Abstract
Twenty strains isolated from sewage sludge were found to degrade various ethers, including alkyl ethers, aralkyl ethers, and dibenzyl ether. In Rhodococcus strain DEE5151, induction of ether degradation needed substrates exhibiting at least one unsubstituted Cα-methylene moiety as the main structural prerequisite. The cleavage reaction observed with anisole, phenetole, and dibenzyl ether indicates that the initial oxidation occurs at such respective Cα positions. Diethyl ether-induced strain DEE5151 degraded dibenzyl ether via intermediately accumulated benzoic acid. Phenetole seems to be subject also to another ether-cleaving enzyme. Other strains of this group showed different enzymatic activities towards the substrate classes investigated.
Ethers are widely used as additives, herbicides, pharmaceuticals, pesticides, and solvents in agriculture, in the production of cosmetics and foods, in petroleum industries, and in polymer syntheses. Because the ether bond has a high dissociation energy (about 360 kJ mol−1), ether compounds are considered recalcitrant (4), and many xenobiotic ether compounds are listed as carcinogens (26, 27). Halogenated alkyl ethers are also used in anesthetics and polymer syntheses, but their use is controversial, because the cytochrome P450-catalyzed degradation metabolites, e.g., trifluoro-substituted aldehydes, are reactive with proteins and cause acute hepatic and immunological disease (6). Significant amounts of ether compounds are released into the environment due to the massive use of chemicals in general.
There is a wealth of information on the degradation of aromatic ethers, including the mechanisms for cleaving the corresponding ether bonds. Also, bacterial catabolism of alkyl ethers has been studied with various microorganisms and enzyme systems, especially for phenoxyacetates (21-23) and lignin-related ethers (16, 18). Bacterial scission of ether bonds has been reviewed by White et al. (28). Cyclic alkyl ether-degrading Rhodococcus strains, which utilize tetrahydrofurane, tetrahydropyrane, and diethyl ether as the sole carbon and energy (C/E) sources, appear to initiate the oxidation of cyclic alkyl ethers at the carbon atom adjacent to the oxygen atom (2, 3). Parales et al. (19) isolated an actinomycete strain, CB1190, which utilized 1,3-dioxane, 1,4-dioxane, 1,3-dioxolane, 2-methyl-1,3-dioxolane, diethyl ether, tetrahydrofurane, tetrahydropyrane, and n-butyl methyl ether as the sole C/E sources. Dimethyl ether was cooxidized to form methanol and formaldehyde by an ammonia monooxygenase of Nitrosomonas europaea (9). Burkholderia cepacia strain G4/PR1, containing a toluene-2-monooxygenase, utilizes diethyl ether as the sole C/E source and oxidizes butyl methyl ether, diethyl sulfide, and 2-chloroethyl ethyl ether (8). Tertiary butyl ethers including tert-butyl methyl ether (MTBE) and tert-butyl ethyl ether, used in gasoline oxygenates, are degraded to tert-butanol and the corresponding aldehydes by several propan-degrading bacteria (25). Mo et al. (17) isolated several MTBE-degrading species of the genera Methylobacterium, Rhodococcus, and Arthrobacter, which utilized tert-butanol slowly as the sole C/E source. There are several reports of the microbial degradation of lignin-related ether compounds under anaerobic conditions (5, 10, 11, 15, 24). A Graphium fungal strain utilizes n-butan and diethyl ether as the sole C/E sources and degrades MTBE by cooxidation with the participation of a P450 enzyme to form tert-butanol via a transient intermediate, tert-butyl formic acid (7). In contrast, lignin-degrading enzymes of a white rot fungus, Phanerochaete chrysosporium, which play a role in the ether scission of a variety of natural and synthetic lignin components, show no activity for alkyl ethers, e.g., MTBE (12).
In this paper, we isolated 18 alkyl ether-, aralkyl ether-, and dibenzyl ether-degrading Rhodococcus strains and two dibenzyl ether-degrading Sphingomonas strains from sewage sludge from Stuttgart-Büsnau, Germany (13). The bacterial isolates were deposited in the strain collection of the Abteilung für Biologische Abluftreinigung, Institut für Siedlungswasserbau, Wassergüte- und Abfallwirtschaft, Universität Stuttgart, Stuttgart, Germany. Among them, Rhodococcus strain DEE5151 was selected for further studies since it was able to utilize a broad range of alkyl ethers including diethyl ether, di-n-propyl ether, di-n-butyl ether, di-n-pentyl ether, di-n-hexyl ether, di-n-heptyl ether, tert-butyl ethyl ether, and a mixture of 2- and 3-isoamylether, phenetole, and dibenzyl ether as sole C/E sources. None of the 20 isolates was able to utilize dimethyl ether, MTBE, isopropyl ether, cyclic alkyl ethers (e.g., tetrahydrofurane, tetrahydropyrane, and 1,4-dioxane), benzyl phenyl ether, and diphenyl ether.
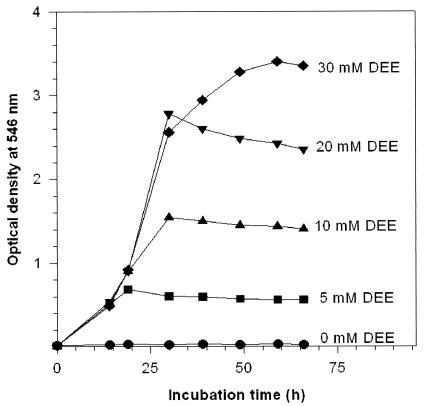
Ethers and other chemicals used in this study were of the highest purities available from Aldrich, Steinheim, Germany, except for di-n-pentyl ether, di-n-hexyl ether, and di-n-heptyl ether (Lancaster, Mülheim am Main, Germany). Before now, there have been several Rhodococcus and actinomycete strains known to take part in the degradation of diethyl ether and cyclic alkyl ethers (2, 3, 19). Compared to those, the strain DEE5151 had different catalytic properties and specificity for the degradation of alkyl ethers, aralkyl ethers, and dibenzyl ether. In addition, this strain utilized 2-chloroethyl ethyl ether as the sole C/E source, but neither 2-chloroethanol nor 2-chloroacetic acid served as the growth substrate, which indicated that this strain was able to metabolize only the unhalogenated ethyl moiety of 2-chloroethyl ethyl ether. This is also indicated by the rather high oxidation rate for 1,2-dichloroethyl ethyl ether (see Table 1). Of the tested ether substrates, diethyl ether was the most promising substrate for growth without substrate inhibition up to 30 mM diethyl ether, when strain DEE5151 was cultivated at 30°C and 108 rpm with diethyl ether (final concentration, 5 to 30 mM) fed in 10 ml of the previously described (14) phosphate-basal minimal medium in 125-ml flasks tightly closed with Teflon-lined septa (Fig. 1).
TABLE 1.
Specific oxygen consumption rates of the diethyl ether-induced strain DEE5151 for ether chemicals
| Ether chemical | Specific O2 consumption ratea | Relative activity (%) (diethyl ether = 100%) |
|---|---|---|
| Diethyl ether | 141 ± 6.0 | 100 |
| Di-n-propyl ether | 95 ± 4.5 | 67.4 |
| Di-n-butyl ether | 66 ± 6.2 | 46.8 |
| Di-n-pentyl ether | 20 ± 1.2 | 14.2 |
| Di-n-hexyl ether | 2.1 ± 0.8 | 1.5 |
| Di-n-heptyl ether | 13 ± 3.5 | 9.2 |
| 2-Chloroethyl ethyl ether | 107 ± 1.0 | 75.9 |
| 2-Bromoethyl ethyl ether | 97 ± 0.5 | 68.8 |
| ±1,2-Dichloroethyl ethyl ether | 98 ± 1.2 | 69.5 |
| Bis-(2-chloroethyl) ether | 34 ± 2.1 | 24.1 |
| Bis-(2-Bromoethyl) ether | 38 ± 1.8 | 27.0 |
| Phenetole (ethoxybenzene) | 18 ± 1.3 | 12.8 |
| 2-Chloroethyl phenyl ether | <1.0 | <0.7 |
Data are expressed as nanomoles of oxygen consumed per milliliter per minute per unit of optical density at 546 nm.
FIG. 1.
Growth of Rhodococcus sp. strain DEE5151 at various concentrations of diethyl ether (DEE) as the sole carbon and energy source.
Oxidation rates for a range of ether chemicals were determined using a Clark-type oxygen electrode (Biolytik, Bochum, Germany) with a glass reaction vessel equipped with a water jacket (30°C) and a magnetic stirrer (200 rpm). To obtain 18 g (wet weight) of the diethyl ether-induced cells, the strain DEE5151 was cultivated in 2 liters of phosphate-basal minimal medium in a 5-liter bioreaction vessel to which diethyl ether (1 to 10 ml day−1) was added in a filter-sterilized airflow (2 liter min−1). The cell culture was conducted under a ventilation hood to remove hazardous waste air, and the pH of medium was maintained at 7.2 with the addition of 5 M NaOH during stirring. The grown cells were harvested by centrifugation at 14,000 × g and 4°C and washed extensively with 50 mM sodium phosphate buffer (pH 7.2) to remove the remaining substrate. For measuring the oxygen consumption rate, the cells were suspended with air-saturated phosphate buffer (50 mM, pH 7.2) to make a total reaction volume of 1 ml and preheated at 30°C for 5 min. The maximum dissolved oxygen concentration in air-saturated buffer at 1 atm and 30°C was determined to be 236 μM (1). Respiration of the resting cells was measured for 5 min, and 10 μl of a substrate stock solution was then added through a needle port with a calibrated syringe. Substrate stock solutions (final concentration, 100 mM) were freshly prepared in acetone, which did not affect the cell respiration. The oxidation rate was determined from the difference between the rates of the cell respiration and the oxygen consumption after addition of the substrate. One unit of the specific rate was defined as 1 nmol of oxygen consumed per ml per min per unit of optical density at 546 nm. The specific rates for alkyl ethers tended to decrease with increasing molecular weights (Table 1). These are closely related to the substrate spectrum for a range of alkyl ethers with various chain lengths, phenetole, and dibenzyl ether. From the tested symmetric alkyl ethers from diethyl ether to di-n-heptyl ether, elongation of the alkyl chains resulted in reduction of the oxidation rate, probably due to the decrease in water solubility or the affinity of the enzyme for substrate. The ether-oxidizing activity was lost transiently, once the strain was cultivated with d-glucose or ethanol (data not shown).
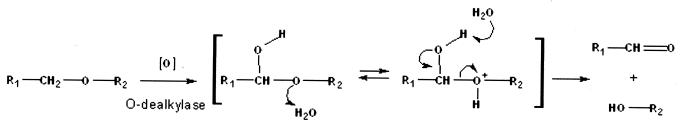
The mechanism of ether cleavage was hypothesized to consist of an O-dealkylase activity. This is indicated by, among other evidence, the production of phenol from anisole (and phenetole, i.e., ethoxybenzene) as the only detectable metabolite, when the DEE5151 cells were pregrown with diethyl ether. Furthermore, when diethyl ether cells were transferred into medium containing diethyl ether, ethanol, or glucose, the remaining oxygen uptake rates of the glucose culture with ethanol (caused by a postulated alcohol dehydrogenase) after 24 h were 25% of that for the ethyl ether culture, whereas the ethanol culture showed a relative uptake rate of 120% (all values compared to the diethyl ether culture). This justifies the assumption that the alcohol dehydrogenase activity may not be identical with the ether-cleaving enzyme, i.e., not be based on a presumptive cytochrome P450 enzyme also exhibiting alcohol-oxidizing activities. Such phenomena have been demonstrated, for example, for toluene side chain monooxygenase of the TOL plasmid. Direct investigations of the above-mentioned enzyme activities in DEE5151 were unsuccessful due to the instability of the etherase and the dehydrogenase activities.
To better understand the putative O-dealkylase activity, the oxidation rate for diethyl ether was compared to those for halogenated ethyl ethers. The rates decreased in the order diethyl ether > 2-chloroethyl ethyl ether ≈ 2-bromoethyl ethyl ether ≈±1,2-dichloroethyl ethyl ether > bis-(2-chloroethyl)-ether ≈ bis-(2-bromoethyl)-ether. The oxidation rates for bis-(2-halogenated ethyl)-ethers were much lower than that for di-n-propyl ether, indicating no significant effect of the molecule size of substituted halogen atom(s) on lowering the oxidation rates. In contrast, the oxidation rates for bis-(2-halogenated ethyl)-ethers were much lower than those of 2-chloroethyl ethyl ether, 2-bromoethyl ethyl ether, and ±1,2-dichloroethyl ethyl ether, suggesting a preference for oxidation of unhalogenated ethyl moieties. The electron density at the Cα position seems to be an important factor determining the oxidation rate for (halogenated) alkyl ethers, since a halogen substitution of the alkyl chain has effects in lowering the electron density. In accordance with this rule, phenetole was oxidized faster than 2-chloroethyl phenyl ether (β-chlorophenetole) was.
Dibenzyl ether, anisole, and phenetole are suitable model chemicals which have one or two unsubstituted Cα-carbon moieties. Concentrations of the model compounds and their metabolites were determined by reversed-phase high-pressure liquid chromatography (HPLC) with UV detection at 254 nm. The HPLC system (Thermo Separation Products, Darmstadt, Germany) consisted of a SpectraSeries P200 pump, a SpectraSystem UV2000 detector, and a SpectraSeries AS100 autosampler installed with a 20-μl injection loop. The analytical column was a LiChrosorb100-RP8, 5-μm, 4- by 125-mm (Merck, Darmstadt, Germany) column installed with a LiChroCART4-4 guard column. A diode array detector was scanned in 2-nm steps between 200 and 360 nm. An isocratic mobile phase consisted of methanol-water-85% phosphoric acid in a ratio of 500:500:1, vol/vol/vol (buffer A), or 700:300:1, vol/vol/vol (buffer B). Compounds eluted from HPLC were identified by means of UV spectra and coelution with authentic chemicals. Benzyl alcohol (retention time, 2.14 min), benzyl aldehyde (retention time, 2.71 min), and benzoic acid (retention time, 2.85 min) were determined with buffer A at 1.5 ml min−1, and dibenzyl ether (retention time, 3.34 min) was determined with buffer B at 1.5 ml min−1. Anisole (retention time, 5.61 min), phenetole (retention time, 8.65 min), and phenol (retention time, 2.58 min) were determined with buffer A at 1 ml min−1.
The diethyl ether-induced strain DEE5151 degraded dibenzyl ether to benzoic acid as the only detectable metabolite in this study. When the strain was cultivated with diethyl ether or dibenzyl ether, the oxidation capacities differed for diethyl ether, dibenzyl ether, benzyl alcohol, and benzoic acid (Table 2). Diethyl ether and dibenzyl ether were oxidized eventually by induction with either diethyl ether or dibenzyl ether, suggesting the involvement of the same enzyme in the oxidation of the two compounds. The conversion of dibenzyl ether to benzoic acid seems to be initiated by the oxidation of a methylene group, and the formed hemiacetal bond spontaneously decomposes to benzyl alcohol and benzaldehyde. This reaction mechanism was adjusted by the chemical Cα-oxidation of benzyl phenyl ether and lignin components in the presence of 2,3-dichloro-5,6-dicyano-1,4-benzoquinone (20). Benzyl alcohol and benzaldehyde may be rapidly oxidized to benzoic acid because of the presence of a high activity for benzyl alcohol-benzaldehyde oxidation. In the absence of initial activity for benzoic acid oxidation, the cells (optical density at 546 nm = 13.6) converted dibenzyl ether (0.52 mM) almost stoichiometrically to benzoic acid (1.08 mM) at 30°C for 1 h.
TABLE 2.
Specific oxygen consumption rates of strain DEE5151 induced after growth with diethyl ether or dibenzyl ether (0.1% [vol/vol])
| Test chemical (1 mM) | Rate with growth substratea:
|
|
|---|---|---|
| Diethyl ether | Dibenzyl ether | |
| Diethyl ether | 73 ± 5.8 | 36 ± 7.1 |
| Dibenzyl ether | 33 ± 4.2 | 33 ± 6.7 |
| Benzoic acid | NDb | 29 ± 3.5 |
| Benzyl alcohol | 152 ± 10 | 63 ± 7.8 |
Data are expressed as nanomoles of oxygen consumed per milliliter per minute per unit of optical density at 546 nm.
ND, not detected.
The diethyl ether-induced DEE5151 was able to degrade anisole and phenetole to phenol as the only detectable metabolite in this study. There existed a balance between the specific rates (micromolar concentration minute−1 per unit of optical density at 546 nm = 1.0) of oxygen consumption (1.3 ± 0.2 for anisole and 7.7 ± 1.1 for phenetole) and phenol production (1.7 ± 0.3 for anisole and 8.0 ± 0.7 for phenetole). Since it was difficult to determine concentrations of volatile anisole and phenetole directly from the culture fluids, the rates of phenol production were determined for 30 min. In this time, the cells displayed no initial activity for phenol oxidation. Phenetole served as a growth substrate as well as an inducer for a putative O-dealkylase, but anisole was neither a growth substrate nor an inducer for that. The cooxidation of anisole implies that a properly induced O-dealkylase cooxidizes methyl ethers, such as dimethyl ether and MTBE. The induction of the ether-degrading enzyme appeared to necessitate an ether compound with an unsubstituted Cα-methylene moiety, such as in diethyl ether, phenetole, and dibenzyl ether. This structural prerequisite for enzyme induction corresponds with the substrate spectrum of alkyl ether-degrading Rhodococcus strain DEE5151 and the proposed degradation pathways for various alkyl ethers, aralkyl ethers, and dibenzyl ether, as illustrated in Fig. 2.
FIG. 2.
Proposed degradation pathways of alkyl ethers (R1, R2 = aliphatic), aralkyl ethers (R1 = H, R2 = phenyl, anisole; R1 = methyl, R2 = phenyl, phenetole), and dibenzyl ether (R1 = phenyl, R2 = benzyl) by a putative O-dealkylase activity from Rhodococcus sp. strain DEE5151. Spontaneous ether scission is schematized in the bracket according to a general acid-catalyzed mechanism of hemiacetal cleavage.
Acknowledgments
This research was supported in part by the DAAD (Deutscher Akademischer Austauschdienst).
REFERENCES
- 1.American Public Health Association. 1985. Standard methods for the examination of water and wastewater, 16th ed. American Public Health Association, Washington, D.C.
- 2.Bernhardt, D., and H. Diekmann. 1991. Degradation of dioxane, tetrahydrofuran and other cyclic ethers by an environmental Rhodococcus strain. Appl. Microbiol. Biotechnol. 36:120-123. [DOI] [PubMed] [Google Scholar]
- 3.Bock, C., R. M. Kroppenstedt, and H. Diekmann. 1996. Degradation and bioconversion of aliphatic and aromatic hydrocarbons by Rhodococcus ruber 219. Appl. Microbiol. Biotechnol. 45:408-410. [Google Scholar]
- 4.Cain, R. B. 1981. Microbial degradation of surfactants and “builder” components, p. 325-370. In T. Leisinger, R. Hütter, A. M. Cook, and J. Nüesch (ed.), Microbial degradation of xenobiotics and recalcitrant compounds. Academic Press, London, United Kingdom.
- 5.Daniel, S. L., E. S. Keith, H. Yang, Y. S. Lin, and H. L. Drake. 1991. Utilization of methoxylated aromatic compounds by the acetogen Clostridium thermoaceticum: expression and specificity of the co-dependent O-demethylating activity. Biochem. Biophys. Res. Commun. 180:416-422. [DOI] [PubMed] [Google Scholar]
- 6.Elliott, R. H., and L. Strunin. 1993. Hepatotoxicity of volatile anaesthetics. Br. J. Anaesth. 70:339-348. [DOI] [PubMed] [Google Scholar]
- 7.Hardison, L. K., S. S. Curry, L. M. Ciuffetti, and M. R. Hyman. 1997. Metabolism of diethylether and cometabolism of methyl-tert-butyl ether by a filamentous fungus, a Graphium sp. Appl. Environ. Microbiol. 63:3059-3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hur, H.-G., L. M. Newman, L. P. Wackett, and M. J. Sadowsky. 1997. Toluene 2-monooxygenase-dependent growth of Burkholderia cepacia G4/PR1 on diethyl ether. Appl. Environ. Microbiol. 63:1606-1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hyman, M. R., C. L. Page, and D. J. Arp. 1994. Oxidation of methyl fluoride and dimethyl ether by ammonia monooxygenase in Nitrosomonas europaea. Appl. Environ. Microbiol. 60:3033-3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kajikawa, H., H. Kudo, T. Kondo, K. Jodai, Y. Honda, M. Kuwahara, and T. Watanabe. 2000. Degradation of benzyl ether bonds of lignin by ruminal microbes. FEMS Microbiol. Lett. 187:15-20. [DOI] [PubMed] [Google Scholar]
- 11.Kaufmann, F., G. Wohlfarth, and G. Diekert. 1998. O-Demethylase from Acetobacterium dehalogenans—substrate specificity and function of the participating proteins. Eur. J. Biochem. 253:706-711. [DOI] [PubMed] [Google Scholar]
- 12.Kay-Shoemake, J. L., and M. E. Watwood. 1996. Limitations of the lignin peroxidase system of the white-rot fungus, Phanerochaete chrysosporium. Appl. Microbiol. Biotechnol. 46:438-442. [Google Scholar]
- 13.Kim, Y.-H. 1999. Ph.D. dissertation. Universität Stuttgart, Stuttgart, Germany.
- 14.Kim, Y.-H., K.-H. Engesser, and C. E. Cerniglia. 2003. Two polycyclic aromatic hydrocarbon o-quinone reductases from a pyrene-degrading Mycobacterium. Arch. Biochem. Biophys. 416:209-217. [DOI] [PubMed] [Google Scholar]
- 15.Kreft, J. U., and B. Schink. 1994. O-Demethylation by the homoacetogenic anaerobe Holophaga foetida studied by a new photometric methylation assay using electrochemically produced cob(I)alamin. Eur. J. Biochem. 226:945-951. [DOI] [PubMed] [Google Scholar]
- 16.Masai, E., Y. Katayama, S. Kawai, S. Nishikawa, M. Yamasaki, and N. Morohoshi. 1991. Cloning and sequencing of the gene for a Pseudomonas paucimobilis enzyme that cleaves beta-aryl ether. J. Bacteriol. 173:7950-7955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mo, K., C. O. Lora, A. E. Wanken, M. Javanmardian, X. Yang, and C. F. Kulpa. 1997. Biodegradation of methyl t-butyl ether by pure bacterial cultures. Appl. Microbiol. Biotechnol. 47:69-72. [DOI] [PubMed] [Google Scholar]
- 18.Odier, E., and C. Rolando. 1985. Catabolism of arylglycerol-beta-aryl ethers lignin model compounds by Pseudomonas cepacia 122. Biochimie 67:191-197. [DOI] [PubMed] [Google Scholar]
- 19.Parales, R. E., J. E. Adamus, N. White, and H. D. May. 1994. Degradation of 1,4-dioxane by an actinomycete in pure culture. Appl. Environ. Microbiol. 60:4527-4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Penn, J. H., and Z. Lin. 1990. Substituent dependence of the π-acceptor induced bond cleavage reactions of benzyl phenyl ethers. J. Org. Chem. 55:1554-1559. [Google Scholar]
- 21.Pieper, D. H., K.-H. Engesser, and H.-J. Knackmuss. 1989. Regulation of catabolic pathways of phenoxyacetic acids and phenols in Alcaligenes eutrophus JMP134. Arch. Microbiol. 151:365-371. [Google Scholar]
- 22.Pieper, D. H., W. Reineke, K.-H. Engesser, and H.-J. Knackmuss. 1988. Metabolism of 2,4-dichlorophenoxyacetic acid, 4-chloro-2-methylphenoxyacetic acid and 2-methylphenoxyacetic acid by Alcaligenes eutrophus JMP 134. Arch. Microbiol. 150:95-102. [Google Scholar]
- 23.Pieper, D. H., K. Stadler-Fritzsche, K.-H. Engesser, and H.-J. Knackmuss. 1990. Metabolism of 2-chloro-4-methylphenoxyacetate by Alcaligenes eutrophus JMP-134. Arch. Microbiol. 160:169-178. [DOI] [PubMed] [Google Scholar]
- 24.Speranza, G., B. Mueller, M. Orlandi, C. F. Morelli, P. Manitto, and B. Schink. 2002. Mechanism of anaerobic ether cleavage: conversion of 2-phenoxyethanol to phenol and acetaldehyde by Acetobacterium sp. J. Biol. Chem. 277:11684-11690. [DOI] [PubMed] [Google Scholar]
- 25.Steffan, R. J., K. McClay, S. Vainberg, C. W. Condee, and D. Zhang. 1997. Biodegradation of the gasoline oxygenates methyl-tert-butylether, ethyl-tert-butylether, and tert-amylmethylether by propane-oxidizing bacteria. Appl. Environ. Microbiol. 63:4216-4222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.U.S. Environmental Protection Agency. 1996. Hazardous waste characteristics scoping study, p. 4.1-4.44. Office of Solid Waste, U.S. Environmental Protection Agency, Washington, D.C.
- 27.U.S. Environmental Protection Agency. 1998. Health effects notebook for hazardous air pollutants about health effects fact sheets. Office of Air Quality Planning and Standards, U.S. Environmental Protection Agency, Washington, D.C.
- 28.White, G. F., N. J. Russell, and E. C. Tidswell. 1996. Bacterial scission of ether bonds. Microbiol. Rev. 60:216-232. [DOI] [PMC free article] [PubMed] [Google Scholar]