Abstract
The class Prasinophyceae (Chlorophyta) contains several photosynthetic picoeukaryotic species described from cultured isolates. The ecology of these organisms and their contributions to the picoeukaryotic community in aquatic ecosystems have received little consideration. We have designed and tested eight new 18S ribosomal DNA oligonucleotide probes specific for different Prasinophyceae clades, genera, and species. Using fluorescent in situ hybridization associated with tyramide signal amplification, these probes, along with more general probes, have been applied to samples from a marine coastal site off Roscoff (France) collected every 2 weeks between July 2000 and September 2001. The abundance of eukaryotic picoplankton remained high (>103 cells ml−1) during the sampling period, with maxima in summer (up to 2 × 104 cells ml−1), and a single green algal species, Micromonas pusilla (Prasinophyceae), dominated the community all year round. Members of the order Prasinococcales and the species Bathycoccus prasinos (Mamiellales) displayed sporadic occurrences, while the abundances of all other Prasinophyceae groups targeted remained negligible.
Several studies have demonstrated the importance of eukaryotic picoplankton (cell size, 0.2- to 3-μm) in terms of biomass and productivity in the euphotic zone of oceanic oligotrophic waters (15), as well as in coastal waters (10). To date, only ∼40 species belonging to nine algal classes (Chlorophyceae, Prasinophyceae, Trebouxiophyceae, Prymnesiophyceae, Bolidophyceae, Eustigmatophyceae, Pinguiophyceae, Bacillariophyceae, and Pelagophyceae) of photosynthetic picoplanktonic eukaryotes have been formerly described (41). However, phylogenetic analyses of sequences retrieved from natural samples in different oceanic regions have demonstrated much higher diversity, since many of these sequences do not correspond to any described taxa (19). The contributions of the different taxonomic groups to the picoplanktonic biomass, diversity, and ecology are poorly known because simple and reliable methods to detect and quantify such organisms in natural samples are lacking. Pigment signatures, scanning electron microscopy, and serial dilution cultures suggest that the classes Prasinophyceae (division Chlorophyta), Pelagophyceae (division Heterokontophyta), and Prymnesiophyceae are major components of the picoplankton biomass in different marine systems (20, 35).
Among these, the class Prasinophyceae contains several photosynthetic picoeukaryote species. This class is considered to be the most primitive in the green lineage and to have given rise to all other green algal classes, as well as to the land plants (34). Members are known to be common in temperate and cold regions and can occur as prominent constituents of marine picoplankton (38). Within these organisms, genera such as Ostreococcus, Bathycoccus, and Micromonas have been described in coastal waters (4b, 6). Micromonas pusilla (the only described species in the genus Micromonas) has been identified as a major component of the picoplanktonic community in several oceanic and coastal regions, such as the Mediterranean Sea (39), the Norwegian Sea (37), and central California waters (35). However, the techniques used to establish these facts (microscopic identification of cells presenting few morphological characteristics or serial dilution cultures) are time-consuming and incompatible with extensive ecological studies. In consequence, the precise distributions and the seasonal dynamics of an apparently very common picoplankter, such as M. pusilla, are poorly known.
The aim of this work was to identify and study the seasonal variations of the dominant taxa in the picoeukaryote community at a coastal site of the western English Channel in the vicinity of a long-term oceanographic observation site (31). Oligonucleotide probes targeting 18S rRNA coupled to fluorescent in situ hybridization associated with tyramide signal amplification were used to detect the picoplanktonic taxa (22). Eight new oligonucleotide probes specific for Prasinophyceae were designed and validated on pure cultures. These probes, as well as more general probes targeting the Chlorobionta and the eukaryotes, allowed the study in detail of the dynamics of the dominant taxa along a seasonal time series between July 2000 and September 2001.
MATERIALS AND METHODS
Cultures.
Nineteen unialgal strains of picoeukaryotes belonging to different phylogenetic clades of the Prasinophyceae were selected (Table 1). They were grown in Nalgene (Rochester, N.Y.) flasks at 20°C in K medium (11). In order to test the new probes designed in this study, cells were harvested during the mid-exponential growth phase. For each culture, 4.5 ml was harvested and fixed with paraformaldehyde (1% final concentration) for 1 h. These cultures were filtered on 0.2-μm-pore-size Anodisc filters (Whatman International Ltd., Maidstone, England). The filters were dehydrated in an ethanol series (50, 80, and 100%; 3 min each) and stored at −80°C until further hybridization tests were performed.
TABLE 1.
Origins and culture conditions of picoplankton strains
| RCCa no. | Genus and species | Strain | Light (μE m−2 s−1) | Cell diameter (μm) | Origin |
|---|---|---|---|---|---|
| 116 | Ostreococcus tauri | OTTH 0595 | 100 | 0.8 | Thau Lagoon |
| 143 | Ostreococcus sp. | EUM 13BBL | 100 | 0.8 | Tropical Atlantic Ocean |
| 141 | Ostreococcus sp. | EUM 16BBL | 100 | 0.8 | Tropical Atlantic Ocean |
| 114 | M. pusilla | CCMPb 490 | 100 | 2 | North Atlantic |
| 299 | M. pusilla | NOUM 17 | 100 | 2 | Equatorial Pacific |
| 372 | M. pusilla | Naples | 100 | 2 | Gulf of Naples |
| 373 | M. pusilla | Skagerrak | 100 | 2 | Skagerrak |
| 417 | Mantoniella squamata | CCMP 480 | 100 | 3-5 | North Sea |
| 113 | B. prasinos | CCMP 1898 | 100 | 2 | Mediterranean Sea |
| 369 | Coccoid | CCMP 1205 | 100 | 2-6 | Sargasso Sea |
| 287 | Coccoid | NOUM 15 | 100 | 2.5 | Equatorial Pacific Ocean |
| 261 | P. marina | TAK 9801 | 40 | 4 | Takapoto Atoll (Pacific Ocean) |
| 370 | P. provasolii | CCMP 1203 | 100 | 2-4 | North Atlantic |
| 135 | P. provasolii | CCMP 1199 | 100 | 2-4 | Gulf of Mexico |
| 251 | Pycnococcus sp. | ROS 9401 | 100 | 2 | English Channel |
| 253 | Pycnococcus sp. | ROS 9404 | 100 | 2 | English Channel |
| 136 | P. capsulatus | CCMP 1407 | 100 | 3-6 | Sargasso Sea |
| 134 | Prasinococcus sp. | CCMP 1194 | 100 | 3-5 | Gulf of Mexico |
| 137 | Prasinoderma sp. | CCMP 1220 | 100 | 3-8 | Gulf of Mexico |
Roscoff Culture Collection (http://www.sb-roscoff.fr/Phyto/RCC/).
CCMP (Provasoli-Guillard National Center for Culture of Marine Phytoplankton, West Boothbay Harbor, Maine [http://ccmp.bigelow.org/]).
Natural samples.
Natural samples were collected twice a month between July 2000 and September 2001 at 0.5-m depth with 5-liter Niskin bottles off Roscoff, France, at the ASTAN station (48°46′ N, 3°57′ W). The water was prefiltered through a 200-μm-pore-size mesh and further processed in the laboratory. Temperature, salinity, and concentrations of phosphate, nitrate, and ammonium were measured by standard oceanographic methods.
For fluorescent in situ hybridization (FISH), 90 ml of seawater was prefiltered through 3-μm-pore-size Nuclepore filters (Whatman International Ltd.) and fixed with 10 ml of 10% paraformaldehyde for 1 h. Samples were then filtered onto 0.2-μm-pore-size Anodisc filters under a maximum pressure of 200 mm Hg and dehydrated in an ethanol series (50, 80, and 100%; 3 min each). The filters were stored at −80°C.
For flow cytometry analyses, 1.5 ml of prefiltered (3-μm pore size) samples was fixed with a mixture of 1% paraformaldehyde and 0.1% glutaraldehyde (final concentrations) and then deep frozen in liquid nitrogen and stored at −80°C.
For high-performance liquid chromatography (HPLC) pigment measurements, the <200-μm seawater fraction (1 liter) was collected on a GF/F filter (Whatman International Ltd.). The 3- to 200-μm fraction was collected on a 3-μm-pore-size Nuclepore filter. Finally, the <3-μm fraction was collected on a GF/F filter. The filtrations were conducted under a pressure of 200 mm Hg, and all of the filters were immediately deep frozen in liquid nitrogen and stored at −80°C.
Flow cytometry.
Total photosynthetic-cell counts were obtained from fixed seawater samples using a FACSsort flow cytometer (Becton Dickinson, San José, Calif.), as described previously (18). Photosynthetic picoeukaryotes were discriminated from cyanobacteria using Cytowin software (available from http://www.sb-roscoff.fr/Phyto/cyto.html).
HPLC.
Pigment analyses were performed using the method of Zapata et al. (43), with minor modifications as described by Latasa et al. (14). The contributions of different algal groups to the total chlorophyll a (Chl a) was estimated using CHEMTAX (17). While the contributions of the Mamiellales, Prasinococcales, and Pseudoscourfieldiales, which possess prasinoxanthin, could be computed, those of the other clades (prasinoxanthinless Prasinophyceae) cannot be distinguished from those of other Chlorophyta.
FISH associated with tyramide signal amplification.
In situ hybridization with horseradish peroxidase-labeled probes, signal amplification, and target cell detection were performed as described previously by Not et al. (22). The only difference was the use of a more viscous antifading reagent, AF1 (Citifluor Ltd., London, United Kingdom), instead of AF3 in order to preserve the hybridized slides longer (up to 2 weeks in the dark at 4°C) without significant loss of fluorescence.
Epifluorescence microscopy and image acquisition.
The hybridized cells were observed with an Olympus (Tokyo, Japan) BX 51 epifluorescence microscope equipped with a mercury light source and an 100× UVFL (Olympus, Tokyo, Japan) objective. Excitation-emission filters were 360/420 for DAPI (4′,6′-diamidino-2-phenylindole) and 490/515 for fluorescein isothiocyanate. For each sample, 10 randomly chosen microscopic fields were counted by eye. For probes with broad taxonomic specificity (e.g., CHLO02), >500 cells were counted. Because of the large number of hybridizations and the time required for each analysis, it was not possible to count replicates for each sample. However, for 33 samples, three replicates (i.e., three hybridizations with the same probe on three different filters) were analyzed, and the average error was 15% (range, 2 to 38%).
Design of 18S rRNA oligonucleotide probes.
The oligonucleotide probes (Table 2) were designed with ARB software (16) using a small-subunit rRNA database containing >30,000 complete and partial sequences. In addition to published sequences, our database also contained unpublished partial eukaryote sequences retrieved from three coastal sites. Although the probes could have been designed based only on the public sequences, the additional sequences allowed us to confirm that the targeted regions were conserved on the corresponding sites of coastal representatives of taxa belonging to the clades targeted. When the probes were designed with the ARB Probe Design function, care was taken to maximize the number of mismatches to nontarget sequences and to position these mismatches near the center of the probe. The theoretical specificities of the new probes were checked using the Probe Match function of the ARB software. Oligonucleotide probes with a 5′ amino link (C6) were purchased from MWG (Courtaboeuf, France). The probes were then labeled with horseradish peroxidase (Roche Diagnostic Boehringer, Meylan, France) as described previously (40). The optimal formamide concentration was determined empirically to be 40% in the hybridization buffer, and specificity tests of the new probes were performed at this concentration. These probes are available in the rRNA probe database for protists and cyanobacteria (http://www.sb-roscoff.fr/Phyto/Databases/RNA_probes_introduction.php). Because probes with broad taxonomic specificity usually do not work equally well for all targeted organisms, the three probes EUK1209R, CHLO01, and NCHLO01 were used in combination to estimate all eukaryotes (22). Finally, probe CHLO02, specific for the subregnum Chlorobionta (Chlorophyta and Streptophyta), was applied (30). To our knowledge, the division Streptophyta has no representative in the pelagic marine systems. Consequently, in this study, cells labeled by the probe CHLO02 were considered to belong to the division Chlorophyta.
TABLE 2.
Novel oligonucleotide probes targeting prasinophycean taxa
| Probe name | Sequence | Target group | Position of 16S rRNA (Escherichia coli) | Closest sequence not targeted
|
|
|---|---|---|---|---|---|
| Taxonomy | No. of mismatches | ||||
| PRAS01 | 5′-ACG GTC CCG AAG GGT TGG-3′ | Pseudoscourfieldiales clade V | 193 | Tilletia caries | 2 |
| PRAS03 | 5′-GCC ACC AGT GCA CAC CGG-3′ | Prasinococcales | 620 | Friedmannia israeliensis | 2 |
| PRAS04 | 5′-CGT AAG CCC GCT TTG AAC-3′ | Mamiellales (except the genus Dolichomatix) | 651 | Choricystis minor | 1 |
| PRAS05 | 5′-GCC AGA ACC ACG TCC TCG-3′ | Clade VIIA, RCC 287, CCMP 1205 | 651 | Pyramimonas olivacea | 3 |
| PRAS06 | 5′-AAT CAA GAC GGA GCG CGT-3′ | Environmental clade VIIB | 651 | Scutopus ventrolineatus | 3 |
| MICRO01 | 5′-AAT GGA ACA CCG CCG GCG-3′ | M. pusilla | 211 | Ostreococcus tauri | 1 |
| BATHY01 | 5′-ACT CCA TGT CTC AGC GTT-3′ | B. prasinos | 651 | Uncultivated bacteria | 3 |
| OSTREO01 | 5′-CCT CCT CAC CAG GAA GCU-3′ | Ostreococcus | 647 | Corynebacterium genitalium | 3 |
Tree construction.
PAUP* version 4.0 beta 10 (33) was used to perform phylogenetic analyses of complete 18S rRNA sequences using neighbor-joining methods. Cyanophora paradoxa, Pavlova gyrans, Mesostigma viride, and Chara foetida were included in the analyses as an outgroup, and the tree was rooted with C. paradoxa. Bootstrapping (1,000 replicates) allowed the evaluation of tree significance. Trees were drawn using TreeView (Roderic Page, University of Glasgow, Glasgow, United Kingdom).
RESULTS
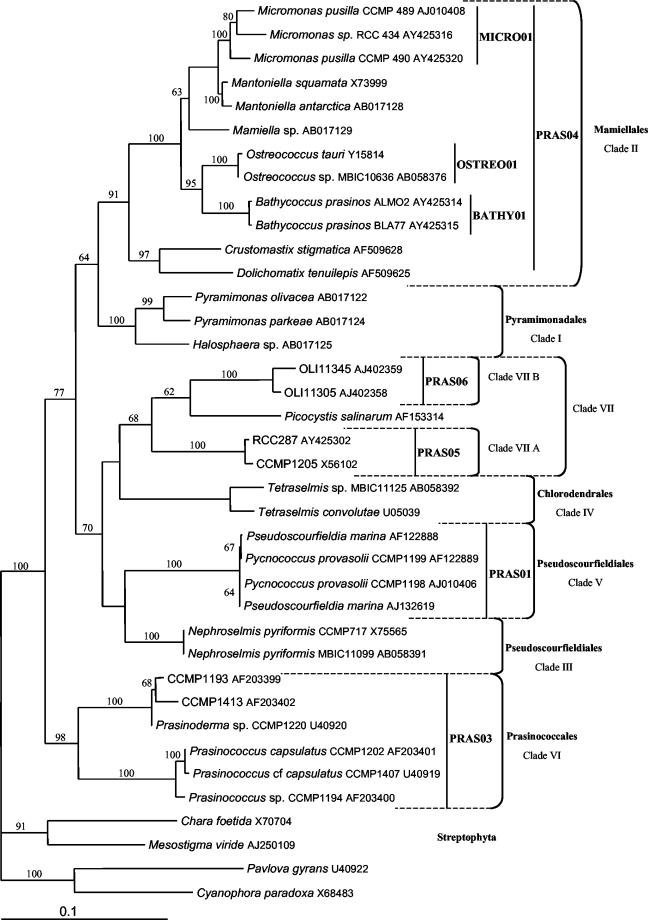
Design and specificity tests of 18S ribosomal DNA (rDNA) probes targeting Prasinophyceae. No unique characteristic exists that unites all Prasinophyceae taxa to the exclusion of other Viridiplantae or members of other algal phyla (34). This is also supported by molecular phylogenies that show the paraphyletic structure of the class Prasinophyceae (21). Therefore, it is not possible to identify a single probe targeting all Prasinophyceae species. Based on comparative 18S rRNA sequence analyses, we designed eight oligonucleotide probes (Fig. 1). The probes PRAS01, PRAS03, PRAS04, PRAS05, and PRAS06 are specific, respectively, for the orders Pseudoscourfieldiales (clade V only); Prasinococcales (clade VI); Mamiellales (clade II, with the exception of Dolichomastix); clade VIIA, containing the coccoid strain CCMP1205; and clade VIIB, composed of sequences retrieved from the Pacific Ocean. The probes OSTREO01, BATHY01, and MICRO01 are specific for the genus Ostreococcus and the species Bathycoccus prasinos and M. pusilla, respectively (Table 2).
FIG. 1.
Phylogenetic tree of the Prasinophyceae obtained by the neighbor-joining method and based on analyses of complete 18S rRNA gene sequences. The specificities of the different probes designed in this study are presented. The clades were named according to the system of Guillou et al. (8). The numbers on the branches correspond to bootstrap values (done on 1,000 replicates).
The specificities of the newly designed probes were evaluated by whole-cell hybridization of well-characterized reference strains (Table 3). Among the probes targeting Prasinophyceae orders and clades (PRAS01 to -06), only PRAS04 hybridized all, and only, target strains. Since no isolate of clade VIIB has yet been established in culture, positive controls could not be performed for PRAS06, but the probe did not show any nonspecific labeling with the strains tested. PRAS01, PRAS03, and PRAS05 hybridized only target strains, but not all of them (Table 3). We encountered two different problems. First, PRAS01 conferred a good fluorescence signal when hybridized with the two strains RCC 261 (Pseudoscourfieldia marina) and RCC 253 (Pycnococcus sp.), but no signal was observed with the strain RCC 135 (Pycnococcus provasolii). Second, RCC 369 (CCMP1205) and RCC 287, targeted by PRAS05, and Prasinoderma strain RCC 137, targeted by PRAS03, presented a heterogeneous signal after hybridization: only ∼50% of the cells showed a positive signal. The problem encountered with these probes was also observed with the general Chlorophyta probe CHLO02 with the same strains. The three genus- and species-specific probes (OSTREO01, BATHY01, and MICRO01) labeled all, and only, the taxa for which they were designed (Table 4).
TABLE 3.
Specificity tests of the oligonucleotide probes for Prasinophyceae order level clades
| Species | RCC no. | Specificitya
|
|||||
|---|---|---|---|---|---|---|---|
| PRAS01 | PRAS03 | PRAS04 | PRAS05 | PRAS06 | CHLO02 | ||
| M. pusilla | 114 | − | − | + | − | − | + |
| Ostreococcus tauri | 116 | − | − | + | − | − | + |
| Mantoniella squamata | 417 | − | − | + | − | − | + |
| B. prasinos | 113 | − | − | + | − | − | + |
| Coccoid strain | 369 | − | − | − | ± | − | ± |
| Coccoid strain | 287 | − | − | − | ± | − | ± |
| P. marina | 261 | + | − | − | − | − | + |
| P. provasolii | 135 | − | − | − | − | − | − |
| Pycnococcus sp. | 253 | + | − | − | − | − | + |
| Prasinoderma sp. | 137 | − | ± | − | − | − | ± |
| Prasinococcus sp. | 134 | − | + | − | − | − | + |
| P. capsulatus | 136 | − | + | − | − | − | + |
Boldface indicates the theoretical specificity, while + and − indicate the results of in situ hybridization tests (+, bright fluorescent signal; −, no detectable fluorescent signal; ±, weak fluorescent signal).
TABLE 4.
Specificities of oligonucleotide probes for Mamiellales genera and species
| Species | RCC no. | Specificitya
|
||
|---|---|---|---|---|
| BATHY01 | OSTREO01 | MICRO01 | ||
| Ostreococcus tauri | RCC 116 | − | + | − |
| Ostreococcus sp. | RCC 143 | − | + | − |
| Ostreococcus sp. | RCC 141 | − | + | − |
| B. prasinos | RCC 113 | + | − | − |
| M. pusilla | RCC 114 | − | − | + |
| M. pusilla | RCC 299 | − | − | + |
| M. pusilla | RCC 373 | − | − | + |
| M. pusilla | RCC 372 | − | − | + |
See Table 3, footnote a, for explanation of symbols.
Hydrological conditions at ASTAN station.
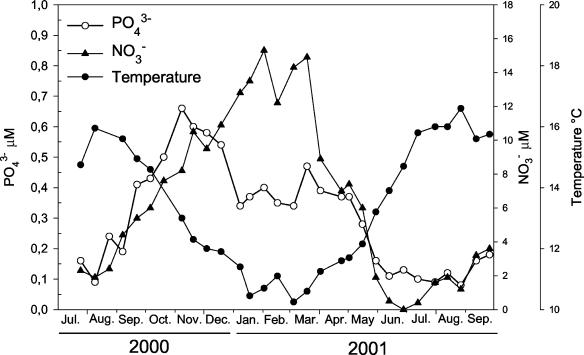
Due to the strong tidal mixing, the coastal waters off Roscoff are permanently mixed all year round (32). During the sampling period, the temperature varied between 10.2 (March 2001) and 16.6°C (August 2001). The nitrate and phosphate concentrations followed a similar pattern of variation, with minima in summer (0.4 and 0.10 μM, respectively) and maxima in late fall and winter (15.3 and 0.66 μM, respectively) (Fig. 2).
FIG. 2.
Variations in temperature and phosphate and nitrate concentrations at the ASTAN station between July 2000 and September 2001.
Photosynthetic pigments.
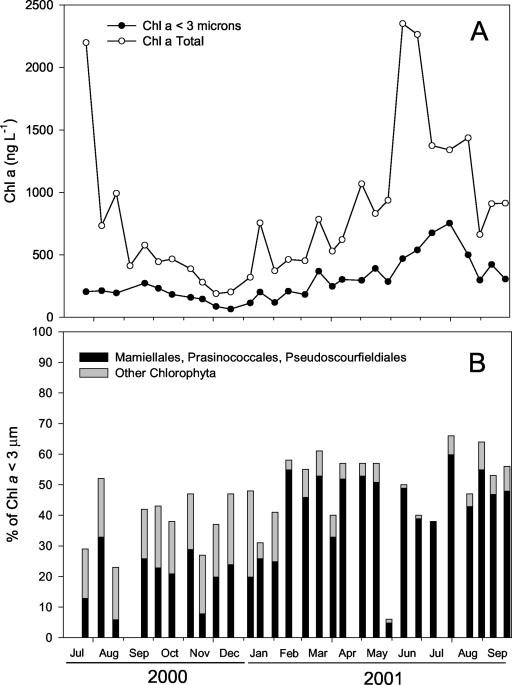
During the period studied, Chl a concentrations in the 0.2- to 200-μm fraction (total phytoplankton) varied according to the classical pattern observed in this area, with minima in winter (0.2 μg liter−1) and maxima in summer (2.5 μg liter−1), corresponding to the diatom bloom (32). The 2001 bloom occurred in late June (Fig. 3A). Maximum concentrations of picoplanktonic Chl a occurred later, at the end of July (753 ng liter−1). Large phytoplankton (>3-μm diameter) and picoplankton (<3-μm diameter) contributed approximately equally to the Chl a biomass under nonbloom conditions. During bloom periods (July 2000 and July 2001), large phytoplankton dominated the total Chl a biomass. Overall, picophytoplankton made up 34% of the total Chl a throughout the study period.
FIG. 3.
(A) Variations in Chl a biomass as measured by HPLC (total <200-μm and fraction <3-μm) at the ASTAN station. (B) Contributions of the division Chlorophyta and of the orders Mamiellales, Prasinococcales, and Pseudoscourfieldiales to the picoplankton fraction (<3-μm diameter) according to the CHEMTAX algorithm applied to HPLC data.
Within the picoplanktonic Chl a fraction, the contribution of the division Chlorophyta was almost always >25% (45%, on average) (Fig. 3B), except at the end of May, when it plummeted below 10%, although picoplanktonic Chl a decreased only slightly (Fig. 3A). Within the Chlorophyta, the contributions of the Mamiellales, Prasinococcales, and some of the Pseudoscourfieldiales (containing prasinoxanthin) to picoplanktonic Chl a, as estimated by CHEMTAX, were significant all year round and largely dominant (>80 and up to 100%) during spring and summer 2001.
Overall composition of the picoeukaryotic community.
The abundances of the photosynthetic picoeukaryotes determined by flow cytometry varied between 1 × 103 and 2 × 104 cells ml−1. Maximum abundances were recorded in May and July 2001 (Fig. 4A). The abundance of picoeukaryotic cells detected by epifluorescence microscopy after in situ hybridization with a combination of the probes EUK1209R, CHLO01, and NCHLO01 varied between 1.3 × 103 and 1.35 × 104 cells ml−1 (Fig. 4A). Cells belonging to the division Chlorophyta (targeted by CHLO02) dominated the picoeukaryotic community all year long (1 × 103 to 1.5 × 104 cells ml−1; 85%, on average, of the total number of picoeukaryotes) (Fig. 4A).
FIG. 4.
Abundances (number of cells ml−1) of photosynthetic picoplankton off Roscoff (western English Channel) between July 2000 and September 2001. The average percentages over the time series of the different groups are represented in pie charts. (A) Picoeukaryotic photosynthetic cells detected by flow cytometry counts (Flow Cytometry); picoeukaryotic cells targeted by the mix of the general probes EUK1209R, CHLO01, and NCHLO01 (EUK1209R+CHLO01+NCHLO01); and cells belonging to the division Chlorophyta detected by the probe CHLO02 (CHLO02). (B) Cells targeted by the probe CHLO02 and cells detected by the probes specific for the clades PRAS01, PRAS03, PRAS04, PRAS05, and PRAS06. (C) Cells targeted by the probe specific for Mamiellales (PRAS04) and cells detected by the probes (MICRO01, BATHY01, and OSTREO01) specific for the species and genera.
Within the Chlorophyta, organisms belonging to the order Mamiellales (Prasinophyceae), targeted by the probe PRAS04, dominated year round (Fig. 4B). Within the Mamiellales, M. pusilla was the dominant species during the sampling period (75% of the cells detected by PRAS04, on average) (Fig. 4C) and accounted for 45% of picoplanktonic eukaryotes. The second most abundant species detected was B. prasinos, which accounted for 12% (range, 1 to 67%) of the Mamiellales and 8% of the picoplanktonic eukaryotes (Fig. 4C). Cells belonging to the genus Ostreococcus were less abundant, with an average of 3% (range, 0 to 19%) of the Mamiellales and 1.4% of the eukaryotes (Fig. 4C).
The second most abundant clade within the division Chlorophyta was the order Prasinococcales, targeted by the probe PRAS03 and accounting for an average of 3.4% (range, 0 to 34%) of the Chlorophyta cells (Fig. 4B). Cells belonging to the other clades were detected at very low abundances all year long. Probes PRAS01, PRAS05, and PRAS06 targeted an average of 0.6, 0.4, and 0.6%, respectively, of cells belonging to the division Chlorophyta (Fig. 4B). Only 16%, on average, of the cells detected by the probe specific for the division Chlorophyta were not targeted by the Prasinophyceae probes used in this study.
Seasonal variation of picoeukaryotic community.
The picoeukaryotic community exhibited a marked seasonal cycle, with minima of abundance in early winter and maxima (∼10 times the winter abundances) in midsummer. The patterns of abundance variation during the sampling period were similar for total picoplanktonic eukaryotes, photosynthetic picoeukaryotes, Chlorophyta, and Mamiellales and for the species M. pusilla. On a smaller temporal scale, three major peaks of abundance were observed in 2001, in late spring and summer (mid-May, mid-June, and the end of July) (Fig. 4A). For the Mamiellales and for M. pusilla, an additional peak was observed in late August (Fig. 4C). The other clades and genera detected in this study showed more sporadic proliferations throughout the year. The species B. prasinos was detected all year long but became more abundant during spring (1.8 × 103 cells ml−1 in March 2001) (Fig. 4C). The cells targeted by the probe PRAS03 (order Prasinococcales) reached significant abundance in early fall (1.2 × 103 cells ml−1) and late spring (600 cells ml−1). In fall 2000, they accounted for up to 34% of the cells targeted by the CHLO02 probe (Fig. 4B). Spectacular decreases in the cell abundances of all the taxa detected by our probes and in flow cytometry counts were observed on May 30, 2001.
DISCUSSION
New probes targeting Prasinophyceae.
Phylogenetic analyses of 18S rDNAs recovered directly from samples collected in different oceanic regions have revealed an unsuspected diversity within eukaryotic picoplankton (20). In most gene libraries analyzed, including libraries constructed from samples collected in the western English Channel (26), prasinophycean sequences were well represented (42). The set of probes presented in this study covers most known clades (Fig. 1) and, compared to the few previously published probes for prasinophycean taxa, have improved specificities. Indeed, PRAS04 has no mismatch with any Mamiellales sequence known to date, in contrast to PRAS02 (2), which has one mismatch with B. prasinos. The probes Micro/Manto1 and Micro/Manto2, designed in 1996 (12) for Micromonas and Mantoniella, present matches with the genus Ostreococcus and mismatches with some strains of Micromonas. They are therefore not specific for their initial targets (data not shown).
When tested against pure cultures, probes PRAS04, MICRO01, OSTREO01, and BATHY01 showed perfect specificity for their target strains and delivered a bright fluorescent signal (Tables 3 and 4). In contrast, we encountered hybridization problems with some strain-probe combinations, i.e., either cells showed no fluorescent signal and a strong red fluorescence under blue excitation or the fluorescent signal was heterogeneous. Given that the corresponding sites on the 18S rRNA sequences of the strains tested showed 100% homology with the probes, the most likely hypothesis is that the probes did not penetrate into the cells. In fact, the red chlorophyll fluorescence remaining in these strains after ethanol treatment suggests that their cell walls are probably very thick and resistant, a fact previously established for P. provasolii and Prasinoderma sp. (9), and therefore prevent probe penetration.
Analysis of the picoeukaryotic community.
The seasonal variation in microphytoplankton diversity and abundance in temperate sea waters has been well described. Off Roscoff, the microphytoplankton bloom occurs in late spring (with cell densities up to 3 × 104 cells liter−1) and is characterized by the succession of a very limited number of diatom species (such as Guinardia delicatula) that occurs at low concentration outside this period (S. Ristori, unpublished data; 32). The present study indicates that the structure and dynamics of the picoplanktonic community are totally different from the classical scheme observed for microphytoplankton. First, picophytoplankton abundance remains high all year round (1 × 103 to 2 × 104 cells ml−1). Hence, this size class contributes an average of 50% of the total Chl a biomass under nonbloom conditions. Second, both FISH and chemotaxonomic analyses show that green algae, and more precisely Prasinophyceae, dominate the community (70% of the cells and 35% of the total picoplanktonic Chl a). Chlorophytes and Prasinophyceae have been previously identified by chemotaxonomic analyses (i.e., the presence of Chl b) as major components of the nano- or picoplankton size fractions in the English Channel (3), as well as in other coastal systems, such as the Faroe-Shetland Channel (20% of total Chl a) and the Galician coast (60% of total Chl a in winter) (24, 25).
Most interestingly, FISH data allowed us to establish unambiguously that a single species, M. pusilla, is dominant all year round and shapes the dynamics of the whole picophytoplankton community. Using published values of Chl a content per cell for Micromonas (0.025 pg cell−1) (5) and cell abundances obtained by FISH, the contribution of Micromonas to the total Chl a can be estimated to range between 2 and 50% (22%, on average) of the picoplanktonic Chl a. M. pusilla has already been detected in different polar and temperate marine systems, including coastal and open-ocean regions, such as the Norwegian coast (38), Bay of Biscay (1), Arctic Sea (36), Mediterranean Sea (44), and Skagerrak (13), with abundances ranging between 1 × 102 and 9 × 103 cells ml−1. The species has also been found to be present year round in the Skagerrak and North Sea (13) and to be abundant during winter-spring in the Mediterranean Sea (39) or spring and summer in the North Atlantic and North Sea (13).
The contributions of other Mamiellales genera to the picoplanktonic community were less important. The contribution of the nonmotile picoplanktonic species B. prasinos, described from the Mediterranean Sea (6) and reported from the Atlantic and Pacific Oceans (28) and Norwegian coastal waters (6), is much more sporadic: for example, in spring 2001, it accounted for 30% of picoeukaryote cells targeted. The genus Ostreococcus was detected, but always at low abundance, off Roscoff. Prior to this study, it had been found in abundance only in a somewhat atypical ecosystem, the Thau lagoon, a shallow ecosystem used intensively for oyster culture (4). The second most abundant group within the Prasinophyceae is the order Prasinococcales, which contains species such as Prasinococcus capsulatus, which has been observed and is suspected to be significant in the western Atlantic and the western Pacific (27).
The persistence of high abundances of picophytoplankton year round could be explained by the fact that picophytoplankton is controlled by small predators with short generation times (29) that prevent any sudden cell proliferation, even when conditions are optimal. This contrasts with what happens for diatoms controlled by copepods with long generation times and complex life cycles, which are able to reduce biomass only after a delay, allowing blooms to develop. The strong dominance of M. pusilla over other species all year in the system studied is very similar to what is observed for picophytoplanktonic prokaryotes, among which single genera, such as Prochlorococcus and Synechococcus, dominate certain types of ecosystems (23). Micromonas may have a broad ability to respond to light and nutrient variations and thus, because of this large environmental spectrum, may be little affected by changes in the environment. However, during the seasonal series studied, a sharp decrease in abundance of picoeukaryotes (and M. pusilla) was recorded at the end of May. This phenomenon was associated with a major peak in the ammonium concentration (data not shown). One explanation for this decrease could be that Micromonas was infected by viruses. Previous studies have suggested that viruses infecting M. pusilla have a profound impact on populations of the species in natural systems (44). Most surprising is the ability of Micromonas populations to recover rapidly to their initial levels only 15 days after the decrease. This upturn could be due to the proliferation of a different Micromonas genotype, resistant to the virus responsible for the decay. Indeed, a recent study based on the analysis of 18S rDNA gene sequences demonstrated the presence of three clades within the species M. pusilla (8).
Our results suggest that the taxonomic structure of the picoplanktonic communities is different from the structure of the microphytoplanktonic community, with one species (M. pusilla) being dominant all year round off Roscoff. Hence, if the diatoms, dinoflagellates, and coccolithophorids (i.e., Chl c-containing lineages) dominate the large eukaryotic phototrophic assemblages in the contemporary oceans (7), a few species of the green (Chl b-containing) lineage may dominate picoplankton and shape the marine microbial food webs. The next step will be to extend these observations to other coastal and oceanic areas in order to determine how widespread M. pusilla dominance is and to understand the factors that regulate the diversity and abundance of picoeukaryotes.
Acknowledgments
We thank Isabelle Biegala and Carlos Pedrós-Alió for constructive discussions, Laure Guillou and Erwan Corre for help with phylogenetic analyses, the MYSIS crew for efficient assistance with sampling, and Florence Le Gall for help with phytoplankton cultures.
This work was funded by the following programs: PICODIV, supported by the European Union (EVK3-CT-1999-00021); PICMANCH, supported by the Région Bretagne; and BIOSOPE, supported by PROOF (CNRS). F.N. was supported by a doctoral fellowship from the French Research Ministry.
REFERENCES
- 1.Ansotegui, A., A. Sarobe, J. M. Trigueros, I. Urrutxurtu, and E. Orive. 2003. Size distribution of algal pigments and phytoplankton assemblages in a coastal-estuarine environment: contribution of small eukaryotic algae. J. Plankton Res. 25:341-355. [Google Scholar]
- 2.Biegala, I. C., F. Not, D. Vaulot, and N. Simon. 2003. Quantitative assessment of picoeukaryotes in the natural environment using taxon-specific oligonucleotide probes in association with tyramide signal amplification-fluorescent in situ hybridization and flow cytometry. Appl. Environ. Microbiol. 69:5519-5529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Breton, E., C. Brunet, B. Sautour, and J. M. Brylinski. 2000. Annual variations of phytoplankton biomass in the Eastern English Channel: comparison by pigment signatures and microscopic counts. J. Plankton Res. 22:1423-1440. [Google Scholar]
- 4.Chrétiennot-Dinet, M. J., and C. Courties. 1997. Biodiversity of unicellular algae: example of pico and ultraplanktonic eucaryotes of the Thau lagoon. Vie Milieu 47:317-324. [Google Scholar]
- 4b.Courties, C., A. Vaquer, M. Trousselier, J. Lautier, M.-J. Chrétiennot-Dinet, J. Neveux, C. Machado, and H. Claustre.1994. Smallest eukaryotic organism. Nature 370:255.
- 5.DuRand, M. D., R. E. Green, H. M. Sosik, and R. J. Olson. 2002. Diel variations in optical properties of Micromonas pusilla (Prasinophyceae). J. Phycol. 38:1132-1142. [Google Scholar]
- 6.Eikrem, W., and J. Throndsen. 1990. The ultrastructure of Bathycoccus gen. nov. and Bathycoccus prasinos sp. nov., a non-motile picoplanktonic alga (Chlorophyta, Prasinophyceae) from the Mediterranean and Atlantic. Phycologia 29:344-350. [Google Scholar]
- 7.Grzebyk, D., O. Schofield, C. Vetriani, and P. G. Falkowski. 2003. The Mesozoic radiation of eukaryotic algae: the portable plastid hypothesis. J. Phycol. 39:259-267. [Google Scholar]
- 8.Guillou, L., W. Eikrem, M.-J. Chrétiennot-Dinet, F. Le Gall, R. Massana, K. Romari, C. Pedros-Aliò, and D. Vaulot. Diversity of picoplanktonic Prasinophytes assessed by direct nuclear SSU rDNA of environmental samples and novel isolates retrieved from oceanic and coastal marine ecosystems. Protist, in press. [DOI] [PubMed]
- 9.Hasegawa, T., H. Miyashita, M. Kawachi, H. Ikemoto, N. Kurano, S. Miyachi, and M. Chihara. 1996. Prasinoderma coloniale gen. et sp. nov., a new pelagic coccoid prasinophyte from the western Pacific ocean. Phycologia 35:170-176. [Google Scholar]
- 10.Joint, I. R., N. J. P. Owen, and A. J. Pomroy. 1986. Seasonal production of photosynthetic picoplankton and nanoplankton in the Celtic sea. Mar. Ecol. Prog. Ser. 28:251-258. [Google Scholar]
- 11.Keller, M. D., R. C. Selvin, W. Claus, and R. R. L. Guillard. 1987. Media for the culture of oceanic ultraphytoplankton. J. Phycol. 23:633-638. [Google Scholar]
- 12.Knauber, D. C., E. S. Berry, and M. W. Fawley. 1996. Ribosomal RNA-based oligonucleotide probes to identify marine green ultraphytoplankton. J. Eukaryot. Microbiol. 43:89-94. [DOI] [PubMed] [Google Scholar]
- 13.Kuylenstierna, M., and B. Karlson. 1994. Seasonality and composition of pico- and nanoplanktonic cyanobacteria and protist in the Skagerrak. Bot. Mar. 37:17-33. [Google Scholar]
- 14.Latasa, M., K. van Lenning, J. L. Garrido, R. Scharek, M. Estrada, F. Rodriguez, and M. Zapata. 2001. Losses of chlorophylls and carotenoids in aqueous acetone and methanol extracts prepared for RPHPLC analysis of pigments. Chromatographia 53:385-391. [Google Scholar]
- 15.Li, W. K. W. 1994. Phytoplankton biomass and chlorophyll concentration across the North Atlantic. Sci. Mar. 58:67-79. [Google Scholar]
- 16.Ludwig, W., O. Strunk, R. Ralf Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Förster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. König, T. Liss, R. Lüβmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. P. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K. H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mackey, M. D., D. J. Mackey, H. W. Higgins, and S. W. Wright. 1996. CHEMTAX—a program for estimating class abundances from chemical markers: application to HPLC measurements of phytoplankton. Mar. Ecol. Prog. Ser. 144:265-283. [Google Scholar]
- 18.Marie, D., C. Brussaard, F. Partensky, and D. Vaulot. 1999. Flow cytometric analysis of phytoplankton, bacteria and viruses, p. 11.11.1-11.11.15. In J. W. Sons (ed.), Current protocols in cytometry. International Society for Analytical Cytology, New York, N.Y.
- 19.Moon-van der Staay, S. Y., R. De Wachter, and D. Vaulot. 2001. Oceanic 18S rDNA sequences from picoplankton reveal unsuspected eukaryotic diversity. Nature 409:607-610. [DOI] [PubMed] [Google Scholar]
- 20.Moon-van der Staay, S. Y., G. W. M. van der Staay, L. Guillou, D. Vaulot, H. Claustre, and L. K. Medlin. 2000. Abundance and diversity of Prymnesiophytes in the picoplankton community from the equatorial Pacific Ocean inferred from 18S rDNA sequences. Limnol. Oceanogr. 45:98-109. [Google Scholar]
- 21.Nakayama, T., B. Marin, H. D. Kranz, B. Surek, V. A. R. Huss, I. Inouye, and M. Melkonian. 1998. The basal position of scaly green flagellates among the green algae (Chlorophyta) is revealed by analyses of nuclear-encoded SSU rRNA sequences. Protist 149:367-380. [DOI] [PubMed] [Google Scholar]
- 22.Not, F., N. Simon, I. C. Biegala, and D. Vaulot. 2002. Application of fluorescent in situ hybridization coupled with tyramide signal amplification (FISH-TSA) to assess eukaryotic picoplankton composition. Aquat. Microb. Ecol. 28:157-166. [Google Scholar]
- 23.Partensky, F., W. R. Hess, and D. Vaulot. 1999. Prochlorococcus, a marine photosynthetic prokaryote of global significance. Microbiol. Mol. Biol. Rev. 63:106-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Riegman, R., and G. W. Kraay. 2001. Phytoplankton community structure derived from HPLC analysis of pigments in the Faroe-Shetland Channel during summer 1999: the distribution of taxonomic groups in relation to physical/chemical conditions in the photic zone. J. Plankton Res. 23:191-205. [Google Scholar]
- 25.Rodriguez, F., Y. Pazos, A. Morono, J. Maneiro, and M. Zapata. 2003. Temporal variation in phytoplankton assemblages and pigment composition in a fixed station of the Ria of Pontevedra (NW Spain). Estuarine Coastal and Shelf Science 58:499-515. [Google Scholar]
- 26.Romari, K., and D. Vaulot. 2004. Composition and temporal variability of picoeukaryote communities at a coastal site of the English Channel from 18S rDNA sequences. Limnol. Oceanogr. 49:784-798.
- 27.Sieburth, J. M., M. D. Keller, P. W. Johnson, and S. M. Myklestad. 1999. Widespread occurrence of the oceanic ultraplankter, Prasinococcus capsulatus (Prasinophyceae), the diagnostic “Golgi-decapore complex” and the newly described polysaccharide “capsulan.” J. Phycol. 35:1032-1043. [Google Scholar]
- 28.Silver, M. W., M. M. Gowing, and P. J. Davoll. 1986. The association of photosynthetic picoplankton and ultraplankton with pelagic detritus through the water column (0-200m). Can. Bull. Aquat. Sci. 214:311-341.
- 29.Simek, K., P. Hartman, J. Nedoma, J. Pernthaler, D. Springmann, J. Vrba, and R. Psenner. 1997. Community structure, picoplankton grazing and zooplankton control of heterotrophic nanoflagellates in a eutrophic reservoir during the summer phytoplankton bloom. Aquat. Microb. Ecol. 12:49-63. [Google Scholar]
- 30.Simon, N., L. Campbell, E. Ornolfsdottir, R. Groben, L. Guillou, M. Lange, and L. K. Medlin. 2000. Oligonucleotide probes for the identification of three algal groups by dot blot and fluorescent whole-cell hybridization. J. Eukaryot. Microbiol. 47:76-84. [DOI] [PubMed] [Google Scholar]
- 31.Sournia, A., and J.-L. Birrien. 1995. La série océanographique côti[grave]ere de Roscoff (Manche occidentale) de 1985 à 1992. Cahier Biol. Mar. 36:1-8. [Google Scholar]
- 32.Sournia, A., J.-L. Birrien, J.-L. Douville, B. Klein, and M. Viollier. 1987. A daily study of the diatom spring bloom at Roscoff (France) in 1985. I. The spring bloom within the annual cycle. Estuarine Coastal and Shelf Science 25:355-367. [Google Scholar]
- 33.Swofford, D. L. 2002. PAUP*: phylogenetic analysis using parsimony (*and other methods), version 4 beta10 ed. Sinauer Associates, Sunderland, Mass.
- 34.Sym, S., and R. Pienaar. 1993. The class Prasinophyceae, p. 281-376. In F. Round and D. Chapman (ed.), Progress in phycological research. Biopress Ltd., Bristol, United Kingdom.
- 35.Thomsen, H. A., and K. R. Buck. 1998. Nanoflagellates of the central California waters: taxonomy, biogeography and abundance of primitive, green flagellates (Pedinophyceae, Prasinophyceae). Deep Sea Res. II 45:1687-1707. [Google Scholar]
- 36.Throndsen, J. 1970. Flagellates from Arctic waters. Nytt Magasin for Botanykk 17:49-57. [Google Scholar]
- 37.Throndsen, J. 1969. Flagellates of Norwegian coastal waters. Nytt Magasin for Botanykk 16:161-214. [Google Scholar]
- 38.Throndsen, J. 1976. Occurrence and productivity of small marine flagellates. Nor. J. Bot. 23:269-293. [Google Scholar]
- 39.Throndsen, J., and A. Zingone. 1994. Micromonads of the Mediterranean sea. G. Bot. Ital. 128:1031-1044. [Google Scholar]
- 40.Urdea, M. S., B. D. Warner, J. A. Running, M. Stempien, J. Clyne, and T. Horn. 1988. A comparison of non-radioisotopic hybridization assay methods using fluorescent, chemiluminescent, and enzyme labeled oligodeoxyribonucleotide probes. Nucleic Acids Res. 16:4937-4956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vaulot, D., F. Le Gall, D. Marie, L. Guillou, and F. Partensky. The Roscoff Culture Collection (RCC): a collection dedicated to marine picoplankton. Nova Hedwigia, in press.
- 42.Vaulot, D., K. Romari, and F. Not. 2002. Are autotrophs less diverse than heterotrophs in marine picoplankton? Trends Microbiol. 10:266-267. [DOI] [PubMed] [Google Scholar]
- 43.Zapata, M., F. Rodriguez, and J. L. Garrido. 2000. Separation of chlorophylls and carotenoids from marine phytoplankton: a new HPLC method using a reversed phase C-8 column and pyridine-containing mobile phases. Mar. Ecol. Prog. Ser. 195:29-45. [Google Scholar]
- 44.Zingone, A., D. Sarno, and G. Forlani. 1999. Seasonal dynamics in the abundance of Micromonas pusilla (Prasinophyceae) and its viruses in the Gulf of Naples (Mediterranean Sea). J. Plankton Res. 21:2143-2159. [Google Scholar]