Abstract
Quorum-sensing (QS) signals (N-acyl homoserine lactones [AHLs]) were extracted and detected from five commercially produced vacuum-packed meat samples. Ninety-six AHL-producing bacteria were isolated, and 92 were identified as Enterobacteriaceae. Hafnia alvei was the most commonly identified AHL-producing bacterium. Thin-layer chromatographic profiles of supernatants from six H. alvei isolates and of extracts from spoiling meat revealed that the major AHL species had an Rf value and shape similar to N-3-oxo-hexanoyl homoserine lactone (OHHL). Liquid chromatography-mass spectrometry (MS) (high-resolution MS) analysis confirmed the presence of OHHL in pure cultures of H. alvei. Vacuum-packed meat spoiled at the same rate when inoculated with the H. alvei wild type compared to a corresponding AHL-lacking mutant. Addition of specific QS inhibitors to the AHL-producing H. alvei inoculated in meat or to naturally contaminated meat did not influence the spoilage of vacuum-packed meat. An extracellular protein of approximately 20 kDa produced by the H. alvei wild-type was not produced by the AHL-negative mutant but was restored in the mutant when complemented by OHHL, thus indicating that AHLs do have a regulatory role in H. alvei. Coinoculation of H. alvei wild-type with an AHL-deficient Serratia proteamaculans B5a, in which protease secretion is QS regulated, caused spoilage of liquid milk. By contrast, coinoculation of AHL-negative strains of H. alvei and S. proteamaculans B5a did not cause spoilage. In conclusion, AHL and AHL-producing bacteria are present in vacuum-packed meat during storage and spoilage, but AHL does not appear to influence the spoilage of this particular type of conserved meat. Our data indicate that AHL-producing H. alvei may induce food quality-relevant phenotypes in other bacterial species in the same environment. H. alvei may thus influence spoilage of food products in which Enterobacteriaceae participate in the spoilage process.
Microbial growth and activity is well supported by fresh meat, and the quality of chill-stored meat products deteriorates due to microbial activity. Factors such as temperature, gaseous atmosphere, and pH affect the shelf life of fresh meat. The microbiota of aerobically packed meat stored at chill temperatures is dominated by Pseudomonas spp., which reach a level of 109 CFU/g at the point of spoilage. The expected shelf life is in the range of days (15, 24, 36, 42). At the time of rejection the typical off odor is putrid and slime is visible at the surface (15). The shelf life of vacuum-packed fresh meat products stored at chill temperatures is extended to weeks or months and the microbiota typically consists of lactic acid bacteria (LAB) and Enterobacteriaceae at levels of 108 and 106 CFU/g, respectively (6, 40, 50, 53, 57). Spoilage off-odors are characterized as dense, sour, and slightly putrid (8).
Meat is a high-value food product, and by elucidating the spoilage mechanisms of fresh meat products it may be possible to control and predict the shelf life. Knowledge of the dominating microbial species is not always sufficient to identify the bacteria that produce the off odors and off flavors (17). To identify the specific spoilage bacteria one must compare sensory, microbiological, and chemical characteristics of naturally spoiling foods and inoculated trials (28). In aerobically packed meat there is a direct relationship between the numbers of Pseudomonas spp. and the degree of spoilage (17). In vacuum-packed meat products, however, it is believed that the spoilage arises from a metabiotic interaction between LAB (the dominating flora) and Enterobacteriaceae (found in lower numbers) (8, 16, 20). Normally, a level of 108 to 109 CFU/g of a spoilage microorganism has to be present for sensory changes to be detectable. The level of Enterobacteriaceae in vacuum-packed meat is in the range of 105 to 107 CFU/g and their contribution to the spoilage process therefore seems speculative. Hafnia alvei and Serratia spp. dominate the Enterobacteriaceae community in vacuum-packed spoiled meat (8, 46). These organisms are often capable of producing N-acyl homoserine lactones (AHLs) (27, 45, 54), which gram-negative bacteria use as signal molecules in quorum-sensing (QS)-regulated control of gene expression. Bacterial cells use the signal molecules to sense their population density and trigger a coordinated expression of particular genes when the density reaches a threshold level. Typically, QS results in an up-regulating of specific phenotypes (56). In food-relevant Enterobacteriaceae, QS controls production of exoenzymes in Serratia proteamaculans (12), Serratia liquefaciens (47), and Erwinia carotovora (44), the swarming ability in S. liquefaciens (18), antibiotic production in E. carotovora (4), and the motility and morphology in Yersinia pseudotuberculosis (3, 19). Some of these AHL-controlled phenotypes may affect the quality of food products (1, 12; M. Rasch, J. B. Andersen, L. R. Floodgard, K. F. Nielsen, M. Givskov, and L. Gram, unpublished data).
On this background it can be hypothesized that members of Enterobacteriaceae contribute to the spoilage of vacuum-packed meat through their ability to use QS systems. Due to uneven distribution and local fluctuations of the concentrations of bacterial cells, QS systems could allow bacteria, present at an average concentration of 106 CFU/g, locally to induce expression of phenotypes that would otherwise require a average of value of 108 CFU/g. Jay et al. (33) recently suggested an involvement of QS in meat spoilage. They hypothesized that the slime formed on aerobically packed meat is a biofilm formed by the Pseudomonas spp. If QS is involved in regulation of phenotypes that are associated with meat spoilage, the application of new food preservatives such as QS inhibitors (QSI), which specifically block AHL regulation (26, 30), can become an area of interest.
The purpose of the present work was to elucidate if bacterial signals (AHLs) and AHL-dependent regulation contribute to the spoilage of fresh meat. We have therefore assessed the presence of AHLs in fresh and spoiled meat as well as the presence and level of AHL-producing bacteria. The effect of AHL and AHL-producing bacteria on spoilage of meat was investigated by comparing spoilage ability of a wild-type AHL-producing H. alvei and a mutant deficient in AHL production (knockout of the AHL synthetase coding gene halI). Subsequently, also the effect of specific QSI molecules (halogenated furanones) was determined.
MATERIALS AND METHODS
Purchase and storage of meat sample.
Five commercially produced chill-stored vacuum-packed roasts from beef were purchased from commercial supermarkets and stored at 5°C. One sample from each of the five roasts was aseptically withdrawn on a weekly basis. Each sample (of approximately 2 by 2 cm2) contained both surface and center of the meat, and samples were taken until the meat was rejected by a sensory panel consisting of two or three people judging odor and off-odor. Samples were analyzed for microbial counts; content of AHL and pH was measured at the start and the end of the trial. After each withdrawal the meat was vacuum-packed in plastic (0.45 cm3/m2/d/atm O2 at 95% relative humidity) and stored at 5°C. One roast, steaks, and diced and minced meat (all beef) packed in modified atmosphere (30% CO2-70%O2) were likewise purchased from commercial supermarkets. Samples were withdrawn three times a week and packed aerobically; otherwise, the conditions and sampling were as for the vacuum-packed meat.
Enumeration of bacteria.
Meat samples were diluted 1:9 with physiological saline with 0.1% peptone, homogenized in a stomacher 400 (Seward Medical, London, United Kingdom) for 2 min, and serially 10-fold diluted. Aerobic counts were made on tryptone soy agar (TSA) (CM131; Oxoid, Basingstoke, England) incubated at 25°C for 3 days. LAB were plated on Man, Rogosa, and Sharpe (MRS) agar (CM361; Oxoid) and incubated at 25°C for 3 days. Enterobacteriaceae were determined by pour plating in TSA, and after 1 h a cover of violet red bile glucose (VRBG; CM485; Oxoid) was added and plates were incubated at 25°C for 2 days. Bacteria presumably producing AHLs were enumerated by replica plating from TSA agar plates onto AHL-indicative plates containing an AHL monitor bacterium (see below). From each piece of meat, five colonies from VRBG were isolated and inoculated in LB (1% peptone, 0.5% yeast extract, 0.5% NaCl) (7) and incubated at 25°C for 24 h. Presumed AHL-producing colonies, identified based on the replica plating procedure, were isolated from the TSA master plates. These strains were also inoculated in LB and incubated at 25°C for 1 day. The LB cultures were sterile filtered (0.2-μm pore size; Sartorius Minisart) and stored at −20°C until tested for presence of AHLs in a well diffusion assay (see below). Bacterial isolates were stored frozen at −80°C in 2% (wt/vol) dried skim milk (23). AHL-producing strains were identified using standard biochemical techniques (5). AHL-producing isolates from replica plating presumed by preliminary phenotypic tests to be Enterobacteriaceae were identified by API E20 biochemical strips (bioMerieux, Marcy l'Etoile, France). To confirm the API E20 identification, five isolates were identified by partial 16S rRNA gene sequencing by Deutsche Sammlung von Mikroorganismen und Zellkulturen. The results were analyzed by multiple alignment comparing the 16S rRNA gene sequences with representative 16S rRNA gene sequences of organisms belonging to the Enterobacteriaceae.
Screening for AHL-producing bacteria by replica plating.
Agrobacterium tumefaciens NT1(pZLR4) (11) and Chromobacterium violaceum CV026 (55) were used for the detection of AHLs in replica plating, in well diffusion assays, and by thin-layer chromatography (TLC). A preculture of the monitor bacteria was grown in LB for 24 h at 25°C with shaking at 200 rpm for aeration. One milliliter of the A. tumefaciens preculture was inoculated in 50 ml of ABT medium [ABT contains, per liter, 0.4 g of (NH4)2SO4, 0.6 g of Na2HPO4, 0.3 g of KH2PO4, 0.3 g of NaCl, 1 mM MgCl2, 0.1 mM CaCl2, 0.01 mM FeCl3, 2.5 mg of thiamine supplemented with 0.5% glucose (Merck, 1.08337), and 0.5% Casamino Acids (Difco, 0230-17-3)] (13) and 0.1 ml of C. violaceum preculture was inoculated in 50 ml of LB. The cultures were grown for 24 h at 25°C with aeration and were mixed with 100 ml of ABT-agar (A. tumefaciens) or 100 ml of LB agar (C. violaceum). Both agars contained 1.2% agar and were kept at 46°C. The agar was immediately poured into five 8.5-cm-diameter petri dishes. Colonies from a TSA dish were copied using a Whatman no. 4 filter (catalog no. 1004 150; Whatman, Maidstone, England) to the replica dish and the plates were incubated at 25°C for 24 h before positive colonies were identified by color change in the surrounding agar.
Screening meat extracts and bacterial supernatants for AHLs.
Meat ethyl acetate extracts were prepared (9), and bacterial supernatants were obtained by sterile filtering (pore size, 0.22 μm) bacterial cultures. Bacterial supernatants or ethyl acetate extracts were tested for possible presence of AHL in a well diffusion assay with A. tumefaciens or C. violaceum as monitor bacteria (45). AHLs in extracts and supernatants were profiled by separating the AHLs using reverse-phase C18 TLC and subsequently developing the plates using an agar containing an AHL monitor strain (45, 51).
LC-MS analysis of bacterial supernatants.
Ethyl acetate extracts of 1 or 50 ml of culture supernatant were evaporated in vacuo, redissolved in ethanol, and analyzed by liquid chromatography-positive-electrospray high-resolution mass spectrometry (LC-MS) on an LCT orthogonal time-of-flight mass spectrometer (Micromass Manchester, United Kingdom) as described by Nielsen and Smedsgaard (41). The column was changed to a Phenomenex (Torrance, Calif.) Luna II C18 column (particle size, 3 μm; 50 by 2 mm) with a Phenomenex 2-mm C18 SecurityGuard precolumn. Separation was performed with water (MilliQ) containing 10 mM ammonium formiate and 20 mM formic acid (both analytical grade) and acetonitrile (AcN) (gradient grade) containing 20 mM formic acid. A flow of 0.3 ml/min was used, starting with 5% AcN going to 100% in 25 min and then holding 3 min, before returning to 5% AcN in 4 min and then equilibrating 7 min.
Construction of H. alvei 718 halI mutant.
H. alvei 718, which was isolated from vacuum-packed meat (8), was used as model organism in meat spoilage experiments. An AHL-lacking mutant of H. alvei 718 was constructed in a similar way to the construction of an AHL-deficient mutant of S. proteamaculans B5a described by Christensen et al. (12). In brief, a bank of random insertion mutants was made by using the mini-Tn5 transposon delivery system described by Herrero et al. (31). The mutant bank was transferred onto selective LB plates containing nalidixic acid (25 μg/ml) and kanamycin (25 μg/ml) and incubated for 12 h at 25°C. Subsequently, the mutant bank was replica plated onto indicative LB plates containing approximately 1% (vol/vol) outgrown culture of the monitor bacterium C. violaceum CV026 (see “Screening for AHL-producing bacteria by replica plating” above). Putative AHL-lacking clones were unable to induce violacin production after 12 h of incubation at 25°C.
Extraction of chromosomal DNA.
Chromosomal DNA was extracted as described by Givskov et al. (25).
Southern blotting.
Southern blot analyses were performed as previously described (25). Chromosomal DNA of the H. alvei 718 halI mutant was digested with KpnI and PstI, and a digoxigenin-labeled kanamycin resistance gene was used as probes against H. alvei 718.
DNA manipulations.
Standard techniques for DNA manipulations were used (49). pAC35 was constructed as follows. Chromosomal DNA of H. alvei 718 halI was digested with BglII and ligated into the BamHI site of pLOW1. Kanamycin-resistant Escherichia coli MT102 clones containing pAC35 were carrying a DNA fragment of approximately 12 kb.
DNA nucleotide sequencing.
Sequencing was performed at a commercial sequencing facility (K. J. Ross-Petersen AS, Holte, Denmark). The oligonucleotide used as sequencing primer for sequencing downstream of the kanamycin resistance gene insertion was AC31/o-end: 5′-CACTTGTGTATAAGAGTCAG. The template used for sequencing was pAC35.
Kinetics of AHL production in H. alvei 718.
H. alvei 718 AHL production in ABT medium was studied at low (5°C) or high (25°C) temperatures. Cultures at 25°C were grown with aeration, whereas cultures at 5°C were grown both aerated and under static conditions. H. alvei 718 was precultured at 10 or 25°C and inoculated at approximately 106 CFU/g in ABT. Growth was monitored by optical density at 450 nm (OD450). The AHL production was measured, in a well diffusion assay with A. tumefaciens NT1(pZLR4), by testing sterile filtered samples and comparing the size of the induction zone to sizes caused by known concentrations of N-3-oxo-hexanoyl-homoserine lactone (OHHL) (catalog no. K3007; Sigma). Growth and possible AHL production of H. alvei strain 718 halI mutant were also investigated.
Effect of AHL or QSIs (furanone C-30) on meat spoilage.
The potential growth-inhibitory effect of furanone C-30 on H. alvei 718 was tested by adding various concentrations (0.1, 1, and 10 μM) of C-30 to freshly inoculated cultures at an initial cell density of 0.01 (OD450) in ABT. The samples were incubated at 25°C, and the OD450 was measured on an hourly basis. Fresh meat (beef) with a low content of fat and sinews was purchased from a slaughterhouse (Slagteriskolen, Roskilde, Denmark). One part of the meat was surface sterilized by submersion in boiling water for 2 min, and dried in a flow cabinet for 15 min. The surface was removed with a sterile knife. Pieces of approximately 15 g were inoculated with a bacterial suspension (10 μl/g) of a preculture grown in LB at 10°C, yielding an initial level of, per gram, about 104 CFU of H. alvei 718 or H. alvei 718 halI mutant. One set of samples inoculated with H. alvei 718 was also treated at the start of the storage trial with furanone C-30 at a final concentration of 1 μmol of furanone C-30 per kg of meat. The remaining (nonsterilized) part of the meat was cut in pieces of approximately 15 g. Half of these were treated with furanone C-30 at a concentration of 1 μM. Half of the nontreated meat pieces were used as control samples. The meat pieces were placed in petri dishes (diameter, 4.5 cm), individually vacuum-packed in plastic (0.45 cm3/m2/d/atm O2 at 95% relative humidity), and stored at 5°C. Changes in bacterial densities and AHL content were determined as describe earlier. A sensory evaluation was performed by five laboratory workers giving the meat samples grades from 1 (no off-odor) to 5 (extreme off-odor). Fresh meat stored frozen and thawed just before evaluation was used as a reference.
SDS-PAGE of extracellular H. alvei 718 extracts.
H. alvei 718, the H. alvei 718 halI mutant, and the H. alvei 718 halI mutant with 10 μM OHHL were grown in ABT for 24 h at 25°C with aeration. OD450 was measured after 24 h and the cells were harvested by centrifugation (10,000 × g for 10 min). The supernatants were sterile filtered, and aliquots of 100 μl were stored at −20°C until analysis. One hundred microliters of sample buffer was added to each sample (1.2 ml of 0.5 M Tris-HCl [pH 6.8], 1 ml of glycerol, 2 ml of 10% sodium dodecyl sulfate [SDS], 25 μl of mercaptoethanol, 0.5 ml of 0.1% bromophenol blue, and 4.8 ml of Millipore water) and treated for 5 min in boiling water. SDS-polyacrylamide gel electrophoresis (PAGE) was carried out on a slab gel apparatus (SE 600; Hoefer Scientific Instruments, San Francisco, Calif.) as described by Laemmli (35). The amounts loaded were adjusted according to the OD450 of the growing bacterial cultures. Samples were electrophoresed in a 5% stacking gel and a 15% separation gel with 0.1% SDS at a current of 20 mA for 90 min for the stacking gel and 40 mA for 210 min for the separation gels. The temperature during electrophoresis was kept at 15°C by a thermal recirculation unit. Gels were silver stained and air dried between two sheets of cellophane.
AHL-mediated interactions between H. alvei 718 and other Enterobacteriaceae.
H. alvei 718, the H. alvei 718 halI mutant, S. proteamaculans B5a, and the S. proteamaculans B5a sprI mutant (12) were grown separately in LB for 24 h at 25°C. The cultures were inoculated individually in commercially produced skim milk at approximately 104 CFU/ml. Furthermore, milk was inoculated with combinations of the S. proteamaculans B5a sprI mutant and H. alvei 718 or the H. alvei 718 halI mutant. All samples were incubated at 5°C for 3 to 4 weeks. Bacterial counts were performed on casein agar (10% skim milk, 1.2% agar), where the protease-negative H. alvei and the protease-deficient S. proteamaculans B5a sprI mutant appear without clearing zones around the colonies, whereas S. proteamaculans B5a caused clearing zones around the colonies. When enumeration of the S. proteamaculans B5a sprI mutant together with H. alvei bacteria was carried out, 1 μM OHHL was added to the agar to allow the S. proteamaculans B5a sprI mutant strain to secrete protease.
Nucleotide sequence accession number.
The full 16S rRNA gene sequence of H. alvei 718 has been submitted to GenBank under the accession number AY572428.
RESULTS
Presence of AHL and AHL-producing bacteria in meat.
Off- odors developed in the purchased vacuum-packed meat samples after 2 to 3 weeks and samples were rejectable after 21 to 28 days and characterized as sour or putrid (Table 1). The aerobic count (TSA) in freshly purchased vacuum-packed meat varied between 3 × 106 and 5 × 108 CFU/g. In general LAB counts (MRS agar) were at the same level as aerobic counts or 0.5 to 1 log unit lower. Counts of Enterobacteriaceae (VRBG) were typically 1 to 10% of the aerobic counts. Levels of AHL-producing bacteria (determined by replica plating from TSA) were similar to the level of Enterobacteriaceae. Bacterial counts increased to (or remained constant at) approximately 108 CFU/g, with the flora being dominated by LAB in all vacuum-packed meat samples. Ethyl acetate extracts from 22 out of 25 vacuum-packed meat samples induced the broad-range AHL-monitor A. tumefaciens NT1(pZLR4), which detects mainly 3-oxo AHLs with acyl chain lengths from 6 to 14 C atoms in a well diffusion assay, whereas none of the extracts induced the narrow-range C. violaceum CV026, which mainly detects nonsubstituted AHLs. The size of the induction zone indicative of AHL concentration in the four to five samples from a batch was almost constant throughout the storage period.
TABLE 1.
Presence of AHLs and AHL-producing bacteria in vacuum-packed meat stored at 5°C
| Sample no. | Storage time (days) | Off-odor | pH | Presence of AHLsb
|
Log CFU/g for culture group
|
||||
|---|---|---|---|---|---|---|---|---|---|
| CV026 | pZLR4 | TSA | MRS | VRBG | AHLc | ||||
| 1 | 0 | 5.5 | − | − | 3.4 × 106 | 4.6 × 105 | 5.2 × 104 | 3 × 104 | |
| 4 | − | − | 1.2 × 107 | 2.0 × 106 | 7.0 × 103 | ||||
| 7 | − | − | 5.8 × 107 | 1.1 × 107 | 2.4 × 104 | 1 × 105 | |||
| 14 | − | (+) | 3.1 × 108 | 9.6 × 106 | 9.0 × 104 | ||||
| 21 | Sour | − | + | 1.8 × 108 | 4.7 × 107 | 3.7 × 104 | |||
| 28 | Soura | 5.0 | − | + | 1.4 × 108 | 8.9 × 107 | 5.5 × 104 | ||
| 2 | 0 | 5.6 | − | ++ | 6.6 × 107 | 5.9 × 107 | 3.6 × 106 | 6 × 106 | |
| 7 | − | +++ | 3.4 × 108 | 1.3 × 108 | 3.8 × 106 | 3 × 106 | |||
| 14 | − | +++ | 2.0 × 108 | 1.7 × 108 | 4.3 × 106 | 7 × 106 | |||
| 21 | − | +++ | 2.8 × 108 | 2.1 × 108 | 1.3 × 107 | 7 × 107 | |||
| 28 | Putrida | 5.3 | − | +++ | 6.0 × 108 | 3.7 × 108 | 1.3 × 107 | 1 × 107 | |
| 3 | 0 | 5.8 | − | + | 2.8 × 106 | 2.8 × 106 | 1.1 × 104 | 1 × 104 | |
| 7 | − | (+) | 3.6 × 108 | 3.1 × 108 | 2.3 × 106 | 6 × 105 | |||
| 14 | − | + | 5.1 × 108 | 1.6 × 108 | 3.7 × 106 | 2 × 106 | |||
| 21 | Putrid | − | (+) | 2.2 × 108 | 9.2 × 107 | 6.2 × 105 | |||
| 28 | Putrida | 5.6 | − | + | 4.7 × 108 | 2.3 × 108 | 6.8 × 106 | 1 × 106 | |
| 4 | 0 | 5.3 | − | ++ | 4.8 × 108 | 1.6 × 108 | 1.9 × 106 | ||
| 7 | − | + | 4.4 × 108 | 1.8 × 108 | 1.3 × 106 | 2 × 106 | |||
| 14 | − | + | 4.5 × 108 | 1.8 × 108 | 1.7 × 106 | ||||
| 21 | Sour | − | + | 3.5 × 108 | 3.1 × 108 | 2.0 × 106 | 3 × 106 | ||
| 28 | Soura | 5.2 | − | + | 4.2 × 108 | 3.1 × 108 | 2.0 × 106 | 8 × 105 | |
| 5 | 0 | 5.7 | − | ++ | 7.8 × 107 | 1.2 × 107 | 1.1 × 107 | 4 × 106 | |
| 7 | − | + | 5.5 × 108 | 7.8 × 107 | 1.9 × 107 | 6 × 107 | |||
| 14 | Putrid | − | ++ | 1.0 × 109 | 6.0 × 108 | 1.4 × 107 | 8 × 107 | ||
| 21 | Putrida | 5.5 | − | + | 3.6 × 108 | 3.0 × 108 | 2.4 × 107 | 2 × 106 | |
Rejectable based on odor evaluation.
Estimated level (OHHL equivalents) of AHL. Symbols: −, monitor bacteria not induced; (+), monitor bacteria induced around the well; +, zone equivalent of 10−9 to 10−8 M OHHL; ++, zone equivalent of 10−8 to 10−7 M OHHL; +++, zone equivalent of 10−7 to 10−6 M OHHL.
The results for AHL-positive bacteria are based on the number of AHL-positive bacteria found by replica plating.
One hundred and ten colonies were isolated from TSA plates as assumed AHL-producing bacteria based on replica plating (Table 2). Culture supernatants from 96 of these isolates induced the A. tumefaciens NT1(pZLR4) monitor. All the 96 AHL-producing bacteria were gram-negative bacteria, and 92 of these were identified as Enterobacteriaceae. H. alvei was isolated as the most common AHL-producing bacterium (67 of 96 strains). Since the group of Enterobacteriaceae was isolated as the most common AHL-producing group, colonies isolated from VRBG were tested for their ability to induce A. tumefaciens NT1(pZLR4). A total of 146 randomly picked isolates were tested, and 83 (57%) were positive.
TABLE 2.
Identification of AHL-producing bacteria isolated from vacuum-packed meat by replica plating
| Sample no. | Storage time (days) | No. of isolates | Total no. of AHL-positive isolates | No. of isolates (AHL positive)
|
|||
|---|---|---|---|---|---|---|---|
| H. alvei | Serratia sp. | Other Enterobacteriaceae | Non-Enterobacteriaceae | ||||
| 1 | 0 | 11 | 3 | 1 | 1 | 1 | |
| 4 | 2 | ||||||
| 7 | 3 | 1 | 1 | ||||
| 14 | |||||||
| 21 | |||||||
| 28 | |||||||
| 2 | 0 | 6 | 6 | 6 | |||
| 7 | 3 | 3 | 3 | ||||
| 14 | 8 | 8 | 8 | ||||
| 21 | 18 | 18 | 17 | 1 | |||
| 28 | 14 | 14 | 14 | ||||
| 3 | 0 | 1 | 1 | 1 | |||
| 7 | 6 | 6 | 6 | ||||
| 14 | 2 | 2 | 2 | ||||
| 21 | |||||||
| 28 | 1 | 1 | 1 | ||||
| 4 | 0 | ||||||
| 7 | 2 | 2 | 2 | ||||
| 14 | |||||||
| 21 | 3 | 3 | 3 | ||||
| 28 | 9 | 8 | 4 | 4 | |||
| 5 | 0 | 5 | 4 | 1 | 2 | 1 | |
| 7 | 6 | 6 | 2 | 3 | 1 | ||
| 14 | 8 | 8 | 6 | 2 | |||
| 21 | 2 | 2 | 1 | 1 | |||
| Total | 110 | 96 | 67 | 20 | 5 | 4 | |
To confirm the API E20 identification, the partial 16S rRNA gene sequence of three H. alvei isolates (all from different samples) and two other strains of Enterobacteriaceae were analyzed. The results of the genetic analyses were in agreement with the API E20 identification (data not shown). Furthermore, a full 16S rRNA gene sequence of H. alvei 718 (8) confirmed its identity as H. alvei.
Eleven out of 20 ethyl acetate extracts from four aerobically packed meat samples induced A. tumefaciens NT1(pZLR4), while none of the extracts induced C. violaceum CV026. AHL-producing bacteria were present at a level of 0.01 to 5% of the aerobic counts (data not shown). A total of 11 AHL-producing bacteria were identified: one Pseudomonas spp. and 10 (91%) Enterobacteriaceae. None of the Enterobacteriaceae were identified as H. alvei. Culture supernatants from 25% of randomly picked isolates from VRBG induced A. tumefaciens NT1(pZLR4) in a well diffusion assay.
TLC profiles and LC-MS.
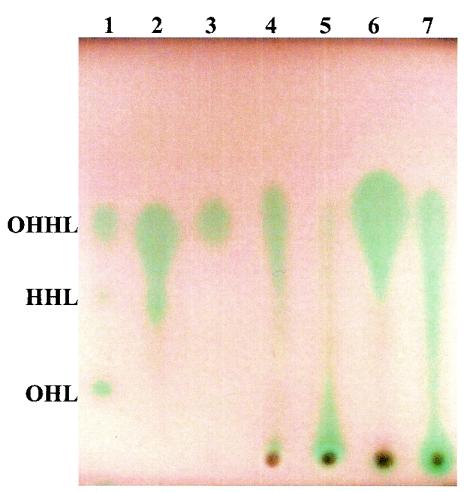
Ethyl acetate extracts from meat samples and sterile filtered culture supernatants from the isolated strains were separated by TLC. All meat extracts that were positive in the well diffusion assay caused AHL induction around the loading area and remains of extracts were clearly visible as brown stains (Fig. 1). Furthermore, a smear appeared in the profile from samples containing fat (Fig. 1, lanes 4, 5, and 7), whereas no smear appeared from samples with low fat content (Fig. 1, lane 6). A possible explanation could be lipid interference with the migration of AHL. To avoid this, meat extracts were loaded, separated by means of TLC, scraped off, and reextracted—leaving the loading stains on the primary TLC plate. A reextracted sample is shown in lane 3 of Fig. 1.
FIG. 1.
TLC profiles of meat extracts from store 2 developed with A. tumefaciens NT1(pZLR4). Lane 1, standards of OHHL, HHL (N-hexanoyl homoserine lactone), and OHL (N-octanoyl homoserine lactone); lane 2, H. alvei 718 grown in LB5; lane 3, reextracted meat extracts after 4 weeks of storage; lane 4, meat extracts after 3 weeks of storage; lane 5, meat extracts after 2 weeks of storage; lane 6, meat extracts after 1 weeks of storage; lane 7, meat extracts at the day of purchase.
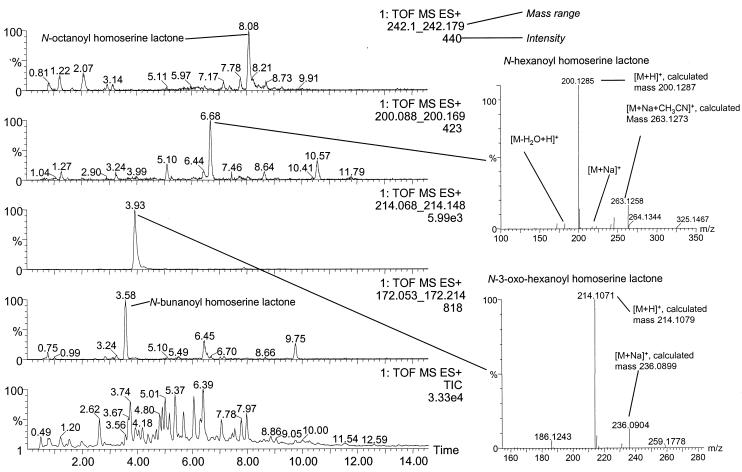
H. alvei strains isolated from different samples in the present study and H. alvei 718 isolated in a previous study (8) produced AHL with almost identical TLC profiles. Compounds with shapes and Rf values similar to those of OHHL were the major AHLs produced and were also tentatively identified in meat extracts (Fig. 1). LC-MS of ethyl acetate extracts from 50 ml of culture supernatant of H. alvei strain 718 unambiguously detected N-3-oxo-hexanoyl homoserine lactone and N-hexanoyl homoserine lactone with correct retention times (deviation < 0.01 min). They were further validated by the accurate masses of the ions not deviating more than 0.015 Da (Fig. 2). N-octanoyl homoserine lactone and N-butanoyl homoserine lactone were tentatively identified (dereplicated) based on the accurate masses (<1 mDa of deviation) of their [M+H]+ and [M+Na]+ adducts as described for fungal metabolites (41) and based on their elution order on C18 phases (39). Further, 1 ml of culture supernatant of six strains of H. alvei, including strain 718, was extracted, and LC-MS analysis identified N-3-oxo-hexanoyl homoserine lactone as the dominating molecule with traces of N-hexanoyl homoserine lactone.
FIG. 2.
High-performance liquid chromatography-positive electrospray ionization (ESI+)-MS chromatograms, with the lower-left panel showing the total ion current (TIC) chromatogram and the upper four panels at left showing extracted ion chromatograms for the [M+H]+ adducts of four detected homoserine lactones. The positive electrospray mass spectra for N-hexanoyl homoserine lactone and N-3-oxo-hexanoyl homoserine lactone are shown with the calculated masses for the predominant adducts. TOF, time of flight.
Effect of AHL production and/or QSIs on spoilage of vacuum-packed meat.
The potential influence of AHL regulation on the spoilage of vacuum-packed meat was studied from two angles: (i) by comparing spoilage of meat inoculated with an AHL-producing or an AHL-negative H. alvei mutant, and (ii) by comparing spoilage of vacuum-packed meat (either naturally contaminated or inoculated with H. alvei 718) with and without the addition of a known QS inhibitor (QSI), furanone C-30.
Construction of H. alvei 718 halI mutant.
An AHL-deficient H. alvei 718 mutant (halI) was constructed by random transposon mutagenesis as described in Materials and Methods. A Southern blot analysis confirmed that only one copy of the transposable element had been integrated into the chromosome of this mutant (data not shown). DNA sequence analysis on the downstream flanking region of the kanamycin insertion of the AHL-deficient H. alvei 718 mutant positioned the insertion approximately 450 bases downstream from a predicted translation start point, in a gene belonging to the family of luxI-homologous AHL synthases.
Growth kinetic of H. alvei 718 and H. alvei 718 halI mutant at 5 and 25°C.
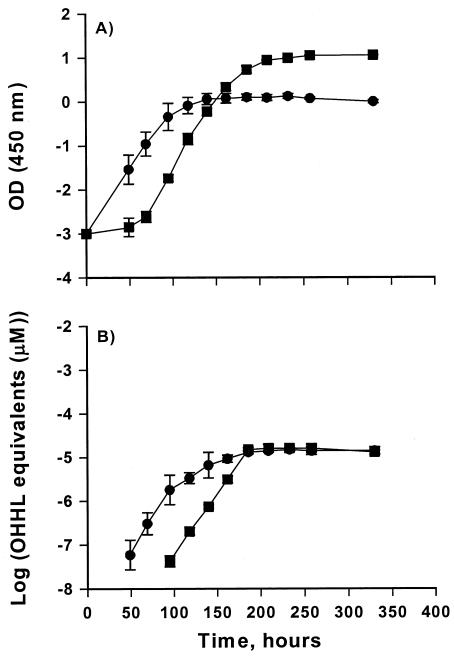
H. alvei 718 and the H. alvei 718 halI mutant grew with similar growth rates in ABT at 5°C (generation time of 24 h) and 25°C (generation time of 45 min) (results not shown). H. alvei 718 produced AHL (assumed to be OHHL) that was detectable when cell numbers reached approximately 106 CFU/g and AHL production did not appear to be up-regulated. No AHL was detected from cultures of the H. alvei 718 halI mutant when using the A. tumefaciens NT1(pZLR4) AHL monitor system. The peak concentration of AHL was approximately 10 μM independent of the maximum cell density. This difference in yield (maximum AHL/maximum cell number) was caused by differences in aeration (Fig. 3). Growth ceased in the static cultures at an OD450 of 1.2, but the AHL production continued to increase for several days, reaching a level of 10 μM. The aerated cultures reached an OD450 of 11. The differences in maximum cell density were statistically significant (P < 0.01), whereas the concentrations of AHLs did not differ significantly. Testing of diluted samples resulted in the same AHL concentration, showing that the peak in AHL concentration was not caused by saturation of the monitor system.
FIG. 3.
Growth and AHL production by H. alvei 718 at 5°C in ABT under static or aerated conditions. (A) Symbols: ▪, growth of H. alvei 718 with aeration; •, growth of H. alvei 718 under static conditions. AHL was measured by OD450. (B) Symbols: ▪, AHL production by H. alvei 718 grown under aerated conditions; •, AHL production by H. alvei 718 grown under static conditions. AHL was measured by well diffusing assay with A. tumefaciens pZLR4 as monitor bacterium.
Meat inoculation experiment.
H. alvei 718 and the corresponding AHL-deficient mutant were inoculated separately on blocks of meat excised under aseptic (albeit not sterile) conditions and stored under vacuum at 5°C. Bacterial counts (on VRBG) increased within 14 days from the inoculated 104 to 5 × 107 CFU/g with identical growth curves. AHL was not detected in control samples (not inoculated) and samples inoculated with the H. alvei 718 halI mutant. AHL was detected in samples inoculated with H. alvei 718 after 7 days when a cell density of 8 × 105 CFU/g was reached. LAB counts increased in all samples to a level of 108 CFU/g within 14 days even though no colonies were detected on MRS agar at the initial withdrawal. These results indicate that LAB were the dominating bacterial species in the control samples and that H. alvei 718, as in naturally contaminated products, grew in coculture with LAB. Five laboratory workers evaluated the off-odor of the meat samples. The grades for all samples increased through the storage period from high quality to spoiled meat. No difference was seen between spoilage of meat by H. alvei 718 and its AHL-negative mutant.
High concentrations (10 μM) of the QSI (furanone C-30) prolonged the lag phase of H. alvei 718, but lower concentrations had no effect on bacterial growth. A concentration of 1 μM furanone C-30 was used in the meat inoculation trials. No difference was detected in development of off odors between samples with or without furanone C-30 addition. Also, the naturally contaminated sample spoiled at the same rate as the treated sample.
AHL-controlled phenotype in H. alvei 718.
Bacteria equipped with AHL-based QS systems are engaged in either symbiotic or pathogenic relationships with eukaryotic hosts. The latter relationship involves the production of extracellular enzymes and other virulence factors, several of which are AHL regulated. Therefore, a range of experiments was conducted to determine a possible regulatory function of AHLs in H. alvei 718. Comparisons were made between the wild type, the AHL-negative mutant, and the OHHL-complemented AHL-negative mutant in a number of assays. In short, no difference was found between the strains in the following tests: production of antibiotics, biogenic amines, adhesion and biofilm formation, motility, starvation survival, resistance to oxidative stress, or virulence as determined using a Drosophila model (data not shown).
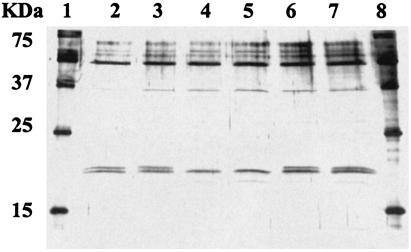
To determine if AHL had any influence on expression of extracellular proteins, SDS-PAGE profiling of H. alvei 718, the H. alvei 718 halI mutant, and the H. alvei 718 halI mutant complemented with OHHL was carried out (Fig. 4). A protein band of approximately 20 kDa was present in cultures from H. alvei 718 that was not found in cultures from the H. alvei 718 halI mutant. When the culture was complemented with OHHL, the band reappeared, indicating that its production or secretion is under AHL control.
FIG. 4.
SDS-PAGE of extracellular protein extracts from H. alvei 718, H. alvei 718 halI mutant, and H. alvei 718 halI mutant complemented with OHHL. Lanes 1 and 8, standard; lanes 2 and 3, H. alvei 718; lanes 4 and 5, H. alvei 718 halI mutant (AHL-deficient mutant); and lanes 6 and 7, H. alvei 718 halI mutant complemented with 10 μM OHHL.
Interaction between H. alvei 718 and AHL receptive bacteria.
AHLs are believed to mediate interactions in a bacterial population and may facilitate communication between different species in a population. Hence, we hypothesized that AHL produced by H. alvei could be utilized by other bacteria to express AHL-controlled phenotypes. Since we could not detect any effect of AHL production on meat spoilage, we tested this hypothesis in another food model system (liquid milk) where growth and metabolism of psychrotrophic Enterobacteriaceae is a known cause of spoilage (37). The spoilage (curdling) of liquid milk by the psychrotrophic S. proteamaculans strain B5a is controlled by AHLs (12), of which OHHL appears to be the dominant molecule. An S. proteamaculans B5a sprI mutant (no AHL production) was inoculated with H. alvei 718 or the H. alvei 718 halI mutant in milk and stored at 5°C. Combinations of these cultures complemented with OHHL were also included. All samples were inoculated at a starting density of approximately 104 CFU/ml. The milk was not spoiled by H. alvei 718, the H. alvei 718 halI mutant, or the S. proteamaculans B5a sprI mutant even when cell densities of 109 CFU/g were reached (3 to 4 weeks) (Table 3). In contrast, the AHL-producing S. proteamaculans B5a spoiled the milk at this cell density. When H. alvei 718 and the S. proteamaculans B5a sprI mutant were coinoculated, the milk was spoiled at a cell density of 109 CFU/g for S. proteamaculans and 108 CFU/g for H. alvei 718. No signs of spoilage were found at comparable cell densities when coinoculating the S. proteamaculans B5a sprI mutant and H. alvei 718 halI mutant in milk. These results indicate that the AHL produced by H. alvei 718 may contribute to the expression of AHL-controlled phenotypes in other bacteria, which in turn can affect the spoilage of food products.
TABLE 3.
Bacterial counts in inoculated skim milk after 3 to 4 weeks of storage at 5°C
| Bacterial straina | Coagulation of milk | Bacterial count (CFU/g)
|
|
|---|---|---|---|
| Serratia | Hafnia | ||
| H. alvei 718 | − | 8.2 × 108 | |
| H. alvei 718 halI mutant | − | 1.8 × 109 | |
| S. proteamaculans B5a | + | 2.2 × 109 | |
| S. proteamaculans B5a sprI mutant | − | 2.1 × 109 | |
| S. proteamaculans B5a sprI mutant H. alvei 718 | + | 2.3 × 109 | 1.9 × 108 |
| S. proteamaculans B5a sprI mutant H. alvei 718 halI mutant | − | 2.5 × 109 | 4.0 × 108 |
| S. proteamaculans B5a sprI mutant with 1 μM OHHL | + | 1.6 × 109 | |
| Negative control (milk only) | − | <104 | <104 |
Bacteria were inoculated into milk at 104 CFU/g.
DISCUSSION
Compounds eliciting a response in microbiological AHL monitors were extracted from vacuum-packed red meats and from strains of Enterobacteriaceae isolated from the meat samples. In pure culture of H. alvei, AHLs were detectable when cell counts reached 106 CFU/ml. Hence, the cell densities acquired in chilled packed meats are sufficient to activate QS systems, and such compounds appear to be common in foods where Enterobacteriaceae grow well, such as smoked fish (27) and bean sprouts (Rasch et al., unpublished data). The production of AHL was not up-regulated during growth as seen in the Vibrio fischeri system.
In agreement with other studies, we found that the microbiota in vacuum-packed meat was dominated by LAB (6, 40, 50, 53, 57), with Enterobacteriaceae accounting for 1 to 10% of the microbiota. The level of Enterobacteriaceae and the level of AHL-producing bacteria as estimated by replica plating onto an AHL monitor strain were similar. This indicates that the majority of the AHL-producing bacteria are Enterobacteriaceae, as observed by Gram et al. (27) and Ravn et al. (45). They found that that H. alvei and Serratia spp. were the dominating species among the AHL-producing Enterobacteriaceae isolated from vacuum-packed meat. Several studies have reported that these bacteria produce AHLs and employ QS systems (12, 18, 32, 54).
The replica plating was useful for isolating potential AHL-producing bacteria from food products. However, the method is probably not applicable to samples where the level of AHL-producing bacteria constitutes less than 1% of the microbiota, since the replica plates will be overgrown with non-AHL-producing organisms.
H. alvei was the dominant member of the Enterobacteriaceae in vacuum-packed meat, which is in agreement with observations of Stanbridge and Davies (52). At the same time changing culture conditions in vitro from aerobic to microaerophilic conditions of H. alvei 718 caused a 10-fold increase in the specific AHL yield. Whether the latter has any influence on the dominance of the organism under vacuum is at present unknown. It has previously been demonstrated that environmental factors such as temperature and growth media affect the type of AHLs produced (3, 38), but only minor differences in specific AHL yields have been reported when testing the influence of different food preservations parameters (22).
All H. alvei strains isolated from different samples in the present study and H. alvei strain 718 isolated in a previous study (8, 27) produced AHLs with almost identical TLC profiles, and the major spot had an Rf value and a shape similar to that of OHHL (51). LC-MS confirmed the presence of this compound in all strains. Swift et al. (54) inserted a lux-based reporter plasmid into H. alvei and demonstrated AHL production; however, the structure of the signaling compound was never elucidated. An AHL spot similar to OHHL was the only AHL detected in the meat extracts, and we therefore believe that the H. alvei strains were producing the majority of the AHLs extracted from meat samples. In agreement with Morin et al. (39) we found LC-MS to be a rapid and very sensitive method for detection of bacterial AHLs. In our study AHLs were detected and identified from as little as 1 ml of bacterial culture supernatant. This technique will greatly facilitate studies of bacterial QS.
Using a range of different AHL monitor systems in combination—including the LuxR-based system in E. coli, the TraR-based system in A. tumefaciens NT1(pZLR4), and the CviR-based systems in C. violaceum CV026—it is possible to detect the entire range of known AHLs (45). When the above-mentioned systems were employed with extracts from the H. alvei 718 halI mutant, no AHL activity could be detected, indicating that halI is the only gene responsible for synthesizing AHLs in H. alvei 718. In support of this observation, no other AHL types could be detected when TLC analyses were performed with extracts from AHL-producing E. coli clones harboring DNA fragments from H. alvei 718 (data not shown).
AHL-regulated bacterial phenotypes may influence the quality of food products, and the enzymatic degradation of bean sprouts (Rasch et al., unpublished data), onions (1), and milk (12) is accelerated by AHL regulation of hydrolytic enzymes. The effect of AHL regulation on the spoilage of vacuum-packed meat was tested using two approaches. First, we compared the spoilage ability of an AHL-producing H. alvei and its I-negative mutant. Second, we attempted to block spoilage by applying a compound (furanone C-30) that specifically interferes with QS in P. aeruginosa (29). Neither approach demonstrated any influence of AHL production on spoilage of the product. The typical spoilage in vacuum-packed meat is probably a result of an interaction between LAB and Enterobacteriaceae (8, 16, 20). We therefore deliberately inoculated the H. alvei strains on meat samples with a background level of LAB; however, this did not reveal any importance of QS systems in meat spoilage. Vacuum-packed cold-smoked salmon may also spoil due to an interaction of gram-negative and gram-positive bacteria (34), and although AHLs can be extracted from the product and from the Enterobacteriaceae isolated from the product (27), we have not been able to demonstrate any importance of the QS systems in the spoilage of cold-smoked salmon (Flodgaard and Christensen, unpublished data).
It has been suggested that Pseudomonas spp. use a QS system to produce slime at the surface of aerobically packed meat (33). Our preliminary screening of aerobically packed meat only resulted in 11 AHL-producing bacteria, of which only one was identified as Pseudomonas. Based on these findings we do not find it likely that Pseudomonas spp. use a QS system in their spoilage of aerobically packed meat.
The off-odors developing during bacterial spoilage of aerobically packed and vacuum-packed meats as well as of cold-smoked fish are caused by a turnover of free amino acids. This metabolism is probably essential to the nutritional status of the bacterial cell. While one could hypothesize that (AHL-regulated) proteolytic activity could increase the pool of amino acids, one could also argue that metabolic traits crucial to the nutritional status are not likely to be influenced by QS systems. One can argue that QS systems were probably not developed to allow bacteria to spoil foods but rather to enhance their ability to colonize (and infect) a living host. Therefore, foods spoiling due to a bacterial behavior, which is important for colonization and infection, are more likely to be influenced by QS. Examples of this are foods spoiling due to hydrolytic degradation (sprouts, onions, and milk).
It is not known how the AHLs produced by H. alvei influence the physiology of the bacteria. In other food-relevant Enterobacteriaceae, QS systems control a range of phenotypes, including production of exoenzymes and antibiotics as well as swarming ability (10, 12, 18). We compared the expression of a vast range of phenotypes between the wild type and the I-negative mutant, and none appeared to be influenced by the AHL system. In some Enterobacteriaceae extracellular introduction of AHL may be needed to express AHL-controlled phenotypes (2). We therefore evaluated if the production of AHLs by H. alvei AHL could influence the behavior of other organisms employing QS systems. Enterobacteriaceae are common contaminants of pasteurized milk and may cause spoilage (21, 37) due to proteolytic degradation of the casein micelles, leading to clotting (“sweat curdling”) (14). We demonstrated that the AHLs produced by H. alvei strain 718 induced the AHL-regulated protease of a food spoilage-related S. proteamaculans. Hence, the ability of H. alvei to produce AHLs might indirectly influence the spoilage of food products by facilitating expression of QS systems in other organisms. Cross talk between bacteria with AHLs has previously been demonstrated (43, 48).
In conclusion, we demonstrate that compounds inducing QS systems are common in spoiling vacuum-packed meat. However, our data indicate that these compounds are not essential to the meat spoilage process but may influence spoilage of other types of foods.
Acknowledgments
This study was partly financed by the Danish Technical Research Council (projects 26-00-0170 and 9901-295).
Hanne Jakobsen, BioCentrum, is thanked for excellent technical assistance on the HLPC-MS.
REFERENCES
- 1.Aguilar, C., I. Bertani, and V. Venturi. 2003. Quorum-sensing system and stationary-phase sigma factor (rpoS) of the onion pathogen Burkholderia cepacia genomovar I type strain, ATCC 25416. Appl. Environ. Microbiol. 69:1739-1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahmer, B. M. M., J. van Reeuwijk, C. D. Timmers, P. J. Valentine, and F. Heffron. 1998. Salmonella typhimurium encodes an SdiA homolog, a putative quorum sensor of the LuxR family, that regulates genes on the virulence plasmid. J. Bacteriol. 180:1185-1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atkinson, S., J. P. Throup, G. S. A. B. Stewart, and P. Williams. 1999. A hierarchical quorum-sensing system in Yersinia pseudotuberculosis is involved in the regulation of motility and clumping. Mol. Microbiol. 33:1267-1277. [DOI] [PubMed] [Google Scholar]
- 4.Bainton, N. J., P. Stead, S. R. Chhabra, B. W. Bycroft, G. P. Salmond, G. S. A. B. Stewart, and P. Williams. 1992. N-(3-oxohexanoyl)-L-homoserine lactone regulates carbapenem antibiotic production in Erwinia carotovora. Biochem. J. 288:997-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barrow, G. L., and R. K. A. Feltham (ed.). 1993. Cowan and Steel's manual for the identification of medical bacteria, 3rd ed. Cambridge University Press, Cambridge, United Kingdom.
- 6.Beebe, S. D., C. Vanderzant, M. O. Hanna, Z. L. Carpenter, and G. C. Smith. 1976. Effect of initial internal temperature and storage temperature on microbial-flora of vacuum packaged beef. J. Milk. Food Technol. 39:600-605. [Google Scholar]
- 7.Bertani, G. 1951. Studies on lysogenesis. 1. The mode of phage liberation by lysogenic Escherichia coli. J. Bacteriol. 62:293-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borch, E., M. L. KantMuermans, and Y. Blixt. 1996. Bacterial spoilage of meat and cured meat products. Int. J. Food Microbiol. 33:103-120. [DOI] [PubMed] [Google Scholar]
- 9.Buch, C., J. Sigh, J. Nielsen, J. L. Larsen, and L. Gram. 2003. Production of acylated homoserine lactones by different serotypes of Vibrio anguillarum both in culture and during infection of rainbow trout. Syst. Appl. Microbiol. 26:338-349. [DOI] [PubMed] [Google Scholar]
- 10.Byers, J. T., C. Lucas, G. P. C. Salmond, and M. Welch. 2002. Nonenzymatic turnover of an Erwinia carotovora quorum-sensing signaling molecule. J. Bacteriol. 184:1163-1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cha, C., P. Gao, Y. C. Chen, P. D. Shaw, and S. K. Farrand. 1998. Production of acyl-homoserine lactone quorum-sensing signals by gram-negative plant-associated bacteria. Mol. Plant-Microbe Interact. 11:1119-1129. [DOI] [PubMed] [Google Scholar]
- 12.Christensen, A. B., K. Riedel, L. Eberl, L. R. Flodgaard, S. Molin, L. Gram, and M. Givskov. 2003. Quorum-sensing-directed protein expression in Serratia proteamaculans B5a. Microbiology 149:471-483. [DOI] [PubMed] [Google Scholar]
- 13.Clark, D. J., and O. Maaloe. 1967. DNA replication and division cycle in Escherichia coli. J. Mol. Biol. 23:99-112. [Google Scholar]
- 14.Craven, H. M., and B. J. Macauley. 1992. Microorganisms in pasteurized milk after refrigerated storage. 1. Identification of types. Aust. J. Dairy Technol. 47:38-45. [Google Scholar]
- 15.Dainty, R. H., R. A. Edwards, and C. M. Hibbard. 1985. Time course of volatile compound formation during refrigerated storage of naturally contaminated beef in air. J. Appl. Bacteriol. 59:303-309. [DOI] [PubMed] [Google Scholar]
- 16.Dainty, R. H., R. A. Edwards, C. M. Hibbard, and S. V. Ramantanis. 1986. Bacterial sources of putrescine and cadaverine in chill stored vacuum-packed beef. J. Appl. Bacteriol. 61:117-123. [DOI] [PubMed] [Google Scholar]
- 17.Dainty, R. H., and B. M. Mackey. 1992. The relationship between the phenotypic properties of bacteria from chill-stored meat and spoilage processes. J. Appl. Bacteriol. 73:103-114. [DOI] [PubMed] [Google Scholar]
- 18.Eberl, L., M. K. Winson, C. Sternberg, G. S. Stewart, G. Christiansen, S. R. Chhabra, B. Bycroft, P. Williams, S. Molin, and M. Givskov. 1996. Involvement of N-acyl-L-homoserine lactone autoinducers in controlling the multicellular behaviour of Serratia liquefaciens. Mol. Microbiol. 20:127-136. [DOI] [PubMed] [Google Scholar]
- 19.Eberl, L. 1999. N-Acyl homoserinelactone-mediated gene regulation in gram-negative bacteria. Syst. Appl. Microbiol. 22:493-506. [DOI] [PubMed] [Google Scholar]
- 20.Edwards, R. A., R. H. Dainty, and C. M. Hibbard. 1985. Putrescine and cadaverine formation in vacuum-packed beef. J. Appl. Bacteriol. 58:13-19. [DOI] [PubMed] [Google Scholar]
- 21.Eneroth, A., A. Christiansson, J. Brendehaug, and G. Molin. 1998. Critical contamination sites in the production line of pasteurised milk, with reference to the psychrotrophic spoilage flora. Int. Dairy J. 8:829-834. [Google Scholar]
- 22.Flodgaard, L. R., A. B. Christensen, S. Molin, M. Givskov, and L. Gram. 2003. Influence of food preservation parameters and associated microbiota on production rate, profile and stability of acylated homoserine lactones from food-derived Enterobacteriaceae. Int. J. Food Microbiol. 84:145-156. [DOI] [PubMed] [Google Scholar]
- 23.Gibson, L. F., and J. T. Khoury. 1986. Storage and survival of bacteria by ultra-freeze. Lett. Appl. Microbiol. 3:127-129. [Google Scholar]
- 24.Gill, C. O., and K. G. Newton. 1977. Development of aerobic spoilage flora on meat stored at chill temperatures. J. Appl. Bacteriol. 43:189-195. [DOI] [PubMed] [Google Scholar]
- 25.Givskov, M., L. Eberl, G. Christiansen, M. J. Benedik, and S. Molin. 1995. Induction of phospholipase- and flagellar synthesis in Serratia liquefaciens is controlled by expression of the flagellar master operon flhD. Mol. Microbiol. 15:445-454. [DOI] [PubMed] [Google Scholar]
- 26.Givskov, M., R. DeNys, M. Manefield, L. Gram, R. Maximilien, L. Eberl, S. Molin, P. D. Steinberg, and S. Kjelleberg. 1996. Eukaryotic interference with homoserine lactone-mediated prokaryotic signaling. J. Bacteriol. 178:6618-6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gram, L., A. B. Christensen, L. Ravn, S. Molin, and M. Givskov. 1999. Production of acylated homoserine lactones by psychrotrophic members of the Enterobacteriaceae isolated from foods. Appl. Environ. Microbiol. 65:3458-3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gram, L., and H. H. Huss. 1996. Microbiological spoilage of fish and fish products. Int. J. Food Microbiol. 33:121-137. [DOI] [PubMed] [Google Scholar]
- 29.Hentzer, M., K. Riedel, T. B. Rasmussen, A. Heydorn, J. B. Andersen, M. R. Parsek, S. A. Rice, L. Eberl, S. Molin, N. Hoiby, S. Kjelleberg, and M. Givskov. 2002. Inhibition of quorum sensing in Pseudomonas aeruginosa biofilm bacteria by a halogenated furanone compound. Microbiology 148:87-102. [DOI] [PubMed] [Google Scholar]
- 30.Hentzer, M., H. Wu, J. B. Andersen, K. Riedel, T. B. Rasmussen, N. Bagge, N. Kumar, M. A. Schembri, Z. Song, P. Kristoffersen, M. Manefield, J. W. Costerton, S. Molin, L. Eberl, P. Steinberg, S. Kjelleberg, N. Hoiby, and M. Givskov. 2003. Attenuation of Pseudomonas aeruginosa virulence by quorum sensing inhibitors. EMBO J. 22:3803-3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Herrero, M., V. De Lorenzo, and K. N. Timmis. 1990. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 172:6557-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Horng, Y. T., S. C. Deng, M. Daykin, P. C. Soo, J. R. Wei, K. T. Luh, S. W. Ho, S. Swift, H. C. Lai, and P. Williams. 2002. The LuxR family protein SpnR functions as a negative regulator of N-acylhomoserine lactone-dependent quorum sensing in Serratia marcescens. Mol. Microbiol. 45:1655-1671. [DOI] [PubMed] [Google Scholar]
- 33.Jay, J. M., J. P. Vilai, and M. E. Hughes. 2002. Profile and activity of the bacterial biota of ground beef held from freshness to spoilage at 5-7°C. Int. J. Food Microbiol. 81:105-111. [DOI] [PubMed] [Google Scholar]
- 34.Jorgensen, L. V., H. H. Huss, and P. Dalgaard. 2000. The effect of biogenic amine production by single bacterial cultures and metabiosis on cold-smoked salmon. J. Appl. Microbiol. 89:920-934. [DOI] [PubMed] [Google Scholar]
- 35.Laemmli, U. K. 1970. Cleavage of structural proteins during assembly of head of bacteriophage-T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 36.Lambropoulou, K. A., E. H. Drosinos, and G. J. E. Nychas. 1996. The effect of glucose supplementation on the spoilage microflora and chemical composition of minced beef stored aerobically or under a modified atmosphere at 4°C. Int. J. Food Microbiol. 30:281-291. [DOI] [PubMed] [Google Scholar]
- 37.Lindberg, A. M., A. Ljungh, S. Ahrne, S. Lofdahl, and G. Molin. 1998. Enterobacteriaceae found in high numbers in fish, minced meat and pasteurised milk or cream and the presence of toxin encoding genes. Int. J. Food Microbiol. 39:11-17. [DOI] [PubMed] [Google Scholar]
- 38.Lithgow, J. K., V. E. Danino, J. Jones, and J. A. Downie. 2001. Analysis of N-acyl homoserine-lactone quorum-sensing molecules made by different strains and biovars of Rhizobium leguminosarum containing different symbiotic plasmids. Plant Soil. 232:3-12. [Google Scholar]
- 39.Morin, D., B. Grasland, K. Vallee-Rehel, C. Dufau, and D. Haras. 2003. On-line high-performance liquid chromatography-mass spectrometric detection and quantification of N-acylhomoserine lactones, quorum sensing signal molecules, in the presence of biological matrices. J. Chromatogr. A 1002:79-92. [DOI] [PubMed] [Google Scholar]
- 40.Newton, K. G., and C. O. Gill. 1978. Development of anaerobic spoilage flora of meat stored at chill temperatures. J. Appl. Bacteriol. 44:91-95. [DOI] [PubMed] [Google Scholar]
- 41.Nielsen, K. F., and J. Smedsgaard. 2003. Fungal metabolite screening: database of 474 mycotoxins and fungal metabolites for de-replication by standardised liquid chromatography-UV detection-mass spectrometry methodology. J. Chromatogr. A 1002:111-136. [DOI] [PubMed] [Google Scholar]
- 42.Nychas, G. J., and J. S. Arkoudelos. 1990. Microbiological and physicochemical changes in minced meats under carbon-dioxide, nitrogen or air at 3-degrees-C. Int. J. Food Sci. Technol. 25:389-398. [Google Scholar]
- 43.Pierson, E. A., D. W. Wood, J. A. Cannon, F. M. Blachere, and L. S. Pierson. 1998. Interpopulation signalling via N-acyl-homoserine lactones among bacteria in the wheat rhizosphere. Mol. Plant-Microbe Interact. 11:1078-1084. [Google Scholar]
- 44.Pirhonen, M., D. Flego, R. Heikinheimo, and E. T. Palva. 1993. A small diffusible signal molecule is responsible for the global control of virulence and exoenzyme production in the plant pathogen Erwinia carotovora. EMBO J. 12:2467-2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ravn, L., A. B. Christensen, S. Molin, M. Givskov, and L. Gram. 2001. Methods for detecting acylated homoserine lactones produced by Gram-negative bacteria and their application in studies of AHL-production kinetics. J. Microbiol. Methods 44:239-251. [DOI] [PubMed] [Google Scholar]
- 46.Riddel, J., and H. Korkeala. 1997. Minimum growth temperatures of Hafnia alvei and other Enterobacteriaceae isolated from refrigerated meat determined with a temperature gradient incubator. Int. J. Food Microbiol. 35:287-292. [DOI] [PubMed] [Google Scholar]
- 47.Riedel, K., T. Ohnesorg, K. A. Krogfelt, T. S. Hansen, K. Omori, M. Givskov, and L. Eberl. 2001. N-Acyl-l-homoserine lactone-mediated regulation of the lip secretion system in Serratia liquefaciens MG1. J. Bacteriol. 183:1805-1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Riedel, K., M. Hentzer, O. Geisenberger, B. Huber, A. Steidle, H. Wu, N. Hoiby, M. Givskov, S. Molin, and L. Eberl. 2001. N-Acylhomoserine-lactone-mediated communication between Pseudomonas aeruginosa and Burkholderia cepacia in mixed biofilms. Microbiology 147:3249-3262. [DOI] [PubMed] [Google Scholar]
- 49.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 50.Schillinger, U., and F. K. Lucke. 1987. Lactic-acid bacteria on vacuum-packaged meat and their influence on shelf-life. Fleischwirtschaft 67:1244-1248. [Google Scholar]
- 51.Shaw, P. D., G. Ping, S. L. Daly, C. Cha, J. E. Cronan, K. L. Rinehart, and S. K. Farrand. 1997. Detecting and characterizing N-acyl-homoserine lactone signal molecules by thin-layer chromatography. Proc. Natl. Acad. Sci. USA 94:6036-6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stanbridge, L. H., and A. R. Davies. 1998. The microbiology of chill-stored meat, p. 174-219. In A. Davies and R. G. Board (ed.), The microbiology of meat and poultry. Blackie Academic & Professional, London, United Kingdom.
- 53.Sutherland, J. P., J. T. Patterson, and J. G. Murray. 1975. Changes in microbiology of vacuum packaged beef. J. Appl. Bacteriol. 39:227-237. [DOI] [PubMed] [Google Scholar]
- 54.Swift, S., M. K. Winson, P. F. Chan, N. J. Bainton, M. Birdsall, P. J. Reeves, C. E. D. Rees, S. R. Chhabra, P. J. Hill, J. P. Throup, B. W. Bycroft, G. P. C. Salmond, P. Williams, and G. S. A. B. Stewart. 1993. A novel strategy for the isolation of LuxI homologs: evidence for the widespread distribution of a LuxR LuxI superfamily in enteric bacteria. Mol. Microbiol. 10:511-520. [DOI] [PubMed] [Google Scholar]
- 55.Throup, J. P., M. Camara, G. S. Briggs, M. K. Winson, S. R. Chhabra, B. W. Bycroft, P. Williams, and G. S. A. B. Stewart. 1995. Characterization of the YenI/YenR locus from Yersinia enterocolitica mediating the synthesis of two N-acylhomoserine lactone signal molecules. Mol. Microbiol. 17:345-356. [DOI] [PubMed] [Google Scholar]
- 56.Whitehead, N. A., A. M. L. Barnard, H. Slater, N. J. L. Simpson, and G. P. C. Salmond. 2001. Quorum-sensing in gram-negative bacteria. FEMS Microbiol. Rev. 25:365-404. [DOI] [PubMed] [Google Scholar]
- 57.Yost, C. K., and F. M. Nattress. 2002. Molecular typing techniques to characterize the development of a LAB community on vacuum-packaged beef. Int. J. Food Microbiol. 72:97-105. [DOI] [PubMed] [Google Scholar]