66 (0.07) | 1.21 (0.05) | 1.35 (1.06, 1.74) | ||||||
| TT | 2.06 (0.16) | 1.41 (0.10) | 2.29 (1.50, 3.49) | ||||||
| P e | 0.123 | 0.016 | 6.69×10−5* | ||||||
| rs3917436 | 1 | 169694619 | Intron | 0.431 | GG | 1.89 (0.09) | 1.36 (0.06) | 1.00 | |
| GA | 1.72 (0.07) | 1.20 (0.05) | 0.73 (0.56, 0.95) | ||||||
| AA | 1.56 (0.11) | 0.99 (0.07) | 0.62 (0.43, 0.88) | ||||||
| P e | 0.021 | 7.22×10−5* | 4.94×10−3 | ||||||
| rs5368 | 1 | 169696946 | Missense | 0.273 | CC | 1.74 (0.07) | 1.18 (0.05) | 1.00 | |
| CT | 1.66 (0.07) | 1.22 (0.05) | 1.38 (1.09, 1.77) | ||||||
| TT | 2.16 (0.18) | 1.40 (0.12) | 2.33 (1.46, 3.73) | ||||||
| P e | 0.207 | 0.067 | 7.64×10−5* |
Abbreviations: BP, blood pressure; DBP, diastolic blood pressure; MAF, minor allele frequency; SBP, systolic blood pressure; SNP, single-nucleotide polymorphism.
aChromosome.
bGRCh37.p12.
cBP changes per year according to genotype.
dRelative risks and their corresponding 95% confidence intervals according to genotype.
e P values of genotype by follow-up time interaction term were presented for SBP and DBP changes, and P values of the genotype term were presented for hypertension incidence.
*Significant after adjustment for multiple testing.
Results of gene-based analyses are shown in Table 4. The SELE gene was significantly associated with DBP change and hypertension incidence (P values were both <1.00×10−6), the EDNRA gene was significantly associated with hypertension incidence (P = 2.39×10−4), and the DDAH1 and COL18A1 genes were significantly associated with SBP change (P < 1.00×10−6 and P = 4.00×10−6, respectively). Further sensitivity analyses revealed associations of the SELE gene with DBP change and hypertension incidence even after exclusion of the significant SNPs identified by single-marker analyses (P = 6.30×10−4 and 1.80×10−4, respectively).
Table 4.
Gene-based associations of endothelial system genes with BP changes and hypertension
| Genea | SBP | DBP | Hypertension incidence |
|---|---|---|---|
| EDN2 | 0.2374 | 0.2192 | 0.3168 |
| DDAH1 | <1.00×10−6* | 0.1039 | 0.4708 |
| VCAM1 | 0.3064 | 0.3686 | 0.2400 |
| SELP | 0.0082 | 0.3682 | 0.3728 |
| SELE | 0.0174 | <1.00×10−6*,** | <1.00×10−6*,*** |
| EDNRA | 0.7230 | 0.5602 | 2.39×10−4* |
| MEF2C | 0.2894 | 0.3497 | 0.7273 |
| EDN1 | 0.6833 | 0.1868 | 0.6838 |
| SERPINE1 | 0.1812 | 0.2668 | 0.3338 |
| NOS3 | 0.0473 | 0.1719 | 0.1999 |
| VWF | 0.6362 | 0.8390 | 0.2935 |
| EDNRB | 0.2124 | 0.5420 | 0.3558 |
| COL18A1 | 4.00×10−6* | 0.0251 | 0.1166 |
Abbreviations: BP, blood pressure; DBP, diastolic blood pressure; SBP, systolic blood pressure; SNP, single-nucleotide polymorphism.
a CYBA and TGFB1 genes are not included since they only have one tag-SNP each.
*Significant after adjustment for multiple testing.
**Significant after excluding SNP rs3917436 that was significant in single-marker analyses (P = 6.30×10−4).
***Significant after excluding the 3 SNPs that were significant in single-marker analyses (P = 1.80×10−4).
DISCUSSION
As the first candidate gene study to investigate the associations of endothelial system genes with longitudinal BP phenotypes in an East Asian population, we identified 4 common variants in the SELE gene that may play a role in hypertension pathogenesis. Compared to their corresponding major alleles, each copy of the minor alleles of SELE markers rs4656704, rs6427212, and rs5368 predicted higher risk of developing hypertension. In addition, each copy of the minor allele of SELE marker rs3917436 was associated with smaller DBP increases over time. Furthermore, gene-based analyses revealed associations of the DDAH1, SELE, EDNRA, and COL18A1 genes with longitudinal BP phenotypes. Our findings may have important clinical and public health implications. Discovery of genetic variants that influence hypertension susceptibility may aid in the identification of individuals who would benefit most from targeted primary prevention efforts. Moreover, these findings contribute additional information toward our understanding of the genetic mechanisms underlying hypertension development.
We identified 4 SELE variants (rs4656704, rs6427212, rs5368, and rs3917436) that were significantly associated with longitudinal BP phenotypes in the GenSalt follow-up study. Although we provide the first evidence of association of these variants with the longitudinal BP measures, previous studies in participants of European ancestry have linked other SELE variants to BP phenotypes.25 SELE encodes the selectin E protein, which is a member of the selectin family and is typically expressed on the surface of endothelial cells activated in response to certain cytokines and other proinflammatory stimuli.26 Selectin E mediates leukocyte adhesion to endothelial cells stimulating inflammatory and immunological events at the interface of the vessel wall, which can result in subsequent endothelial cell injury.27 Such injury is purported to influence BP by impairing endothelium-dependent vascular relaxation and reconstruction of smooth muscle cells.28,29 Markers rs4656704, rs6427212, and rs5368, which associated with hypertension incidence, are moderately correlated with pairwise r 2 values ranging from 0.60 to 0.77 (see Supplementary Figure 1). These data suggest that the 3 variants may reflect the same causal association signal. In addition, we are the first to identify an association of independent marker rs3917436 with DBP change, although our previous study has identified an association of this marker with the correlated BP salt-sensitivity phenotype.18 With no known functional relevance, markers rs4656704 and rs6427212 are located in the 5′-flanking region of the SELE gene and rs3917436 represents an intronic SELE SNP. In contrast, marker rs5368 represents a missense variant within exon 9 of the SELE gene. The C/T polymorphism results in a potentially damaging amino acid substitution of histidine with tyrosine.30 Although confirmation by functional study is needed, structural change to the selectin E protein resulting from this variant represents a plausible biological mechanism linking rs5368 to hypertension.31,32
In support of our single-marker analysis, gene-based study revealed significant associations of the SELE gene with BP phenotypes for the first time. It is worth pointing out that the association with hypertension incidence was not completely driven by the 3 significant SNPs identified in single-marker analyses since results remained after the simultaneous removal of these variants from the gene-based analysis. These data suggest that there may be multiple variants causally associated with BP phenotypes in the SELE gene, which could not be detected by less powerful single-marker analyses.
In addition, the DDAH1, EDNRA, and COL18A1 genes predicted longitudinal BP changes though no significant single marker was identified in the current study. The DDAH1 gene is hypothesized to influence BP regulation by encoding the critical enzyme for degrading asymmetrical dimethylarginine, an endogenous and competitive inhibitor of NOS.33 The EDNRA gene encodes endothelin receptor type A, which leads to long-lasting vasoconstriction after binding with endothelin 1.34 The COL18A1 gene encodes a potent antiangiogenic protein, whose deficiency is known to enhance inflammatory response and vascular endothelial cell damage.35,36 Although we present the first evidence of gene-based associations of these genes, previous studies have associated DDAH1, EDNRA, and COL18A1 variants with BP-related phenotypes or atherosclerotic risk, including associations with essential hypertension, BP salt sensitivity, preeclampsia, or preclinical stages of atherosclerosis.12,17,37,38 Thus, we add to the accumulating evidence suggesting a role for these genes in BP regulation. Future studies will be needed to identify the causal variant(s) underlying the observed gene-based signals.
Our study has several important strengths. To our knowledge, this is the first investigation to examine associations of endothelial system genes with longitudinal BP change and hypertension incidence in the Chinese Han population. In addition, the current study had a very high follow-up rate (92.28%). Stringent quality control procedures were employed for genotyping and data collection, including BP measurement and measurement of other important covariates. We employed a conservative Bonferroni correction to account for multiple testing. However, some potential limitations of our study should be noted. Further studies in larger population are still warranted to replicate our findings. In addition, although our genotyping platform should provide good coverage of common genetic variants,38 some important rare, low-frequency, and structural variants may be missed by the current study. Moreover, since we modeled the linear relation between SNPs and BP change over time, the current study may not have the statistical power to detect nonlinear relations of SNPs or genes with BP change.
Our study has documented novel and significant associations of SELE markers rs4656704, rs6427212, rs3917436, and rs5368 with longitudinal BP phenotypes. Furthermore, results of gene-based analyses revealed important joint effects of SNPs in the DDAH1, SELE, EDNRA, and COL18A1 genes on BP progression among the Chinese Han population. In aggregate, these findings highlight the potential role of endothelial system genes in BP progression and contribute important information toward elucidating the genomic mechanisms underlying BP regulation.
SUPPLEMENTARY MATERIAL
Supplementary materials are available at American Journal of Hypertension (http://ajh.oxfordjournals.org).
DISCLOSURE
The authors declared no conflict of interest.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by a cooperative agreement project grant (U01HL072507, R01HL087263, and R01HL090682) from the National Heart, Lung and Blood Institute, National Institutes of Health, Bethesda, MD. F.L. was supported by a research training grant (D43TW009107) from National Institutes of Health John E Fogarty International Center, Bethesda, MD.
REFERENCES
- 1.Kearney PM, Whelton M, Reynolds K, Muntner P, Whelton PK, He J. Global burden of hypertension: analysis of worldwide data. Lancet. 2005;365:217–223. doi: 10.1016/S0140-6736(05)17741-1. [DOI] [PubMed] [Google Scholar]
- 2.Lawes CM, Vander Hoorn S, Rodgers A. Global burden of blood-pressure-related disease, 2001. Lancet. 2008;371:1513–1518. doi: 10.1016/S0140-6736(08)60655-8. [DOI] [PubMed] [Google Scholar]
- 3.Lifton RP, Gharavi AG, Geller DS. Molecular mechanisms of human hypertension. Cell. 2001;104:545–556. doi: 10.1016/s0092-8674(01)00241-0. [DOI] [PubMed] [Google Scholar]
- 4.Levy D, DeStefano AL, Larson MG, O’Donnell CJ, Lifton RP, Gavras H, Cupples LA, Myers RH. Evidence for a gene influencing blood pressure on chromosome 17. Genome scan linkage results for longitudinal blood pressure phenotypes in subjects from the Framingham heart study. Hypertension. 2000;36:477–483. doi: 10.1161/01.hyp.36.4.477. [DOI] [PubMed] [Google Scholar]
- 5.Furchgott RF, Zawadzki JV. The obligatory role of endothelial cells in the relaxation of arterial smooth muscle by acetylcholine. Nature. 1980;288:373–376. doi: 10.1038/288373a0. [DOI] [PubMed] [Google Scholar]
- 6.Vallance P, Collier J, Moncada S. Effects of endothelium-derived nitric oxide on peripheral arteriolar tone in man. Lancet. 1989;2:997–1000. doi: 10.1016/s0140-6736(89)91013-1. [DOI] [PubMed] [Google Scholar]
- 7.Lüscher TF, Raij L, Vanhoutte PM. Endothelium-dependent vascular responses in normotensive and hypertensive Dahl rats. Hypertension. 1987;9:157–163. doi: 10.1161/01.hyp.9.2.157. [DOI] [PubMed] [Google Scholar]
- 8.Panza JA, Quyyumi AA, Brush JE, Jr, Epstein SE. Abnormal endothelium-dependent vascular relaxation in patients with essential hypertension. N Engl J Med. 1990;323:22–27. doi: 10.1056/NEJM199007053230105. [DOI] [PubMed] [Google Scholar]
- 9.Salvi E, Kutalik Z, Glorioso N, Benaglio P, Frau F, Kuznetsova T, Arima H, Hoggart C, Tichet J, Nikitin YP, Conti C, Seidlerova J, Tikhonoff V, Stolarz-Skrzypek K, Johnson T, Devos N, Zagato L, Guarrera S, Zaninello R, Calabria A, Stancanelli B, Troffa C, Thijs L, Rizzi F, Simonova G, Lupoli S, Argiolas G, Braga D, D’Alessio MC, Ortu MF, Ricceri F, Mercurio M, Descombes P, Marconi M, Chalmers J, Harrap S, Filipovsky J, Bochud M, Iacoviello L, Ellis J, Stanton AV, Laan M, Padmanabhan S, Dominiczak AF, Samani NJ, Melander O, Jeunemaitre X, Manunta P, Shabo A, Vineis P, Cappuccio FP, Caulfield MJ, Matullo G, Rivolta C, Munroe PB, Barlassina C, Staessen JA, Beckmann JS, Cusi D. Genomewide association study using a high-density single nucleotide polymorphism array and case-control design identifies a novel essential hypertension susceptibility locus in the promoter region of endothelial NO synthase. Hypertension. 2012;59:248–255. doi: 10.1161/HYPERTENSIONAHA.111.181990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Conen D, Glynn RJ, Buring JE, Ridker PM, Zee RY. Association of renin-angiotensin and endothelial nitric oxide synthase gene polymorphisms with blood pressure progression and incident hypertension: prospective cohort study. J Hypertens. 2008;26:1780–1786. doi: 10.1097/HJH.0b013e3283077eef. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Larsson E, Wahlstrand B, Hedblad B, Hedner T, Kjeldsen SE, Melander O, Lindahl P. Hypertension and genetic variation in endothelial-specific genes. PLoS One. 2013;8:e62035. doi: 10.1371/journal.pone.0062035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Benjafield AV, Katyk K, Morris BJ. Association of EDNRA, but not WNK4 or FKBP1B, polymorphisms with essential hypertension. Clin Genet. 2003;64:433–438. doi: 10.1034/j.1399-0004.2003.00148.x. [DOI] [PubMed] [Google Scholar]
- 13.Zhu H, Wang X, Dong Y, Treiber FA, Snieder H. Influence of the eNOS gene on development of blood pressure and left ventricular mass: longitudinal findings in multiethnic youth. Pharmacogenet Genomics. 2005;15:669–675. doi: 10.1097/01.fpc.0000172244.65417.7a. [DOI] [PubMed] [Google Scholar]
- 14.Chen W, Srinivasan SR, Li S, Boerwinkle E, Berenson GS. Gender-specific influence of NO synthase gene on blood pressure since childhood: the Bogalusa Heart Study. Hypertension. 2004;44:668–673. doi: 10.1161/01.HYP.0000145474.23750.2b. [DOI] [PubMed] [Google Scholar]
- 15.Kishimoto T, Misawa Y, Kaetu A, Nagai M, Osaki Y, Okamoto M, Yoshida S, Kurosawa Y, Fukumoto S. eNOS Glu298Asp polymorphism and hypertension in a cohort study in Japanese. Prev Med. 2004;39:927–931. doi: 10.1016/j.ypmed.2004.03.030. [DOI] [PubMed] [Google Scholar]
- 16.GenSalt Collabration Research Group. GenSalt: rationale, design, methods and baseline characteristics of study participants. J Hum Hypertens. 2007;21:639–646. doi: 10.1038/sj.jhh.1002207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perloff D, Grim C, Flack J, Frohlich ED, Hill M, McDonald M, Morgenstern BZ. Human blood pressure determination by sphygmomanometry. Circulation. 1993;88:2460–2470. doi: 10.1161/01.cir.88.5.2460. [DOI] [PubMed] [Google Scholar]
- 18.Defagó MD, Gu D, Hixson JE, Shimmin LC, Rice TK, Gu CC, Jaquish CE, Liu DP, He J, Kelly TN. Common genetic variants in the endothelial system predict blood pressure response to sodium intake: the GenSalt study. Am J Hypertens. 2013;26:643–656. doi: 10.1093/ajh/hps099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shi G, Rice TK, Gu CC, Rao DC. Application of three-level linear mixed-effects model incorporating gene-age interactions for association analysis of longitudinal family data. BMC Proc. 2009;3(Suppl 7:):S89. doi: 10.1186/1753-6561-3-s7-s89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Singer JD. Using SAS PROC MIXED to fit multilevel models, hierarchical models, and individual growth models. J Educ Behav Stat. 1998;23:323–355. [Google Scholar]
- 21.Law MR, Morris JK, Wald NJ. Use of blood pressure lowering drugs in the prevention of cardiovascular disease: meta-analysis of 147 randomised trials in the context of expectations from prospective epidemiological studies. BMJ. 2009;338:b1665. doi: 10.1136/bmj.b1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schabenberger O. Introducing the GLIMMIX procedure for generalized linear mixed models. SUGI 2005; 30:196. [Google Scholar]
- 23.Zaykin DV, Zhivotovsky LA, Westfall PH, Weir BS. Truncated product method for combining P-values. Genet Epidemiol. 2002;22:170–185. doi: 10.1002/gepi.0042. [DOI] [PubMed] [Google Scholar]
- 24.Sheng X, Yang J. Panel unit root test by combining dependent p-values: a comparative study. J Prob Stat. 2011;2011:617652. [Google Scholar]
- 25.Sass C, Pallaud C, Zannad F, Visvikis S. Relationship between E-selectin L/F554 polymorphism and blood pressure in the Stanislas cohort. Hum Genet. 2000;107:58–61. doi: 10.1007/s004390000325. [DOI] [PubMed] [Google Scholar]
- 26.Kansas GS. Selectins and their ligands: current concepts and controversies. Blood. 1996;88:3259–3287. [PubMed] [Google Scholar]
- 27.Bevilacqua MP, Stengelin S, Gimbrone MA, Jr, Seed B. Endothelial leukocyte adhesion molecule 1: an inducible receptor for neutrophils related to complement regulatory proteins and lectins. Science. 1989;243:1160–1165. doi: 10.1126/science.2466335. [DOI] [PubMed] [Google Scholar]
- 28.Lusis AJ. Atherosclerosis. Nature. 2000;407:233–241. doi: 10.1038/35025203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Z, Xu Y, Chen S, Wang L, Ding H, Lu G, Wang D, Zhai Z, Duan J, Zhang W. A common missense single nucleotide polymorphism in the E-selectin gene is significantly associated with essential hypertension in the Han population but only weakly associated in the Uygur population. Hypertens Res. 2012;35:413–417. doi: 10.1038/hr.2011.204. [DOI] [PubMed] [Google Scholar]
- 30.Chen H, Cui B, Wang S, Zhao Z, Sun H, Gu X, Zhao Y, Li X, Ning G. The common variants of E-selectin gene in Graves’ disease. Genes Immun. 2008;9:182–186. doi: 10.1038/sj.gene.6364452. [DOI] [PubMed] [Google Scholar]
- 31.Zhang Y, Thompson AM, Tong W, Xu T, Chen J, Zhao L, Kelly TN, Chen CS, He J. Biomarkers of inflammation and endothelial dysfunction and risk of hypertension among Inner Mongolians in China. J Hypertens. 2010;28:35–40. doi: 10.1097/HJH.0b013e3283324650. [DOI] [PubMed] [Google Scholar]
- 32.De Caterina R, Ghiadoni L, Taddei S, Virdis A, Almerigogna F, Basta G, Lazzerini G, Bernini W, Salvetti A. Soluble E-selectin in essential hypertension: a correlate of vascular structural changes. Am J Hypertens. 2001;14:259–266. doi: 10.1016/s0895-7061(00)01276-0. [DOI] [PubMed] [Google Scholar]
- 33.Hu X, Atzler D, Xu X, Zhang P, Guo H, Lu Z, Fassett J, Schwedhelm E, Böger RH, Bache RJ, Chen Y. Dimethylarginine dimethylaminohydrolase-1 is the critical enzyme for degrading the cardiovascular risk factor asymmetrical dimethylarginine. Arterioscler Thromb Vasc Biol. 2011;31:1540–1546. doi: 10.1161/ATVBAHA.110.222638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lüscher TF, Barton M. Endothelins and endothelin receptor antagonists: therapeutic considerations for a novel class of cardiovascular drugs. Circulation. 2000;102:2434–2440. doi: 10.1161/01.cir.102.19.2434. [DOI] [PubMed] [Google Scholar]
- 35.Hamano Y, Okude T, Shirai R, Sato I, Kimura R, Ogawa M, Ueda Y, Yokosuka O, Kalluri R, Ueda S. Lack of collagen XVIII/endostatin exacerbates immune-mediated glomerulonephritis. J Am Soc Nephrol. 2010;21:1445–1455. doi: 10.1681/ASN.2009050492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sunshine SB, Dallabrida SM, Durand E, Ismail NS, Bazinet L, Birsner AE, Sohn R, Ikeda S, Pu WT, Kulke MH, Javaherian K, Zurakowski D, Folkman JM, Rupnick M. Endostatin lowers blood pressure via nitric oxide and prevents hypertension associated with VEGF inhibition. Proc Natl Acad Sci USA. 2012;109:11306–11311. doi: 10.1073/pnas.1203275109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Akbar F, Heinonen S, Pirskanen M, Uimari P, Tuomainen TP, Salonen JT. Haplotypic association of DDAH1 with susceptibility to pre-eclampsia. Mol Hum Reprod. 2005;11:73–77. doi: 10.1093/molehr/gah116. [DOI] [PubMed] [Google Scholar]
- 38.Nishida N, Koike A, Tajima A, Ogasawara Y, Ishibashi Y, Uehara Y, Inoue I, Tokunaga K. Evaluating the performance of Affymetrix SNP Array 6.0 platform with 400 Japanese individuals. BMC Genomics. 2008;9:431. doi: 10.1186/1471-2164-9-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


