Abstract
Mutations in components of the molecular motor dynein/dynactin lead to neurodegenerative diseases of the motor system or atypical parkinsonism. These mutations are associated with prominent accumulation of vesicles involved in autophagy and lysosomal pathways, and with protein inclusions. Whether alleviating these defects would affect motor symptoms remain unknown. Here, we show that a mouse model expressing low levels of disease linked-G59S mutant dynactin p150Glued develops motor dysfunction >8 months before loss of motor neurons or dopaminergic degeneration is observed. Abnormal accumulation of autophagosomes and protein inclusions were efficiently corrected by lowering dietary protein content, and this was associated with transcriptional upregulations of key players in autophagy. Most importantly this dietary modification partially rescued overall neurological symptoms in these mice after onset. Similar observations were made in another mouse strain carrying a point mutation in the dynein heavy chain gene. Collectively, our data suggest that stimulating the autophagy/lysosomal system through appropriate nutritional intervention has significant beneficial effects on motor symptoms of dynein/dynactin diseases even after symptom onset.
Introduction
Neurodegenerative diseases are characterized by multiple alterations observed in brain tissue of patients. Loss of neuronal cell bodies as well as loss of synapses is generally called neurodegeneration, and is accompanied by other neuronal alterations, in particular protein inclusions and abnormal protein recycling pathways such as autophagy/lysosomal pathway. In many cases, the physician considers that a period for ‘neuronal dysfunction’ occurs before degenerative changes, especially in slowly progressive neurodegenerative diseases, such as spastic paraplegia or juvenile motor neuron diseases. The use of post-mortem tissue precludes, however, to determine which pathological alterations occur first in patients and which one(s) actually cause the neurological symptoms. Getting access to pre-symptomatic events should in principle be easier in animal models, and their careful analysis might help us to determine the sequence of pathological alterations and their relationships with symptoms. However, most commonly used mouse models develop very rapid progression and severe disease, which is not always the case in patients. Thus, slowly progressive mouse models of disease could help to understand the sequence of events upon longitudinal follow-up.
Dysfunction of vesicle trafficking is thought to be an early event in many neurodegenerative diseases. Recently, genetic mutations in two components of dynein, the molecular motor carrying cargoes in the retrograde direction in axons were linked to familial neurodegenerative disease (1,2). Mutations in dynein heavy chain or its associated protein dynactin, cause diseases affecting either motor neurons (3–6), sensory neurons (7,8), medium spiny neurons (9), nigrostriatal neurons (10) and cortical development (5,11,12). Germline mutations in the murine dynein heavy chain lead to proprioceptive neuropathy and striatal dysfunction, but do not trigger neurodegeneration. On the contrary, overexpression of disease-linked G59S mutation in dynactin p150Glued (G59S mice) leads to motor neuron degeneration in end-stage mice (13,14) while knock-in of this same mutation did not lead to observable motor phenotypes (15). Interestingly, G59S mice also develop protein inclusions, and abnormal autophagy/lysosomal pathways (13,14). The slow progression of their disease makes these mice a suitable model to study the sequence of events leading to neurodegeneration and symptoms.
Here, we performed a longitudinal analysis of behavioural symptoms and neurodegenerative features in the M2 line of Laird et al. (13). This M2 line, later referred as G59S mice, is the mouse strain with the slowest disease progression that these authors generated. We observed that loss of neurons and synapses occurs extremely late, months after onset of motor symptoms. Lowering dietary protein content concomitantly mitigated symptoms, reverted autophagosome defects and decreased protein inclusions. Our results suggest that nutritional therapies stimulating autophagy/lysosomal pathway might be useful for symptomatic relief and not only as neuroprotective strategies.
Results
Late and selective neurodegeneration in transgenic G59S mice
We sought to correlate motor symptoms and neuronal loss in G59S mice, as these mice have only been characterized at end-stage in the initial study (13). Reduced muscle grip strength of G59S mice (13) occurred after 6 months of age (Fig. 1A), similar to decreased rotarod performance (Fig. 1B). Body weight loss (Fig. 1C) as well as cramping and tremors (Fig. 1D) occurred after 8 months of age, and disease end-stage was reached at ∼18 months of age. Consistent with the initial study, we observed denervation like spontaneous muscle electrical activity using electromyography (EMG) in end-stage animals (Supplementary Material, Fig. S1). However, these EMG abnormalities were never observed in mice younger than 9 months of age (Supplementary Material, Fig. S1) and neuromuscular junctions (NMJs) of 10-month-old G59S mice were normally innervated (Fig. 2A and B). Thus, the obvious motor deficit observed at 10 months of age is not due to motor neuron degeneration or NMJ dysfunction. Mutations in DCTN1 gene have also been linked with atypical Parkinson's disease (10). However, the number of dopaminergic neurons in the substantia nigra in 10- and 15-month-old G59S mice was similar to wild-type littermate mice (Fig. 2C and D). Dopaminergic neurons were, however, affected late in the disease, since dopamine (DA) levels dropped in 15 month-old G59S mice (Fig. 2E), while DA turnover increased (Fig. 2F) and D1 receptor mRNA decreased (Fig. 2G). Thus, G59S mutation also affects dopaminergic neurons, although in late stages and in the absence of neuronal loss. To more broadly detect a potential degenerative phenotype in G59S, we performed longitudinal magnetic resonance imaging (MRI) analysis. As shown in Figure 2H and I, there was no obvious degeneration of specific brain region nor was there global brain atrophy in these animals. In all, G59S mice neuronal loss appears restricted to motor neurons, and to some extent to nigrostriatal dopaminergic neurons, and occurs several months after motor symptoms.
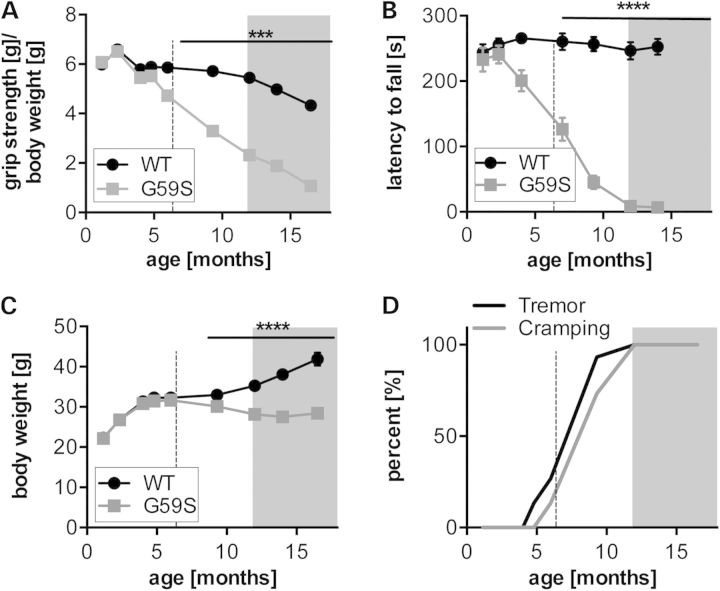
Figure 1.
Early motor dysfunction in G59S mice: (A) grip muscle strength of all limbs in wild-type (black) and G59S-mice (grey) from 1 to 18 months of age, normalized to body weight. ***P < 0.001 versus corresponding wild type (n = 15 mice per group). (B) latency to fall in an accelerating rotarod test in wild-type (black) and G59S-mice (grey) from 1 to 18 months of age. ****P < 0.0001 versus corresponding wild type (n = 15 mice per group). (C) Body weight of wild-type (black) and G59S-mice (grey) from 5 to 70 weeks of age. ****P < 0.0001 versus corresponding wild type (n = 15 mice per group). (D) Kaplan–Meier plot showing incidence of tremor and cramping in G59S-mice, tremor (black line) and cramping (grey line) (n = 15 mice per group). The shaded grey part of each graph indicates time points at which muscle denervation is observed (see Fig. 2). The dashed line indicates the time point at which LPD was initiated in Figure 3.
Figure 2.
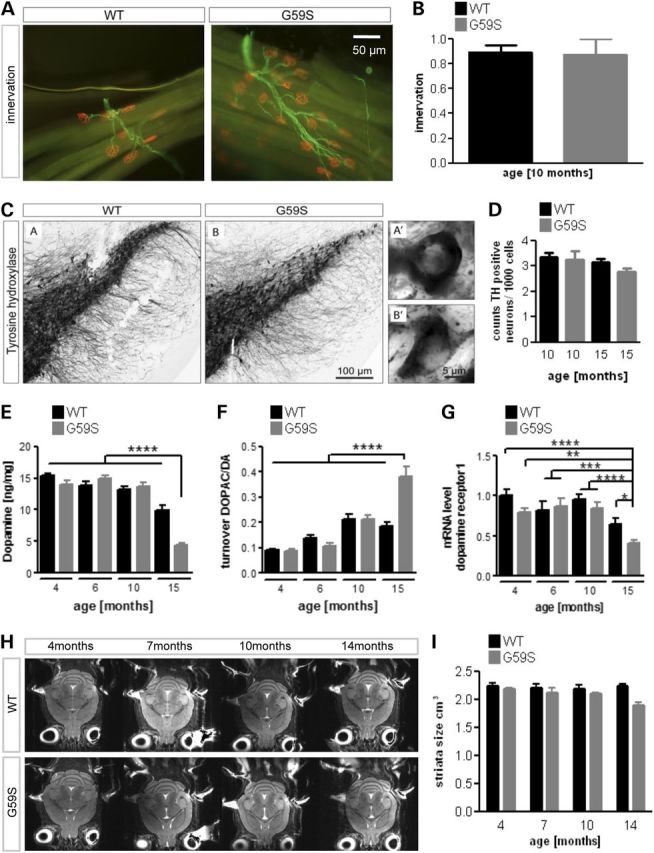
Late neurodegeneration in G59S mice. (A) Representative immunohistochemistry of neuromuscular junctions (NMJs) in the tibialis anterior muscle of wild-type and G59S-mice at the age of 10 months of age (n = 5 mice per group). The post-synaptic part of the NMJ was labelled with bungarotoxin (red), and innervating axons were labelled with anti-neurofilament antibodies (green). Note that all post-synaptic structures are properly innervated at that age. (B) Quantification of previous experiment. The percentage of innervated NMJs is shown. (C) Immunohistological staining of tyrosine hydroxylase (TH)-positive neurons in wild-type and G59S-mice at the age of 15 months. (D) Stereological estimation of the number of TH-positive neurons in the substantia nigra in wild-type and G59S-mice at the age of 10 and 15 months (n = 5 mice per group). (E and F) Dopamine levels (E) and DA turnover (F) as judged on DOPAC/DA ratio from 4 to 15 months of age in wild-type (black) and G59S-mice (grey) (n = 12 mice per group). (G) mRNA levels of DA receptor1 in 4–15-month-old wild-type (black) and G59S-mice (grey) (n = 12 mice per group). (H) Representative horizontal T2-weighted MRI slices of wild-type and G59S-mice at 4, 7, 10 and 14 months of age. (I) Striatal volume of wild-type (black) and G59S-mice (grey) at 4, 7, 10 and 14 months of age. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001 versus corresponding wild type.
Low protein diet partially rescues motor deficits in G59S and Cramping mice
If motor symptoms occur months before actual neuronal loss, then they are not the consequence of neuronal loss. Previous studies had indicated that mutant dynactin overexpression leads to proliferation of membrane bound vesicles including lysosomes (14) and autophagosomes (13) in motor neurons. Lowering protein content of the diet increases lysosomal and autophagic turnover in the periphery (16), and we hypothesized that such dietary treatment could also ameliorate symptoms of G59S mice. Two groups of 6-month-old G59S mice (n = 16–18) and wild-type littermates (n = 11–12) were trained to rotarod during 1 week, assessed for motor coordination for the next 2 weeks and then fed with either chow or low protein diet [LPD, composition in Supplementary Material, Table S1 identical to that of 16). From Week 3 of LPD, we observed that LPD increased rotarod performance of G59S mice as compared both with baseline and their G59S littermates under chow diet (Fig. 3A). The amelioration plateaued after 6 weeks of LPD, and as a whole, LPD rescued about half of the motor defect. Grip strength also tended to increase upon LPD in G59S animals (Fig. 3B). As expected from previous studies, LPD also decreased the final body weight of the animals of 3–5 g. To provide further evidence that LPD might correct established motor symptoms caused by defective axonal transport, we performed similar experiments in Cramping mice. These mice carry a mutant allele of Dync1h1 (17), the gene encoding the heavy chain of dynein, and this mutation lies in the same domain than human mutations associated with motor and sensory neuropathies (4,5,8), and triggers early sensory-motor defects without neurodegeneration (7,9,18). This mouse strain is maintained on a pure C3H background, different from the Bl6 SJL background of G59S mice. In this mouse line, motor defects are established within the first weeks after birth and remain essentially stable through life (9,17). In this completely independent mouse model, we also observed an increased motor performance of Cramping mice upon LPD either in rotarod performance or in grip strength (Fig. 3C and D). Thus, decreasing protein content of the diet was sufficient to mitigate established motor symptoms in these two mouse models, at a time point of disease when neuronal loss is not established.
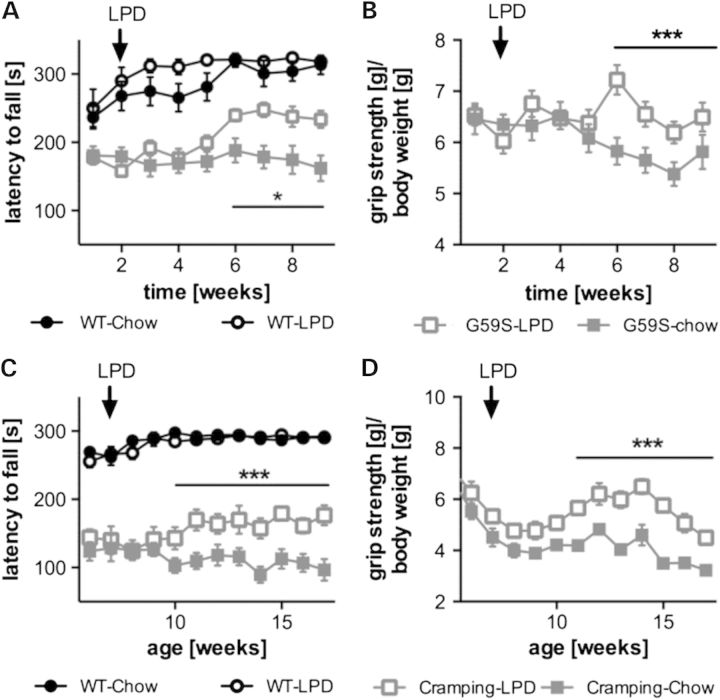
Figure 3.
Low protein diet partially rescues motor deficits in G59S and Cramping mice. (A and B) latency to fall in an accelerating rotarod test (A) or muscle grip strength normalized to body weight (B) in wild-type (black) and G59S-mice (grey) fed with chow diet (filled symbols) or low protein diet (LPD, open symbols). (C and D) latency to fall in an accelerating rotarod test (C) or muscle grip strength normalized to body weight (D) in wild-type (black) and Dync1h1Cra/+ mice (grey). Arrows show the time point mice were switched from chow to LPD. *P < 0.05; ***P < 0.001 versus corresponding wild type. In (B) and (D) grip strength of wild-type mice under chow or LPD were omitted for clarity.
Low protein diet normalizes autophagosomal defects in G59S mice
LPD might be protective towards motor symptoms through multiple mechanisms, including improved axonal transport, direct effects on autophagic machinery or indirect peripheral effects. To control that LPD could directly activate relevant long-term responses in the central nervous system, we monitored mRNA levels of Snat2, the primary amino acid transporter that is induced at the level of transcription when mammalian cells are deprived of amino acids (19,20). Twelve weeks of LPD increased Snat2 mRNA levels in the cerebral cortex (Fig. 4A) and spinal cord (Fig. 4B) of G59S mice as well as in spinal cord of Cramping mice (Fig. 4C). Similar trends were observed in wild-type littermates. Since LPD had effects on the CNS, we hypothesized that it could improve axonal transport, thereby alleviating symptoms (21). To test this hypothesis, we injected fluorogold in hind limb muscles of G59S mice either chow diet or LPD fed, and sacrificed mice 24 h later to quantify retrograde transport of the dye in spinal motor neurons. We observed that G59S mutation led to drastic loss of fluorogold labelling in motor neurons (Fig. 5). LPD did not modify this axonal transport defect.
Figure 4.
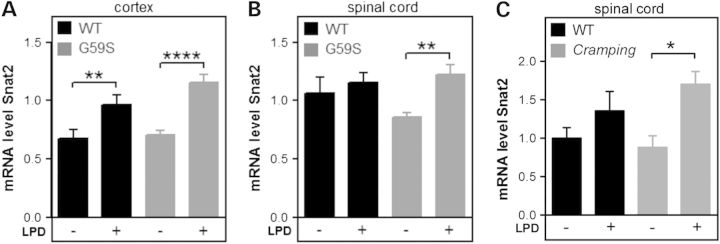
LPD increases expression of Snat2 in the CNS. mRNA levels of the amino acid transporter Snat2 (Slc38a2) in the cortex (A) and spinal cord (B) of wild-type (black) and G59S-mice (grey) and spinal cord of Cramping mice and their controls (C), fed with chow diet (−) or LPD (LPD, +).
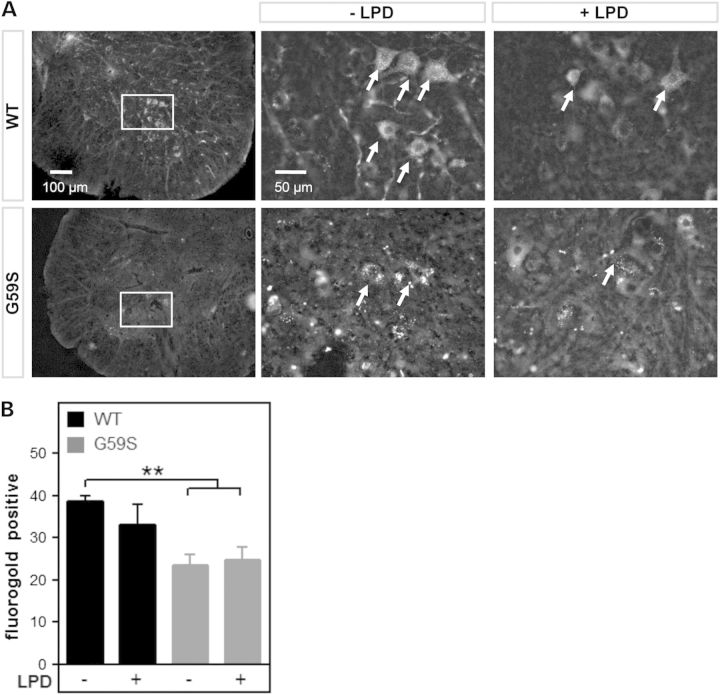
Figure 5.
Low protein diet does not modify axonal transport defect in G59S mice. (A) representative microphotographs of fluorogold fluorescence in lumbar spinal cord of wild-type (upper row) and G59S-mice (lower row) fed with chow diet (−LPD) or low protein diet (+LPD). Motor neurons of wild type mice are filled with fluorogold-positive vesicles (arrows), while motor neurons of G59S mice show less distinct staining, indicative of a decreased rate of fluorogold transport to the spinal cord. The left images show lower magnifications of images in the middle column (box). (B) Number of fluorogold-positive lumbar motor neurons (L3–L5) per ventral horn in wild-type (black columns) and G59S-mice (grey columns) fed with chow diet (−LPD) or low protein diet (+LPD). **P < 0.01 versus wild type, chow diet.
LPD could also alleviate motor symptoms through effects on autophagy, and we thus studied protein levels of the autophagosome marker LC3-II. Consistent with a correction of defective autophagy by LPD, increased LC3-II levels observed in chow-fed G59S mice were reverted by LPD (Fig. 6A). To directly demonstrate effects on autophagic flux in vivo, we used electron microscopy to unambiguously identify autophagosomes and blocked lysosomal proteolysis using chloroquine in a subset of mice (22). Consistent with increased LC3-II, G59S mice displayed many more autophagosomes in neurons than wild-type littermates (Fig. 6B–D). Chloroquine administration led to significantly more autophagosomes in G59S as compared with wild types, showing that G59S mice displayed increased autophagosome formation in basal conditions (Fig. 6B–D). LPD completely abolished increased autophagosome counts in G59S mice. Importantly, blocking autophagosomal clearance with chloroquine prevented the decrease in number of autophagosomes elicited by LPD showing that autophagosome clearance was increased upon LPD. However, autophagosome counts remained higher in the presence of chloroquine in chow fed G59S mice as compared with LPD fed G59S mice, demonstrating that LPD also decreased autophagosome formation. Thus, LPD was able to correct autophagosome defects through both decreased autophagosome generation and increased autophagosome clearance. In the course of the analysis of electron microscopy (EM) images, we noticed the occurrence of prominent protein inclusions in G59S neurons (Fig. 6C–E). LPD decreased the frequency of these inclusions in G59S neurons, an effect prevented by chloroquine. Thus, autophagic flux is impaired in G59S neurons, and LPD restores normal autophagosome numbers and resolves protein aggregates.
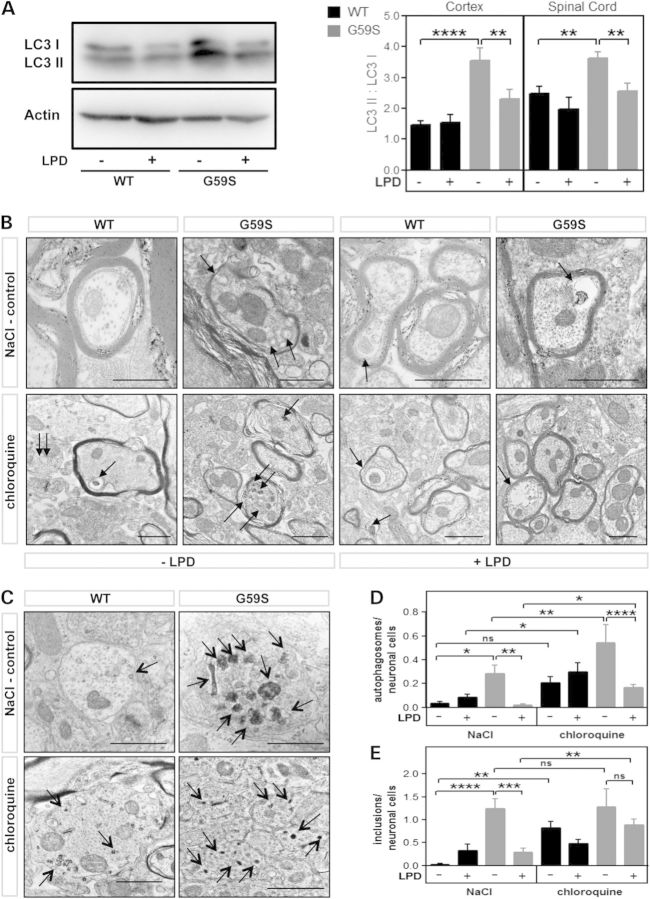
Figure 6.
Low protein diets corrected autophagosome defects in G59S-mice. (A) Left: representative western blot of LC3-I (upper band) and LC3-II (lower band) in the cerebral cortex of wild type or G59S mice, fed with chow or low protein diet. Actin western blot is provided as a standard. Right: LC3 II/LC3 I ratio, as measured using western blotting in the cortex and spinal cord of wild-type (black columns) and G59S-mice (grey columns) fed with chow diet (−LPD) or low protein diet (+LPD). **P < 0.01; ****P < 0.0001 versus corresponding wild type. (B and C) electron micrographs of neuronal cells in spinal cord from age-matched wild-type (WT) and G59S-mice (G59S) under chow or LPD, and either injected with saline (NaCl) or chloroquine. Arrows indicate autophagosomes including degradative organelles (B) or protein inclusions (C); scale bar is 1000 nm. (D) quantitative analysis of autophagosomes (D), and protein inclusions (E) per spinal neuronal cells in G59S-mice and wild-type age-matched controls under the different treatment groups, *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001 versus corresponding wild type.
Low protein diet differentially reprogrammes gene expression in different genetic backgrounds
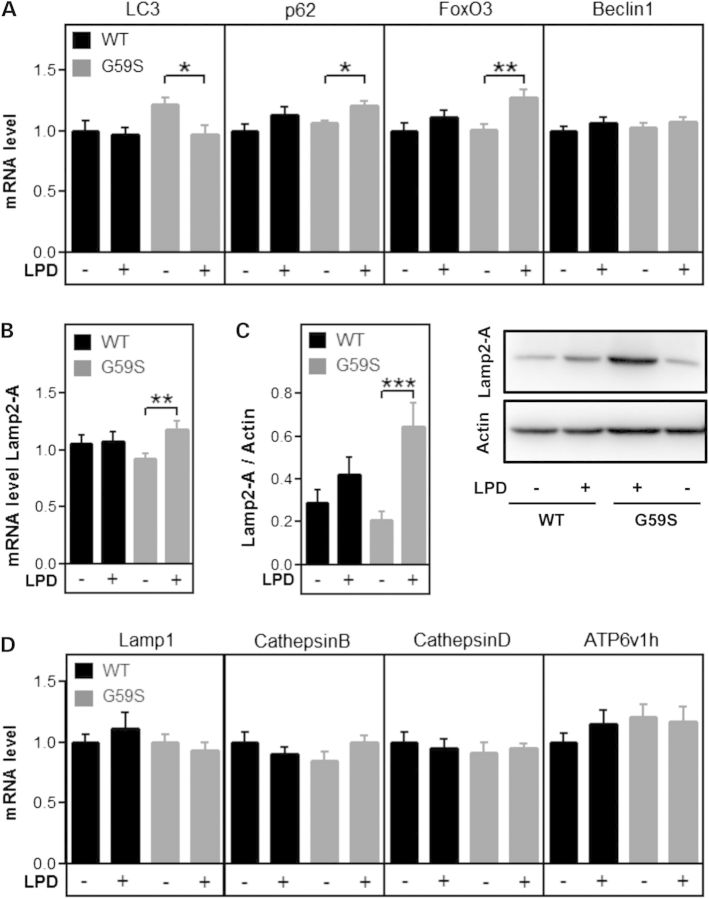
Long-term dietary changes, such as calorie restriction, induce activation of cellular adaptative responses in a process called hormesis (23). We hypothesized that this would also stand true for LPD, and that LPD would stimulate expression of the cellular systems to recycle amino acids in particular macro-autophagy, chaperone mediated autophagy (CMA) and/or lysosomal biogenesis. Consistent with chronic autophagy impairment (24), LC3 expression was slightly increased in chow fed G59S mice as compared with wild-type chow fed mice. Consistent with EM quantifications, LC3 gene expression was decreased in G59S mice fed with LPD. mRNA levels of autophagy key genes, such as p62 and FoxO3 were unchanged in chow diet, but slightly increased upon LPD selectively in G59S mice (Fig. 7A). mRNA levels of key CMA player Lamp2-A were increased (Fig. 7B) and this was mirrored in protein levels (Fig. 7C). However, there was no coordinated upregulation of genes involved in lysosomal biogenesis (Fig. 7D).
Figure 7.
Gene expression in response to LPD in G59S mice. (A) mRNA levels of genes involved in macro-autophagy (LC3, p62, FoxO3 and Beclin1) in the spinal cord of wild-type (black) and G59S-mice (grey) fed with chow diet (−) or low protein diet (LPD, +). (B and C) mRNA (B) and protein (C) levels of Lamp2-A in the spinal cord of wild-type (black) and G59S-mice (grey) fed with chow diet (−) or low protein diet (LPD, +). (D) mRNA levels of genes involved in lysosomal biogenesis (Lamp1, Cathepsin B, Cathepsin D and ATP6v1h) in the spinal cord of wild-type (black) and G59S-mice (grey) fed with chow diet (−) or low protein diet (LPD, +).
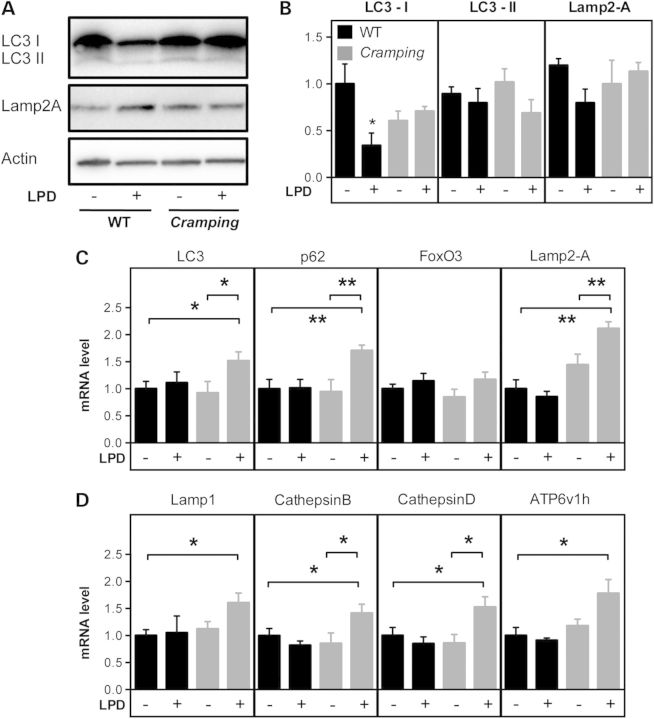
The lysosomal/autophagy pathway was also altered in Cramping mice upon LPD, although with different features. LPD did not change Lamp2-A protein levels, but led to decreased LC3-I levels in C3H control littermates but not in Cramping mice (Fig. 8A and B). Gene expression was dramatically altered in Cramping mice upon LPD, with increased expression of LC3 and p62 but also of key lysosomal proteins such as Lamp1, Cathepsins B and D, and Atp6v1h and key CMA player Lamp2-A (Fig. 8C and D). All of these regulations did not occur in C3H control littermates. Summarizing, LPD activates a transcriptional programme in diseased mice with different features as a function of the genetic background. In Bl6SJL mice, key players of macro-autophagy and of CMA, but not lysosomal biogenesis are upregulated, while in C3H mice, transcriptional upregulation is broader.
Figure 8.
Low protein diet effects in Cramping mice. (A) Left: representative western blot of LC3-I (upper band), LC3-II (lower band), Lamp2-A and actin in the spinal cord of wild type or Cramping mice, fed with chow or low protein diet. (B) Quantification of the western blots shown in A. *P < 0.05 versus wild-type chow fed mice. N = 3–4 per group. (C) mRNA levels of genes involved in macro-autophagy (LC3, p62, FoxO3) and in chaperone-mediated autophagy (Lamp2-A) and Beclin1) in the spinal cord of wild-type (black) and Cramping mice (grey) fed with chow diet (−) or low protein diet (LPD, +). (D) mRNA levels of genes involved in lysosomal biogenesis (Lamp1, Cathepsin B, Cathepsin D and ATP6v1h) in the spinal cord of wild-type (black) and Cramping mice (grey) fed with chow diet (−) or low protein diet (LPD, +).
Discussion
Here, we show that motor symptoms of dynein/dynactin diseases occur long before actual neuronal or axonal loss and are alleviated by low dietary protein content. This is likely mediated through correction of dysfunctional autophagy/lysosomal systems.
A first major result is that motor symptoms in G59S mice are not caused by neuronal death or synaptic loss. We chose to study the correlation between features of neurodegeneration, such as muscle denervation, and motor symptoms in the slowly progressive M2 line (13). Indeed, this mouse strain develops very slow motor symptoms, which are accompanied by detectable motor neuron degeneration, as shown by EMG, molecular and histological endpoints (13 and this study). Thus, these mice recapitulate as closely as possible the symptoms observed in patients carrying the G59S mutation. In our G59S mice, we observed strong motor phenotypes in the complete absence of NMJ denervation or EMG abnormalities, thus demonstrating that degeneration of motor neurons and NMJs is uncoupled from motor symptoms. Similarly, dopaminergic involvement occurs late, and neither DA levels, nor tyrosine hydroxylase (TH)-positive cell counts are changed at 10 months of age despite obvious motor symptoms. MRI imaging did neither show specific atrophy of the brain nor obvious lesions, and we consider thus unlikely that degeneration of other neurons than motor neurons or dopaminergic neurons cause disease. Summarizing, we observe a complete dissociation between motor symptoms and neuronal loss in this model. Interestingly, other mouse models of dynein/dynactin inhibition also show dissociation between degeneration of neurons and symptoms. For instance, Cramping and Legs at odd angles (Loa) mice develop motor and behavioural symptoms while neurodegeneration is absent except for developmental death of proprioceptive neurons (7,9,25). Thus, the motor behaviour caused by inhibition of dynein/dynactin is dissociated from neurodegeneration, and cannot be caused by it. In patients, EMG abnormalities were reported in individuals with dynein (5,26) and dynactin (27) mutations, but these patients were examined in their fifth decade while some displayed symptoms already during childhood (26). Indeed, EMG was found normal in several patients with H306R DYNC1H1 mutation in their third decade or during childhood (6,8) suggesting that, consistent with observations in mice, symptoms might precede neurodegeneration for a long time.
Our study provides insights into underlying pathomechanisms. A major pathological finding in various mouse strains expressing mutant dynactin was that lysosomes and/or autophagosomes were in abnormal numbers. Chevalier-Larsen et al. (14) showed lysosomal proliferation in their G59S line, while we previously observed a progressive and abnormal accumulation of vesicles, and in particular of autophagosomes in another G59S line (line M1) (13). Lai et al. (15) observed increased synaptic vesicles proteins at NMJs. This was corroborated by increased LC3-II levels, and by increased LC3 gene expression in the mouse line studied here. Many mechanisms involving dynactin could underlie increased accumulation of lysosomal/autophagic vesicles. First, increased autophagosome numbers could be a consequence of increased biogenesis. Indeed, a recent study showed that motor neurons with Loa mutation display aberrantly increased signalling response to serum starvation, a well-known autophagy inducer (28). Such a response could in principle alter autophagosome generation. A second, non-mutually exclusive, hypothesis is that G59S mutation impairs autophagic clearance through defective vesicle trafficking. Indeed, the G59S mutation decreases axonal transport rate in vitro (28–30), and we provide here similar evidence in vivo using retrograde fluorogold labelling in motor neurons. Since maturation of autophagosomes to autophagolysosomes occurs during their transport in the axons (21), the accumulation of autophagosomes we observe could be the consequence of decreased transport rate. Dynactin is required for transport initiation at distal neurites, and G59S mutation also affects this function (29,30). Last, dynactin is also required for the stability of microtubule cytoskeleton (31). In our study, we provide direct evidence for both increased autophagosome generation and decreased clearance in G59S mice. Indeed, chloroquine administration increased much more dramatically autophagosomes in wild types than in G59S neurons, showing that at least part of the increased autophagosome levels were due to impaired clearance. However, the difference between these two groups remained significant, consistent with a higher activity of autophagosome generation in G59S mice. That G59S mutation impairs autophagic flux in neurons is consistent with previous work showing that dynein mutations impair autophagic clearance of aggregate prone proteins (32) and autophagic perinuclear clustering of defective mitochondria (18). The downstream mechanisms linking impaired autophagic flux to actual motor symptoms remain to be identified. Among many other possibilities, we would like to speculate that impaired autophagy/lysosomal pathways perturbs axonal homeostasis, for example, by modifying retrograde signalling pathways or impairing mitochondrial bioenergetics.
Our results demonstrate that lowering dietary protein content mitigates motor symptoms after their establishment, as well as restores autophagosome defects. About 50% of rotarod defects were compensated by LPD, while grip strength, less affected at that age, was also increased. Similar observations were done in Cramping mice, showing that low dietary content generally protected animal models of dynein/dynactin diseases. It might seem surprising a priori that a dietary modification leads to such drastic alterations in the CNS. However, calorie restriction also leads to significant adaptative changes in the CNS (33), and, LPD is long known to trigger effects in the CNS on various neurochemical parameters such as tyrosine levels (34) or on transduction pathways of amino acid sensing (35).
How could LPD correct autophagosome defects? The effect of LPD on autophagy were complex, and depending on the genetic background. Measurement of autophagic flux in G59S mice showed that LPD both decreased autophagosome generation, while increasing autophagosome clearance. G59S mice also displayed several transcriptional upregulations, such as p62 or Lamp2-A that might underlie the resolving of protein aggregates. In Cramping mice, LPD maintained the pool of LC3-I, likely through a transcriptional mechanism, and led to a generalized transcriptional upregulation of genes involved in macro-autophagy, CMA and lysosomal biogenesis.
We propose that LPD, as a mild, chronic, nutritional stress, leads to an adaptative hormetic response, leading to a coordinated upregulation of genes helping in resolving the initial stimulus that is decreased amino acid availability. This is consistent with the simultaneous upregulation of amino acid transporters, such as Snat2, as well as of various actors of protein recycling in both G59S and Cramping mice fed with LPD. A potential candidate transcription factor stimulated by LPD would be TFEB, that is critically involved in coordinating autophagy and lysosomal gene expression (36), and was found inhibited by mutant androgen receptor in SBMA, another motor neuron disease and causing decreased autophagic flux (37). In Cramping mice, the overall picture observed would be reminiscent of TFEB activation. However, only one target of TFEB (p62), was overexpressed in G59S mice. Whether the different genetic backgrounds of the mice display different sensitivities to TFEB stimulation is a possibility. It should also be mentioned that Cramping mice display a number of neuronal and non-neuronal phenotypes (7,9,18,38) that could each contribute to the effects of LPD. In general, the transcriptional mechanisms accounting for the adaptative response elicited by LPD, as well as the potential involvement of TFEB, deserve further investigation.
Our study shows that lowering dietary content is protective against symptoms of dynein/dynactin diseases, even after the onset of motor symptoms. This expands the clinical spectrum of diseases in which such diet could be beneficial (16), and other neurodegenerative diseases that display strong lysosomal/autophagy dysfunction such as Huntington's disease or Parkinson's diseases, could represent other conditions in which to test this easy nutritional intervention (39–41). Consistently, LPD is currently suggested in advanced PD, as it is postulated to improve L-DOPA bioavailability. Our results suggest further that increased autophagic flux could indeed also contribute to the benefits of this dietary intervention in advanced PD patients (42,43).
Materials and Methods
Animals
Heterozygous G59S mice in B6 SJL background and Cramping mice in C3H background were maintained and genotyped as described (7,13). Mice were maintained at 22°C with a 12 h light/dark cycle and had food and water ad libitum. Unless otherwise mentioned, mice were fed with chow diet (Supplementary Material, Table S1). Mice were regularly monitored to assess onset and progression of symptoms and were considered at end stage when they could not right themselves within 20 s when placed on their back, or when they developed eye infections. For histological analysis, animals were deeply anaesthetized with 1 mg/kg body weight ketamine chlorhydrate and 0.5 mg/kg body weight xylazine, and transcardially perfused with 4% paraformaldehyde in 0.1 m pH 7.4 phosphate buffer. Tissues were then quickly dissected, post-fixed for 24 h in 4% paraformaldehyde and cryoprotected for 48 h. For the biochemical analysis, animals were sacrificed and tissues were quickly dissected, frozen in liquid nitrogen. All animal experiments in this study were approved by the Ethical Committee of Baden-Württemberg under number 1077.
Dietary treatments
Mice were fed either a LPD (no S0052-E712 EF M by Ssniff) or normal standard food (CHOW) as indicated (Supplementary Material, Table S1 and (16)). Mice were randomly distributed per diet, with at least one mouse per genotype per litter allocated to each experimental condition. For practical reasons in the animal facility, experimentators could not be blinded to the diet.
Behaviour and motor performance
One week before starting experiments, male mice were brought to the behaviour facility and handled every day. Once per week, starting at the age of 4 weeks, body weight and fore and all limb grip strength were measured for a cohort of 15 each of transgenic and wild-type mice. Grip strength measurements were taken using a grip strength meter (Bioseb gripmeter, Vitrolles, France). To take into account that the maximal performance of the animal is strictly dependent upon its body mass, we divided the measure obtained by the body weight of the animal, according to the provider's recommendations. Each measurement was performed in triplicates and data were averaged. To test the motor coordination and balance, rotarod test was performed twice per month (Rotarod Version 1.2.0. MED Associates Inc. St. Albans, VT). Mice were placed onto a rotating rod with auto acceleration from 4 to 40 rpm in 300 s. The length of time the mouse stayed on the rotating rod was recorded. Every mouse had to perform three trials separated by 15 min each other, the three trials were averaged. Identical protocols were performed for G59S and Cramping experiments.
Electromyography
Mice were anesthetized with ketamine/xylazine and electrical activity was recorded using a monopolar needle electrode (diameter 0.3 mm; 9013R0312; Medtronic, Minneapolis, MN, USA) inserted into the tail of the mouse. Recordings were made with a concentric needle electrode (diameter 0.3 mm; 9013S0011; Medtronic). Electrical activity was monitored in both gastrocnemius and tibialis anterior on both legs for at least 2 min. Spontaneous activity was differentiated from voluntary activity by visual and auditory inspection.
Immunohistochemistry
Brain and spinal cord 10 µm cryosections were prepared on gelatin coated slides and processed according to previously published protocols (7). Neuromuscular junction histology was performed on muscle bundles as previously described (7). Primary antibodies were incubated overnight at room temperature. Secondary antibodies (donkey anti-goat antibody coupled with Alexa 555, A21432; Invitrogen, Carlsbad CA, USA; donkey anti-mouse coupled with Alexa 488, A21204; Invitrogen, Carlsbad CA, USA) were incubated 90 min at room temperature. Slides were mounted using Aqua-Poly/Mount (Polysciences, Warrington PA, USA). Images were acquired with a laser scanning microscope (LSM 510; Carl Zeiss, Thornwood, NY, USA) equipped with a Plan-Apochromat 63× oil DIC immersion lens (numerical aperture 1.4).
Stereological analyses of TH-positive cells
Stereological assessment of the number of A9 TH-positive cells in the substantia nigra was performed on a Nikon 80i microscope equipped with an XY motorized stage (Märzhauser, Wetzlar, Germany), a Z-axis motor and a high precision linear encoder (Heidenhain, Traunreut, Germany). The delineation of the areas of interest was done under the 4× objective and the counting was performed at high magnification with a 60× Plan-Apo oil immersive objective (NA = 1.4). The unbiased stereological quantification principles were applied to estimate the numbers of immunopositive cells by using the optical dissector method (44). The assessment was performed in a blinded fashion and the procedure was carried out with a random start systematic sampling routine (New Cast Module in VIS software; Visiopharm A/S, Horsholm, Denmark). To minimize the coefficient of error, the sampling interval was adjusted to count at least 100 cells for each animal.
Dopamine measurements
To measure striatal DA levels 4-, 6-, 10-month-old mice and end-stage mice were sacrificed and striata were quickly dissected, frozen in liquid nitrogen and stored at −80°C. Tissues were then homogenized by ultrasonification for 10 s in 0.4 m perchloric acid and centrifuged at 14 000g for 15 min at 4°C. Afterwards, the supernatants were passed through a 0.2 μm filter (MinisartRC4, Sartorius AG, Göttingen, Germany). Measurements of DA and its metabolites 3,4-dihydroxyphenylacetic acid (DOPAC), 3-methoxytyramine (3-MT) and homovanillic acid were performed using high-performance liquid chromatography (HPLC) combined with electrochemical detection under isocratic conditions as previously described (45). For data acquisition and calculation, Chromeleon™ version 7.1 HPLC software (Thermo Fisher Scientific Inc., USA) was used.
Striatal volume by MRI
Ten G59S mice and 10 age- and gender-matched wild types underwent MRI at an 11.7T small bore animal scanner (Biospec 117/16, Bruker, Ettlingen, Germany) with a 4-element receive-only surface brain array coil at four time points with a time interval of 3 months (T2-weighted protocol, TE/TR 41.6/2000 ms, matrix 512 × 512, in-plane resolution 68 µm × 68 µm, 30 coronar slices of 250 µm). Analysis procedures followed (9). Image analysis was performed blinded to the genotype.
RT-qPCR
Total RNA was extracted using RNeasy Lipid Tissue Mini Kit (Qiagen). RT-qPCR was performed using the iScript cDNA Synthesis Kit (Bio-Rad) and the iQ SYBR Green SupermixR (Bio-Rad) using a CFX96 thermocycler (Bio-Rad). Primer sequences are provided in Supplementary Material, Table S2. Data were analysed using the Cycler software and normalized to the normalization factor calculated from the reference genes (Pol2, β-actin and HPRT).
Western blotting
For western Blot analysis mouse tissue were homogenized in RIPA-Buffer containing protease inhibitor. The following primary antibodies were used: LC3 (Sigma; 1 : 2500 buffered in 1% BSA; 0.02% NaN3 in PBS containing 0.05% Tween 20); Lamp2-A (abcam; 1 : 1000 buffered in 1% BSA; 0.02% NaN3 in PBS containing 0.05% Tween 20); overnight at 4°C. After washing in PBS/0.05% Tween 20, membranes were incubated at room temperature for 1 h with the second antibody (Bio-Rad; 1 : 10 000 in 2.5% Milk Goat Anti-Rabbit IgG-HRPconjugated) and washed again. The blot was visualized (ECL-immunodetection) using Image Quant LAS4000. Samples were corrected for background and quantified using Image Quant LAS 4000. All values were normalized to housekeeping protein (β-actin).
Autophagic flux measurements in vivo
For investigation of autophagy, only female mice were taken. At the age of 30 days, half of the mice get a LPD instead (Supplementary Material, Table S1) of regular chow. At 50 days of age, mice were daily injected (intra peritoneal) with 50 mg/kg chloroquine or 0.9% NaCl. After 10 days of injection, mice were taken for biochemical or histological analysis as previously described.
Electron microscopy
Mice were perfused with 4% paraformaldehyde, spinal cord was dissected and fixed in 2.5% glutaraldehyde for 24 h. Samples were post-fixed in 2% osmiumtetroxide for 1 h and dehydrated in graded series of ethanol. Fully dehydrated samples were then embedded into epon. The embedded samples were cut, collected on copper grids, stained with uranyl acetate and analyzed in a TEM (Zeiss EM 10) equipped with digital camera. Three animals per group and at least 10 fields per animal were analysed. The numbers of lysosomes, inclusions and autophagosomes were counted using Axiovision Rel.4.8. and correlated to the numbers of neuronal cells. Image analysis was performed blinded to the genotype and treatment groups.
Retrograde transport evaluations
Fluorogold (7 µl of a 10 mg/ml solution in PBS, 10% DMSO) was slowly injected in 1 min in the gastrocnemius muscle using a 10 µl Hamilton syringe (Hamilton, Reno, NV, USA). The syringe was gently withdrawn and any leakage tracer was removed from the surface of the muscle. After 24 h, mice were euthanized and L3–L5 spinal cord was processed for histology and cut in 40 µm sections using a vibratome. Fluorogold fluorescence was then acquired using regular epifluorescence microscopy. The number of motor neurons with fluorogold was quantified by an observer blinded to the genotype and treatment in at least 10 sections per animal (n = 5–6 per group).
Statistical analysis
For the experimental data all statistical analysis was done using Prism, version 5.0 (GraphPad Software). For comparison of groups without repeated measurements, ANOVA was used. Differences between means were determined by post hoc comparisons using the Fisher's LSD Test. For longitudinal analysis, we used two-way ANOVA, with genotype and age as co-variates. Differences were considered statistically significant if P < 0.05. Values are presented as means ± SEM.
Supplementary Material
Funding
This work was supported by the Agence Nationale de la Recherche (Dynemit to L.D.); Association pour la recherche sur la SLA et les autres maladies du motoneurone (to L.D.); Thierry Latran Foundation (SpastALS to L.D.); Helmholtz Institute “RNA dysmetabolism in ALS and FTD”(to L.D. and A.C.L.); ALS association (Grant #2235 and 3209 to L.D.); the Frick foundation for ALS research (to L.D.); National Institute of Neurological Disorder and Stroke (Grant R01 NS40014 to P.C.W.); Department of Defense (Grant AL100078 to P.C.W.) and the Robert Packard Center for ALS Research (to P.C.W.).
Supplementary Material
Acknowledgements
We thank Renate Kunz and Reinhard Weih from the Central Facility for Electron Microscopy, Ulm University, Aram Kobalyan, and the imaging platform (Neuropole de Strasbourg) for their help.
Conflict of Interest statement. None declared.
References
- 1.Eschbach J., Dupuis L. (2011) Cytoplasmic dynein in neurodegeneration. Pharmacol. Ther., 130, 348–363. [DOI] [PubMed] [Google Scholar]
- 2.Schiavo G., Greensmith L., Hafezparast M., Fisher E.M. (2013) Cytoplasmic dynein heavy chain: the servant of many masters. Trends Neurosci., 36, 641–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Puls I., Jonnakuty C., LaMonte B.H., Holzbaur E.L., Tokito M., Mann E., Floeter M.K., Bidus K., Drayna D., Oh S.J., et al. (2003) Mutant dynactin in motor neuron disease. Nat. Genet., 33, 455–456. [DOI] [PubMed] [Google Scholar]
- 4.Harms M.B., Ori-McKenney K.M., Scoto M., Tuck E.P., Bell S., Ma D., Masi S., Allred P., Al-Lozi M., Reilly M.M., et al. (2012) Mutations in the tail domain of DYNC1H1 cause dominant spinal muscular atrophy. Neurology, 78, 1714–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fiorillo C., Moro F., Yi J., Weil S., Brisca G., Astrea G., Severino M., Romano A., Battini R., Rossi A., et al. (2014) Novel dynein DYNC1H1 neck and motor domain mutations link distal spinal muscular atrophy and abnormal cortical development. Hum. Mutat., 35, 298–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsurusaki Y., Saitoh S., Tomizawa K., Sudo A., Asahina N., Shiraishi H., Ito J., Tanaka H., Doi H., Saitsu H., et al. (2012) A DYNC1H1 mutation causes a dominant spinal muscular atrophy with lower extremity predominance. Neurogenetics, 13, 327–332. [DOI] [PubMed] [Google Scholar]
- 7.Dupuis L., Fergani A., Braunstein K.E., Eschbach J., Holl N., Rene F., Gonzalez De Aguilar J.L., Zoerner B., Schwalenstocker B., Ludolph A.C., et al. (2009) Mice with a mutation in the dynein heavy chain 1 gene display sensory neuropathy but lack motor neuron disease. Exp. Neurol., 215, 146–152. [DOI] [PubMed] [Google Scholar]
- 8.Weedon M.N., Hastings R., Caswell R., Xie W., Paszkiewicz K., Antoniadi T., Williams M., King C., Greenhalgh L., Newbury-Ecob R., et al. (2011) Exome sequencing identifies a DYNC1H1 mutation in a large pedigree with dominant axonal Charcot-Marie-Tooth disease. Am. J. Hum. Genet., 89, 308–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Braunstein K.E., Eschbach J., Rona-Voros K., Soylu R., Mikrouli E., Larmet Y., Rene F., De Aguilar J.L., Loeffler J.P., Muller H.P., et al. (2010) A point mutation in the dynein heavy chain gene leads to striatal atrophy and compromises neurite outgrowth of striatal neurons. Hum. Mol. Genet., 19, 4385–4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farrer M.J., Hulihan M.M., Kachergus J.M., Dachsel J.C., Stoessl A.J., Grantier L.L., Calne S., Calne D.B., Lechevalier B., Chapon F., et al. (2009) DCTN1 mutations in Perry syndrome. Nat. Genet., 41, 163–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Poirier K., Lebrun N., Broix L., Tian G., Saillour Y., Boscheron C., Parrini E., Valence S., Pierre B.S., Oger M., et al. (2013) Mutations in TUBG1, DYNC1H1, KIF5C and KIF2A cause malformations of cortical development and microcephaly. Nat. Genet., 45, 639–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Willemsen M.H., Vissers L.E., Willemsen M.A., van Bon B.W., Kroes T., de Ligt J., de Vries B.B., Schoots J., Lugtenberg D., Hamel B.C., et al. (2012) Mutations in DYNC1H1 cause severe intellectual disability with neuronal migration defects. J. Med. Genet., 49, 179–183. [DOI] [PubMed] [Google Scholar]
- 13.Laird F.M., Farah M.H., Ackerley S., Hoke A., Maragakis N., Rothstein J.D., Griffin J., Price D.L., Martin L.J., Wong P.C. (2008) Motor neuron disease occurring in a mutant dynactin mouse model is characterized by defects in vesicular trafficking. J. Neurosci., 28, 1997–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chevalier-Larsen E.S., Wallace K.E., Pennise C.R., Holzbaur E.L. (2008) Lysosomal proliferation and distal degeneration in motor neurons expressing the G59S mutation in the p150Glued subunit of dynactin. Hum. Mol. Genet., 17, 1946–1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lai C., Lin X., Chandran J., Shim H., Yang W.J., Cai H. (2007) The G59S mutation in p150(glued) causes dysfunction of dynactin in mice. J. Neurosci., 27, 13982–13990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grumati P., Coletto L., Sabatelli P., Cescon M., Angelin A., Bertaggia E., Blaauw B., Urciuolo A., Tiepolo T., Merlini L., et al. (2010) Autophagy is defective in collagen VI muscular dystrophies, and its reactivation rescues myofiber degeneration. Nat. Med., 16, 1313–1320. [DOI] [PubMed] [Google Scholar]
- 17.Hafezparast M., Klocke R., Ruhrberg C., Marquardt A., Ahmad-Annuar A., Bowen S., Lalli G., Witherden A.S., Hummerich H., Nicholson S., et al. (2003) Mutations in dynein link motor neuron degeneration to defects in retrograde transport. Science, 300, 808–812. [DOI] [PubMed] [Google Scholar]
- 18.Eschbach J., Sinniger J., Bouitbir J., Fergani A., Schlagowski A.I., Zoll J., Geny B., Rene F., Larmet Y., Marion V., et al. (2013) Dynein mutations associated with hereditary motor neuropathies impair mitochondrial morphology and function with age. Neurobiol. Dis., 58C, 220–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Palii S.S., Kays C.E., Deval C., Bruhat A., Fafournoux P., Kilberg M.S. (2009) Specificity of amino acid regulated gene expression: analysis of genes subjected to either complete or single amino acid deprivation. Amino Acids, 37, 79–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gazzola R.F., Sala R., Bussolati O., Visigalli R., Dall'Asta V., Ganapathy V., Gazzola G.C. (2001) The adaptive regulation of amino acid transport system A is associated to changes in ATA2 expression. FEBS Lett., 490, 11–14. [DOI] [PubMed] [Google Scholar]
- 21.Maday S., Wallace K.E., Holzbaur E.L. (2012) Autophagosomes initiate distally and mature during transport toward the cell soma in primary neurons. J. Cell. Biol., 196, 407–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iwai-Kanai E., Yuan H., Huang C., Sayen M.R., Perry-Garza C.N., Kim L., Gottlieb R.A. (2008) A method to measure cardiac autophagic flux in vivo. Autophagy, 4, 322–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mattson M.P. (2008) Hormesis defined. Ageing Res. Rev., 7, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karim M.R., Kawanago H., Kadowaki M. (2014) A quick signal of starvation induced autophagy: transcription versus post-translational modification of LC3. Anal. Biochem., 465C, 28–34. [DOI] [PubMed] [Google Scholar]
- 25.Chen X.J., Levedakou E.N., Millen K.J., Wollmann R.L., Soliven B., Popko B. (2007) Proprioceptive sensory neuropathy in mice with a mutation in the cytoplasmic Dynein heavy chain 1 gene. J. Neurosci., 27, 14515–14524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harms M.B., Allred P., Gardner R., Jr, Fernandes Filho J.A., Florence J., Pestronk A., Al-Lozi M., Baloh R.H. (2010) Dominant spinal muscular atrophy with lower extremity predominance: linkage to 14q32. Neurology, 75, 539–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Puls I., Oh S.J., Sumner C.J., Wallace K.E., Floeter M.K., Mann E.A., Kennedy W.R., Wendelschafer-Crabb G., Vortmeyer A., Powers R., et al. (2005) Distal spinal and bulbar muscular atrophy caused by dynactin mutation. Ann. Neurol., 57, 687–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garrett C.A., Barri M., Kuta A., Soura V., Deng W., Fisher E.M., Schiavo G., Hafezparast M. (2014) DYNC1H1 mutation alters transport kinetics and ERK1/2-cFos signalling in a mouse model of distal spinal muscular atrophy. Brain, 137, 1883–1893. [DOI] [PubMed] [Google Scholar]
- 29.Lloyd T.E., Machamer J., O'Hara K., Kim J.H., Collins S.E., Wong M.Y., Sahin B., Imlach W., Yang Y., Levitan E.S., et al. (2012) The p150(Glued) CAP-Gly domain regulates initiation of retrograde transport at synaptic termini. Neuron, 74, 344–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moughamian A.J., Holzbaur E.L. (2012) Dynactin is required for transport initiation from the distal axon. Neuron, 74, 331–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lazarus J.E., Moughamian A.J., Tokito M.K., Holzbaur E.L. (2013) Dynactin subunit p150(Glued) is a neuron-specific anti-catastrophe factor. PLoS Biol., 11, e1001611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ravikumar B., Acevedo-Arozena A., Imarisio S., Berger Z., Vacher C., O'Kane C.J., Brown S.D., Rubinsztein D.C. (2005) Dynein mutations impair autophagic clearance of aggregate-prone proteins. Nat. Genet., 37, 771–776. [DOI] [PubMed] [Google Scholar]
- 33.Mattson M.P. (2012) Energy intake and exercise as determinants of brain health and vulnerability to injury and disease. Cell Metab., 16, 706–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fernstrom M.H., Fernstrom J.D. (1995) Effect of chronic protein ingestion on rat central nervous system tyrosine levels and in vivo tyrosine hydroxylation rate. Brain Res., 672, 97–103. [DOI] [PubMed] [Google Scholar]
- 35.Gallinetti J., Harputlugil E., Mitchell J.R. (2013) Amino acid sensing in dietary-restriction-mediated longevity: roles of signal-transducing kinases GCN2 and TOR. Biochem. J., 449, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Settembre C., Di Malta C., Polito V.A., Garcia Arencibia M., Vetrini F., Erdin S., Erdin S.U., Huynh T., Medina D., Colella P., et al. (2011) TFEB links autophagy to lysosomal biogenesis. Science, 332, 1429–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cortes C.J., Miranda H.C., Frankowski H., Batlevi Y., Young J.E., Le A., Ivanov N., Sopher B.L., Carromeu C., Muotri A.R., et al. (2014) Polyglutamine-expanded androgen receptor interferes with TFEB to elicit autophagy defects in SBMA. Nat. Neurosci., 17, 1180–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eschbach J., Fergani A., Oudart H., Robin J.P., Rene F., Gonzalez de Aguilar J.L., Larmet Y., Zoll J., Hafezparast M., Schwalenstocker B., et al. (2011) Mutations in cytoplasmic dynein lead to a Huntington's disease-like defect in energy metabolism of brown and white adipose tissues. Biochim. Biophys. Acta, 1812, 59–69. [DOI] [PubMed] [Google Scholar]
- 39.Wong E., Cuervo A.M. (2010) Autophagy gone awry in neurodegenerative diseases. Nat. Neurosci., 13, 805–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nixon R.A. (2013) The role of autophagy in neurodegenerative disease. Nat. Med., 19, 983–997. [DOI] [PubMed] [Google Scholar]
- 41.Dehay B., Martinez-Vicente M., Caldwell G.A., Caldwell K.A., Yue Z., Cookson M.R., Klein C., Vila M., Bezard E. (2013) Lysosomal impairment in Parkinson’s disease. Mov. Disord., 28, 725–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tsui J.K., Ross S., Poulin K., Douglas J., Postnikoff D., Calne S., Woodward W., Calne D.B. (1989) The effect of dietary protein on the efficacy of L-dopa: a double-blind study. Neurology, 39, 549–552. [DOI] [PubMed] [Google Scholar]
- 43.Cereda E., Barichella M., Pedrolli C., Pezzoli G. (2010) Low-protein and protein-redistribution diets for Parkinson’s disease patients with motor fluctuations: a systematic review. Mov. Disord., 25, 2021–2034. [DOI] [PubMed] [Google Scholar]
- 44.West M.J., Slomianka L., Gundersen H.J. (1991) Unbiased stereological estimation of the total number of neurons in the subdivisions of the rat hippocampus using the optical fractionator. Anat. Rec., 231, 482–497. [DOI] [PubMed] [Google Scholar]
- 45.Buck K., Ferger B. (2008) Intrastriatal inhibition of aromatic amino acid decarboxylase prevents l-DOPA-induced dyskinesia: a bilateral reverse in vivo microdialysis study in 6-hydroxydopamine lesioned rats. Neurobiol. Dis., 29, 210–220. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.