Abstract
1,2-Dichloropropane (1,2-D), a widespread groundwater contaminant, can be reductively dechlorinated to propene by anaerobic bacteria. To shed light on the populations involved in the detoxification process, a comprehensive 16S rRNA gene-based bacterial community analysis of two enrichment cultures derived from geographically distinct locations was performed. Analysis of terminal restriction fragments, amplicons obtained with dechlorinator-specific PCR primers, and enumeration with quantitative real-time PCR as well as screening clone libraries all implied that Dehalococcoides populations were involved in 1,2-D dechlorination in both enrichment cultures. Physiological traits (e.g., dechlorination in the presence of ampicillin and a requirement for hydrogen as the electron donor) supported the involvement of Dehalococcoides populations in the dechlorination process. These findings expand the spectrum of chloroorganic compounds used by Dehalococcoides species as growth-supporting electron acceptors. The combined molecular approach allowed a comparison between different 16S rRNA gene-based approaches for the detection of Dehalococcoides populations.
The short-chain chlorinated aliphatic compound 1,2-dichloropropane (1,2-D) has been used as a fumigant to fight root-parasitic nematodes in the production of high-value crops, as an insecticide, and in numerous industrial applications (17). Although 1,2-D is no longer employed in agricultural applications and its industrial use has been reduced, these practices lead to substantial groundwater contamination. Thus, many sites with 1,2-D concentrations exceeding the U.S. Environmental Protection Agency enforced maximum concentration level of 5 ppb (www.epa.gov/safewater/dwh/c-voc.html) exist. Due to the toxicity (and potential for carcinogenicity) of 1,2-D to the liver, kidneys, adrenal glands, bladder, and gastrointestinal and respiratory tracts, its presence in the environment is of genuine public concern, and many of the existing contaminated sites require remedial action.
No naturally occurring aerobic microbial pathways that efficiently transform 1,2-D to nontoxic products have been described (11), although enzymes that transform polychlorinated propanes were generated by in vitro molecular evolution (3, 32). Most of the contamination with 1,2-D has occurred in oxygen-limited or anaerobic subsurface environments, where reductive processes are more probable (17, 34). In a previous study, the reductive dechlorination (i.e., hydrogenolysis) of 1,2-D to monochlorinated propanes and subsequent dehydrochlorination to propene was demonstrated in microcosms derived from river sediment (17). Interestingly, in sediment-free, nonmethanogenic enrichment cultures derived from these microcosms, only vicinal reduction (dichloroelimination) of 1,2-D to propene without the intermediate formation of monochlorinated propanes occurred (17). Hydrogen consumption threshold measurements provided circumstantial evidence for the presence of populations in the enrichment cultures that gain energy from the reductive dechlorination process (e.g., chlororespiration) (17, 21). Thus far, attempts to isolate pure cultures of anaerobic, 1,2-D-dechlorinating populations have proven to be difficult, and the key players involved in the dechlorination process have remained elusive. Some evidence that Dehalobacter populations may contribute to 1,2-D dechlorination in a bioreactor established with Saale River sediments was obtained by using 16S rRNA gene-based approaches (28, 29). To discover the nature of the 1,2-D-dechlorinating population(s), and to elucidate the microbial community structures that were established after long-term enrichment with 1,2-D, two sediment-derived enrichment cultures were thoroughly characterized. 16S rRNA gene-based approaches, along with physiological studies, provided insight into the identities of the dechlorinating populations. A detailed analysis and comparison between communities allowed us to attribute 1,2-D dechlorination to Dehalococcoides populations and to explain the presence of other community members in the enrichment cultures.
MATERIALS AND METHODS
Chemicals and enzymes.
1,2-D, ampicillin, proteinase K, lysozyme, and achromopeptidase were obtained from Sigma (St. Louis, Mo.). DNA extraction and purification kits were purchased from QIAGEN (Valencia, Calif.), Taq DNA polymerase and PCR buffer were from Applied Biosystems (Foster City, Calif.), and bovine serum albumin was from Roche (Mannheim, Germany). The TOPO TA cloning kit and restriction enzymes were obtained from Invitrogen (Carlsbad, Calif.), the oligonucleotide primers for PCR and sequencing were purchased from Integrated DNA Technologies (Coralville, Iowa), and the R2A agar was from Fisher Scientific (Pittsburgh, Pa.).
Enrichment cultures and growth conditions.
A series of microcosms established with aquifer and sediment materials collected from both pristine locations and sites contaminated with 1,2-D, chloroethenes, and/or petroleum hydrocarbons was established with 1,2-D as an electron acceptor and lactate (2.5 mM), pyruvate (2.5 mM), or acetate (2 mM) plus hydrogen (20% by volume) as electron donors as described previously (17) Consecutive transfers to fresh mineral salts medium (17, 31) yielded sediment-free, nonmethanogenic enrichment cultures with robust 1,2-D dechlorination activity from the Red Cedar River in Okemos, Mich. (RC), and the King Salmon River in Alaska (KS). The sampling site in Michigan had no known anthropogenic contamination with 1,2-D but is located in an agricultural area impacted by farmland runoff. The sampling area of KS was contaminated with petroleum hydrocarbons. Sediment-free, nonmethanogenic enrichment cultures were derived from 1,2-D-dechlorinating microcosms by consecutive transfers to fresh mineral salts medium as described previously (17, 19). The RC and KS cultures were transferred 35 times in 160-ml serum bottles containing 100 ml of bicarbonate-buffered basal salts medium amended with acetate (5 mM), hydrogen (17 kPa, approximately 400 μmol), and 1,2-D (17, 21). Cultures were fed 20 μmol of undiluted 1,2-D or 50 μmol of 1,2-D dissolved in 0.2 ml of hexadecane to yield initial aqueous 1,2-D concentrations of approximately 0.2 and 0.4 mM, respectively. The 1,2-D partitioning in the aqueous and gas phases was calculated by using an octanol-water coefficient (Klow) of 105 and a Henry constant of 0.00231 (23). When the initial amount of chlorinated electron acceptor was consumed, cultures were fed with the same amount of 1,2-D. Transfers (1%, vol/vol) to fresh medium occurred before all of the 1,2-D was converted to propene. Serum bottles were sealed with black butyl rubber stoppers (Geo-Microbial Technologies, Inc., Ochelata, Okla.) that were secured with aluminum crimp seals and incubated at room temperature (22 to 25°C). Culture performance was tested in the presence of the peptidoglycan biosynthesis inhibitor ampicillin; 0.25 to 0.5 mg of the antibiotic per ml was added from an aqueous, anoxic, filter-sterilized stock solution (50 mg per ml) prior to inoculation. To ascertain whether heterotrophic bacteria could be isolated from the 1,2-D-dechlorinating communities, 10-fold serial dilutions were made to half-strength tryptic soy broth prepared anaerobically in serum vials, and 200 μl of culture fluid was spread onto complex medium R2A agar plates inside an anaerobic glove box (N2 atmosphere with 4.5% [vol/vol] H2). Replicate agar plates were removed from the glove box for incubation under aerobic conditions. Incubation was done at room temperature for 2 months.
Analytical methods.
Concentrations of chlorinated alkenes and alkanes were determined with a Hewlett Packard model 6890 gas chromatograph equipped with an HP-624 column (60-m length, 0.32-mm diameter, 1.8-μm film thickness) and a flame ionization detector as described previously (10, 17, 21). Headspace samples (200 μl) were withdrawn with gas-tight 250-μl glass syringes (model number 1725; Hamilton Co., Reno, Nev.) and manually injected into a split injector operated at a split ratio of 2:1. The following temperature program was applied: an initial hold at 50°C for 3.5 min, an increase to 200°C at a rate of 50°C per min, and a final hold at 200°C for 2.5 min. Ultra-high-purity nitrogen (99.998%; AGA Specialty Gas, Maumee, Ohio) that was further purified with a gas purification column (Chromatography Research Supplies, Louisville, Ky.) to remove residual moisture, oxygen, and hydrocarbons was used as the carrier gas at a flow rate of 3 ml min−1. Nonchlorinated C1 to C3 alkenes and alkanes were separated and quantified with an HP-PLOT Q column (30-m length, 0.53-mm diameter, 40-μm film thickness) coupled with flame ionization detection as described previously (17).
DNA extraction.
Cells were collected from 20 or 40 ml of actively dechlorinating liquid cultures or from cultures that had consumed all 1,2-D on 0.2-μm-pore polycarbonate membranes by vacuum filtration under aseptic conditions. The cake of biomass was removed from the filter by aseptically placing the filter into a plastic 2-ml microcentrifuge tube and adding 1 ml of sterile phosphate-buffered saline (5 mM potassium phosphate, 0.85% NaCl, pH 7.0) followed by horizontal shaking at high speed on a vortex mixer for 10 min. After removing suspended cells by centrifugation (8,160 × g for 15 min), each filter was transferred to a new microcentrifuge and rinsed twice with 100 μl of saline by vortexing to increase cell recovery. The suspensions were combined, and cells were collected by centrifugation (8,160 × g for 15 min). Genomic DNA was extracted from the pellet by using a QIAGEN tissue kit according to the manufacturer's recommendations, with the following modifications. Cell lysis was achieved by the addition of 20 μl of lysozyme (100 mg/ml) and 10 μl of achromopeptidase (7,500 U/ml) and incubation for 1 h at 37°C followed by the addition 45 μl of proteinase K (25 mg/ml) and incubation at 55°C for 1 h. The tube was inverted twice during each incubation period until lysis was complete, as verified by microscopic examination. DNA was eluted in 200 μl of 10 mM Tris buffer, pH 8.5, and the concentration of isolated DNA was determined spectrophotometrically (27). Isolated DNA was stored at −20°C.
PCR and clone library analysis.
DNA extracted from each of the 1,2-D-dechlorinating communities was screened for the presence of known reductively dechlorinating populations (e.g., Dehalococcoides, Desulfuromonas, Desulfitobacterium, and Dehalobacter spp.) with 16S rRNA gene primers targeting regions that are specific for each individual group (9, 12, 20, 28). PCRs contained 1 to 2 ng of community DNA per μl of reaction mix. For increased sensitivity, an initial amplification of the communities' 16S rRNA genes was performed by using universal bacterial primers 8F and 1541R and under conditions described previously (20). A 1:50 dilution of amplified 16S rRNA gene product (approximately 109 community 16S rRNA gene copies per reaction mix) was used as a template for a second (nested) PCR with the group-specific primers. The number of 16S rRNA gene amplicons was estimated by using twofold dilutions of the PCR products and comparing the band intensities to a fragment of similar size and known concentration. Based on the size of the amplicons (∼1,500 bp), their concentrations (∼75 ng/μl), and the average weight of a base pair (∼660 g/mol), the number of 16S rRNA gene copies present following the initial round of PCR amplification was estimated at about 7 × 1010. Positive controls included genomic DNA of Dehalococcoides sp. strain FL2, Desulfuromonas michiganensis strain BB1, Desulfitobacterium sp. strain Viet-1 (GenBank accession number AF357919), and Dehalobacter restrictus (DSM 9455), which yield amplicons of 620, 815, 1,199, and 823 bp for the Dehalococcoides-, Desulfuromonas-, Desulfitobacterium-, and Dehalobacter-targeted primers, respectively (9, 12, 20, 28).
Clone libraries were constructed for both RC and KS cultures prior to ampicillin treatment by using an Invitrogen pCR2.1-TOPO cloning kit as described previously (20). The community-derived 16S rRNA gene PCR amplicons were ligated into the pCR2.1-TOPO cloning vector and introduced into competent Escherichia coli cells according to the manufacturer's recommendations. Luria Bertani agar plates containing 50 μg of ampicillin ml−1 selected for transformants (27). Template DNA from 72 white colonies from each library was prepared by touching an isolated colony with a sterilized toothpick and suspending the cells in 50 μl of TE buffer (10 mM Tris, 1 mM EDTA, pH 8.0) and then heating the samples in a thermocycler to 95°C for 10 min. Cell debris was removed by centrifugation (8,160 × g for 10 min), and 2 μl of the supernatant was used as a template for PCR with primers TA5′ and TA3′ targeting regions of the cloning vector flanking the inserted 16S rRNA gene fragment (35). PCR was performed as described above except that the reaction mix contained 2 mM MgCl2 and primer annealing occurred at 68°C for 1 min. The resulting PCR products were screened with the specific primer pairs as described above to identify clones that contained 16S rRNA gene inserts of bacterial populations known for their reductive dechlorination ability. The remaining clones in the libraries were characterized by restriction fragment length polymorphism (RFLP) analysis of the TA-vector-flanked 16S rRNA genes of cloned fragments with the restriction endonuclease MspI. Identical restriction patterns were grouped by visual comparison. Clones that yielded amplicons with the dechlorinator-targeted primer pairs were further analyzed by RFLP analysis performed individually with RsaI, MspI, and HhaI under optimum conditions for the respective restriction enzyme as per the manufacturer's recommendations. The resulting DNA fragments were resolved on 2% (wt/vol) Metaphor agarose gels (FMC Bioproducts, Rockland, Maine) and TAE buffer (27) at 4°C, and fragment sizes were estimated by using a 50- to 2,000-bp ladder (Bio-Rad, Hercules, Calif.).
Sequence analysis.
Nearly full-length 16S rRNA gene sequences were obtained from clones that yielded amplicons with the dechlorinator-targeted primer pairs and from clones representing groups of distinct MspI restriction patterns. A combination of six primers that target universal bacterial regions {1392R [5′-ACGGGCGGTGTGT], 907R [5′-CCGTCAATTC(AC)TTT(AG)AGTTT], 529R [CGCGGCTGCTGGCAC], 1114F [GCAACGAGCGCAACCC], 533F [5′-CAGCAGCCGCGGTAA], and 8F [5′-AGAGTTTGATCCTGGCTCAG]} obtained nearly complete 16S rRNA gene sequences for both DNA strands (http://www.genomics.msu.edu). Phylogenetically related populations were identified with BLAST analysis (http://www.ncbi.nlm.nih.gov/BLAST/BLAST.cgi) and included Dehalococcoides ethenogenes strain 195 (GenBank accession number AF004928.2), Dehalococcoides sp. strain CBDB1 (AF230641), Dehalococcoides sp. strain FL2 (AF357918.2), Dehalococcoides sp. strain BAV1 (AY165308), and the environmental clones DCEH2 (AJ249262), DHC-vic (AF388550), and DHC-dll (AF388536). Desulfuromonas sequences included Desulfuromonas michiganensis (accession numbers AF357914.2 and AF357915.2) and Desulfuromonas chloroethenica (U49748). Sequences were initially assembled by using the alignment feature of the MegAlign software of the Lasergene sequence analysis software (DNAstar Inc., Madison, Wis.), ambiguous bases were verified by visual analysis of sequencing chromatograms, and the contiguous sequence was saved in Fasta file format. Each sequence was examined to determine whether it was chimeric by using the ChimeraCheck analysis tool of the Ribosomal Database Project (24).
T-RFLP.
Terminal RFLP (T-RFLP) with the restriction enzymes HhaI, MspI, and RsaI was used to monitor community changes over time and to estimate the microbial diversity in each enrichment culture. By using a reaction volume of 100 μl and conditions described previously (16), 16S rRNA genes of community DNA were amplified with a hexachloro-fluorescein (Hex)-labeled primer, 11F-Hex (5′-GTTTGATCCTGGCTCAG), for detection by a fluorescent automated sequence detector and the unlabeled reverse primer 1492R (5′-ACGGTTACCTTGTTACGACTT). Amplicons generated in two replicate PCRs for each of the enrichment cultures were combined. The combined PCR products (200 μl) were purified by using a QIAGEN PCR Spin Prep purification kit, recovered in a volume of 50 μl, and quantified on agarose gels by comparison with a comparably sized DNA fragment of known concentration. Clean PCR products (approximately 500 ng of DNA) were combined with 2.5 μl of reaction buffer and 1 μl of restriction enzyme solution (10 U) in a total aqueous reaction volume of 25 μl. The samples were incubated for 3.5 h at 37°C followed by denaturation of the enzyme by heating to 65°C for 10 min before freezing and storage at −20°C. Sequence and T-RFLP analyses were performed at Michigan State University's Genomics Center by using an ABI 377 vertical gel DNA sequencer (www.genomics.msu.edu). A culture of Dehalococcoides sp. strain FL2 and a representative clone of each restriction pattern were also examined by the above T-RFLP protocol to experimentally verify the predicted in silico fragment size for the contributing population. To validate successful restriction digests, 15 μl of each sample was analyzed on 2% agarose gels run at 100 V for 45 min (27).
Quantifying Dehalococcoides populations with real-time (RTm) PCR.
The number of Dehalococcoides 16S rRNA gene copies was estimated by using the RTm PCR approach described by He et al. (12). PCR was carried out in a spectrofluorimetric thermal cycler (ABI Prism 7700 sequence detection system; Applied Biosystems). A calibration curve (log DNA concentration versus an arbitrarily set cycle threshold value [CT]) was obtained by using serial dilutions of plasmid DNA carrying a cloned 16S rRNA gene of Dehalococcoides sp. strain BAV1 (13). The CT values obtained for each sample were compared with the standard curve to determine 1,2-D-dependent growth of Dehalococcoides populations in the enrichment cultures. Analyses were performed in triplicate with replicate DNA extractions from 20 ml of fluid obtained from 1,2-D-dechlorinating cultures before and after ampicillin treatment. The total amounts of DNA extracted (about 1.5 μg of DNA per 20 ml of culture fluid) and quantity used per PCR (200 ng) were consistent among all the samples.
RESULTS
Physiological characterization.
Consecutive transfers with 1,2-D as an electron acceptor yielded two stably dechlorinating, sediment-free, nonmethanogenic enrichment cultures from RC and KS sediment materials. Both cultures performed similarly, reducing a single feeding of 20 μmol of 1,2-D to stoichiometric amounts of propene (20.1 ± 1.7 μmol) in 5 days without the apparent accumulation of intermediates. Monochlorinated propanes were not transformed by either culture, indicating that in both cultures, a dichloroelimination pathway was active. Reductive dechlorination was supported by different electron donors, including lactate, pyruvate, or hydrogen. Visible growth was observed within 1 week of inoculation and occurred prior to the onset of dechlorination, which typically started after a lag period ranging from 1 to 2 weeks. Cultures that were transferred twice through defined medium lacking 1,2-D lost the ability to dechlorinate, indicating that 1,2-D was required to maintain the dechlorinating population(s) or the genetic elements encoding the dichloroelimination pathway. Although early enrichments of the RC culture dechlorinated tetrachloroethene (PCE) to cis-dichloroethene (cis-DCE), chloroethenes were not dechlorinated by either culture following 35 successive transfers in medium amended with 1,2-D. No colonies of heterotrophic bacteria were obtained after a 2-month incubation period on complex medium agar plates under anoxic or aerobic conditions or in dilution-to-extinction series in half-strength tryptic soy broth medium.
16S rRNA gene-based community analysis and identification of the dechlorinating populations.
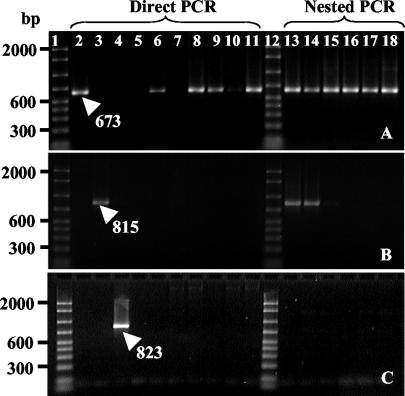
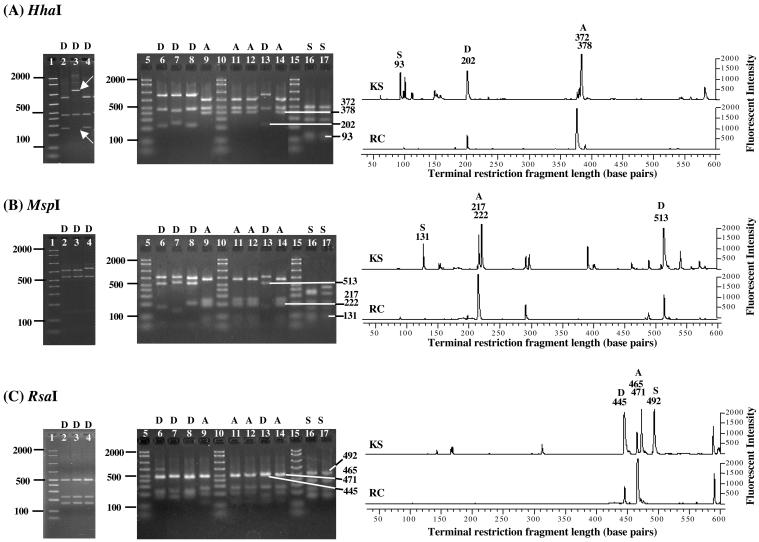
Primer pairs targeting 16S rRNA gene sequences of Dehalococcoides populations yielded amplicons of the expected size and sequence when community DNA extracted from RC and KS dechlorinating cultures was used as a template (Fig. 1A). Desulfuromonas populations were detected in the KS culture by using nested PCR (Fig. 1B). In contrast, Dehalobacter (Fig. 1C) and Desulfitobacterium (data not shown) 16S rRNA gene sequences were never detected in either community, even when a sensitive, nested PCR approach was used. Following two transfers without 1,2-D, Dehalococcoides populations were no longer detected in RC and KS cultures even when the more sensitive nested PCR approach was used (data not shown). Restriction analysis (i.e., RFLP) of 16S rRNA gene clone libraries generated from 72 cloned fragments corroborated that both cultures were highly enriched and that RC culture contained fewer distinct populations than KS culture. Two and four distinct clone types were identified in the RC and KS libraries, respectively. The dominant clones in both libraries belonged to Acetobacterium species that constituted 94 and 89% of the clone patterns from the RC and KS communities, respectively. The clone libraries were comprised of two groups of Acetobacterium 16S rRNA gene sequences, both types of which were found in the KS enrichment and only one of which was found in the RC enrichment. All four RC clones sequenced were 98% similar to Acetobacterium malicum across 1,300 bases of the 16S rRNA gene and exhibited identical restriction profiles with 56 other clones from the RC library. One of the eight sequenced Acetobacterium clones from the KS culture was 99% similar to the RC clone sequences and shared the same restriction pattern with 21 clones from the KS library. These five sequences were identical to the 16S rRNA gene sequence of A. malicum (GenBank accession number X96957) at positions 77 to 90 (E. coli numbering) and therefore possessed the signature region that distinguished them from Acetobacterium wieringae. Seven sequenced KS clones representing 30 restriction patterns in the KS library were 98% similar to the sequence of A. wieringae (accession number X96955) across 1,300 bp of the 16S rRNA gene. The seven KS clone sequences matched the 16S rRNA gene sequence of A. wieringae and A. wieringae isolate HAAP-1 (accession number AF479584) at positions 77 to 90. All eight KS clones differed from A. malicum, A. wieringae isolate HAAP-1, and all of the RC clones at E. coli positions 1,004 to 1,020, where a T-to-C transition (position 1,010) and an A-to-G transition (position 1,019) were consistent with changes in stem pairing in the E. coli 16S rRNA secondary structure (4). Additionally, two restriction patterns representing members of the Desulfuromonas group were identified in the KS community clone library (Fig. 2). Both cloned 16S rRNA genes were sequenced and shared 96% identity with the 16S rRNA gene of Desulfuromonas michiganensis (accession numbers AF357915.2 and AF357914.2). Represented by a single clone sequence, clone AKS21 (from KS culture) was most similar (90%) to environmental clone BSV90 (accession number AJ229233), which was obtained from anoxic rice paddy soil (15). The closest cultured relative was Anaeroarcus burkinensis [corrig.] comb. nov. (accession number AJ010960), which shared 89% sequence similarity (2, 26, 30). Dehalococcoides-like 16S rRNA gene inserts contributed 6% (four clones) and 4% (two clones) of the total clones analyzed in the RC and KS communities, respectively, and no clones representing other known reductively dechlorinating populations were detected. Analysis confirmed that the six Dehalococcoides 16S rRNA gene sequences retrieved from the clone libraries were most similar to 16S rRNA gene sequences of Dehalococcoides isolates FL2, CBDB1, and BAV1, which all belong to the Pinellas group (1, 13, 14, 18). Interestingly, all Dehalococcoides 16S rRNA gene sequences retrieved from the 1,2-D-dechlorinating cultures displayed the identical sequence as isolate BAV1, including the transition (G to A) at position 148 (E. coli numbering), which distinguishes BAV1 from strains FL2 and CBDB1 (1, 13, 14, 18). This transition occurred in variable region 2, which includes many of the signature bases that distinguish the Pinellas, Cornell, and Victoria groups (14). Complete sequence analysis of the amplicons verified their identity and explained slight variations in RFLP banding patterns. For instance, lanes 6 and 7 of Fig. 2 represent two cloned Dehalococcoides 16S rRNA gene fragments inserted in opposite directions into the cloning vector.
FIG. 1.
Identification of Dehalococcoides (A), Desulfuromonas (B), and Dehalobacter (C) populations in KS and RC cultures with 16S rRNA gene-targeted primer pairs specific for each group. Samples from three successional cultures of the 1,2-D-dechlorinating cultures RC and KS were tested. Lanes 2 to 4 and 6 to 11, 50 to 200 ng of extracted genomic DNA was used as a template; lanes 13 to 18, 1:50 dilutions of 16S rRNA gene amplicons were used as a template (nested PCR); lanes 1 and 12, Bio-Rad 50- to 2,000-bp molecular marker; lane 2, Dehalococcoides sp. strain FL2; lane 3, Desulfuromonas michiganensis; lane 4, Dehalobacter restrictus; lane 5, sterile water; lanes 6 and 13, actively dechlorinating KS culture; lanes 7 and 14, 1,2-D-consumed KS culture; lanes 8 and 15, actively dechlorinating, ampicillin-treated KS culture; lanes 9 and 16, actively dechlorinating RC culture; lanes 10 and 17, 1,2-D-consumed RC culture; lanes 11 and 18, actively dechlorinating, ampicillin-treated RC culture.
FIG. 2.
Restriction digest analysis of selected cloned 16S rRNA gene fragments with HhaI (A), MspI (B), and RsaI (C) and detection of community members in T-RFLP profiles. The letters D, A, and S indicate Dehalococcoides, Acetobacterium, and Desulfuromonas clones, respectively. Lanes 1, 5, 10, and 15, 50- to 2,000-bp ladder; lane 2, Dehalococcoides sp. strain FL2; lane 3, Dehalococcoides ethenogenes strain 195; lane 4, Dehalococcoides sp. strain CBDB1; lane 6, ARC13 (D); lane 7, ARC18 (D); lane 8, ARC61 (D); lane 9, ARC62 (A); lane 11, AKS04 (A); lane 12, AKS17 (A); lane 13, AKS31 (D); lane 14, AKS48 (A); lane 16, AKS67 (S); lane 17, AKS68 (S). The white arrows (A) point to the unique band obtained for Dehalococcoides ethenogenes strain 195 when the cloned 16S rRNA gene was digested with enzyme HhaI as well as the missing diagnostic band at 202 bp. The numbers to the right of the gel images indicate the sizes of the corresponding fragments also detected in the T-RFLP profiles. The numbers indicated are the actual T-RF lengths and not the longer restriction fragments obtained due to an additional portion (27 or 55 bp) of the TA cloning vector.
The terminal restriction fragments (T-RFs) determined by in silico digestion of cloned 16S rRNA genes from KS and RC cultures were compared with experimentally obtained T-RFs from RC and KS cultures and the expected T-RFs obtained from in silico digestion of 16S rRNA genes of the nearest relatives identified by BLAST analysis in GenBank. Table 1 shows the correlation between predicted fragment sizes for the cloned nearly complete 16S rRNA genes and the fragments that were detected in the T-RFLP profiles generated for each community. Restriction fragment lengths correlated with 16S rRNA sequences identified by the clone library analysis (Table 1), and when cloned Dehalococcoides-derived or Acetobacterium-derived 16S rRNA gene fragments were used as a template for T-RFLP analysis, a fragment corresponding to a T-RF from the enrichment community was obtained. This careful analysis allowed direct comparisons of the T-RFLP profiles and the RFLP patterns for RC and KS cultures (Fig. 2). All different clone types represented in the 16S rRNA gene libraries (Acetobacterium, Dehalococcoides, Desulfuromonas, and Anaeroarcus) corresponded to a particular T-RF in the T-RFLP profiles and to an RFLP fragment (Fig. 2). Two different 16S rRNA gene sequences belonging to Acetobacterium populations were identified in the KS clone libraries, and these sequences generated two different T-RF signals, correlating with the observed differences at positions 77 to 90 (E. coli numbering) of the 16S rRNA genes. In addition, T-RFs corresponding to known Dehalococcoides isolates of the Pinellas group were observed in cultures RC and KS (Fig. 2). T-RFLP profiles generated from three consecutive transfers of actively dechlorinating RC and KS cultures revealed no apparent changes in community composition, suggesting that climax communities had formed and that the culture conditions used would not result in further enrichment of the 1,2-D-dechlorinating population(s). T-RFLP analysis with three different restriction endonucleases corroborated that the RC culture consisted of fewer populations than the KS culture (Fig. 2).
TABLE 1.
Characteristics of experimentally determined T-RFs in KS and RC communitiesa
| Population or clonea | Length (bases) of T-RF generated by in silico digestion withb
|
Matched T-RFs observed in T-RFLP profiles of cultures (KS/RC) following digestion withc
|
||||
|---|---|---|---|---|---|---|
| HhaI | MspI | RsaI | HhaI | MspI | RsaI | |
| Dehalococcoides ethenogenes strain 195 | 1,073 | 515 | 445 | |||
| Dehalococcoides sp. strains CBDB1, FL2, BAV1 | 202 | 513 | 444 | |||
| Clones ARC13, ARC18, ARC61, ARC31, AKS31, and AKS14 | 202 | 513 | 444 | +/+ | +/+ | +/+ |
| A. malicum, | 373 | 217 | 466 | |||
| A. carbinolicum | 373 | 217 | 468 | |||
| Clones ARC01, ARC03, ARC05, and ARC44 | 373 | 217 | 465 | +/+ | +/+ | +/+ |
| Clones AKS55 and AKS42 | 372 | 217 | 465 | +/+ | +/+ | +/+ |
| A. wieringae | 378 | 222 | 471 | |||
| A. wieringae strain HAAP-1 | 377 | 222 | 471 | |||
| Clones AKS02, AKS04, AKS17, AKS48, AKS70, and AKS29 | 378 | 222 | 471 | +/− | +/− | +/− |
| Desulfuromonas michiganensis | 93 | 81 | 491 | |||
| Desulfuromonas chloroethenica | 93 | 461 | 491 | |||
| Clones AKS67 and AKS68 | 93 | 131 | 492 | +/− | +/− | +/− |
| Anaeroarcus burkinabenensis | 583 | 291 | 488 | |||
| Clone AKS21 | 581 | 293 | 489 | +/− | +/+ | −/− |
T-RFs in KS and RC communities correlate with T-RFs obtained by in silico digestion of cloned 16S rRNA gene fragments and sequences of phylogenetically related populations.
Lengths of T-RFs were calculated from the 3′ forward primer binding site used for T-RFLP analysis.
A +, indicates the presence of and a − indicates the absence of a T-RF matching the in silico determined length in the community T-RFLP profile generated for cultures KS and RC (KS/RC).
Ampicillin treatment.
The addition of ampicillin to the growth medium did not prohibit the reductive dechlorination of 1,2-D to propene, as long as hydrogen was provided as the electron donor. The presence of the antibiotic resulted in further enriched communities with a relative increase in abundance of the Dehalococcoides population(s) in both RC and KS cultures. This conclusion is supported by the intensification of the 673-bp fragment when 200 ng of KS community DNA was used as a template for the Dehalococcoides-targeted PCR (Fig. 1A, lanes 6 and 8 [KS] and lanes 9 and 11 [RC]). Further, T-RFLP profiles developed a more pronounced signal correlating with T-RFs representing Dehalococcoides populations of the Pinellas group as well as a much simpler profile (data not shown). In contrast, the intensity of the Acetobacterium-generated T-RFs decreased relative to the fragment contributed by Dehalococcoides spp., and T-RFs indicative of Desulfuromonas spp. disappeared in the KS enrichment (data not shown). RTm PCR provided additional evidence for the increase in Dehalococcoides populations following transfer in ampicillin-amended medium. In the RC culture, the final number of Dehalococcoides-generated 16S rRNA gene copies in a VC-grown culture increased from (1.0 ± 0.09) × 105 cells per ml prior to the ampicillin treatment to (0.9 ± 0.5) × 107 cells per ml following treatment, an increase in population size of nearly two orders of magnitude. In culture KS, Dehalococcoides increased from (2.3 ± 0.07) × 103 cells per ml to (1.1 ± 0.03) × 107 cells per ml, a 4-order-of-magnitude increase in population size following ampicillin treatment. In a separate experiment, RTm PCR also confirmed 1,2-D-dependent growth in the ampicillin-treated cultures. The number of 16S rRNA gene copies per milliliter of culture fluid increased from (2 ± 0.2) × 103 (transferred with the inoculum) to (0.9 ± 0.5) × 107 and from (2.0 ± 0.4) × 102 to (1.1 ± 0.03) × 107 in RC and KS cultures, respectively. In contrast, RTm PCR failed to detect an increase in the Dehalococcoides population size in cultures that were not amended with 1,2-D.
Detection of Dehalococcoides in cultures of different successional stage.
Dehalococcoides-specific T-RFs (Fig. 2) and amplicons (Fig. 1A) were consistently identified when community DNA was extracted from actively dechlorinating enrichment cultures RC and KS. However, when template DNA was extracted from cultures that had consumed all 1,2-D, direct PCR with Dehalococcoides-specific primers sometimes yielded false-negative results (Fig. 1A, lanes 7 and 10), and resorting to the sensitive nested PCR approach was necessary to detect Dehalococcoides populations (Fig. 1A, lanes 14 and 17). Further, the T-RFLP profiles generated from 1,2-D-depleted cultures showed diminished or absent T-RFs where fragments correlating to Dehalococcoides populations were found in the actively growing cultures.
DISCUSSION
Despite substantial efforts to isolate 1,2-D-dechlorinating populations with culture-based approaches (e.g., dilution-to-extinction series, streak plates, and roll tubes), these attempts were not successful in identifying the bacterial populations responsible for the observed dechlorination process in RC and KS cultures. Hence, 16S rRNA gene-based approaches were used to describe the microbial community structure of two highly enriched, 1,2-D-dechlorinating cultures and to shed light on the identity of the dechlorinating population(s). Detailed community analysis of RC and KS cultures provides evidence that Dehalococcoides populations are responsible for the reductive dechlorination of 1,2-D to propene. Physiological evidence (e.g., resistance to ampicillin and a strict requirement for H2 as the electron donor) supported the involvement of Dehalococcoides populations in the dichloroelimination process. The major clone type found in the 16S rRNA gene libraries belonged to Acetobacterium species. Acetobacterium species grow acetogenically with H2 and CO2 and have been implicated in cometabolic reductive dechlorination of chlorinated alkanes (8) and alkenes (33). Their involvement in 1,2-D dechlorination in RC and KS cultures, however, is unlikely. Transfers in medium without 1,2-D caused no apparent change in the Acetobacterium community but caused a decline in Dehalococcoides numbers to below the detection limit of the nested PCR approach and resulted in the complete loss of 1,2-D dechlorination activity (data not shown). All known Dehalococcoides populations are strictly hydrogenotrophic chlororespiring populations and cannot grow in the absence of a chloroorganic compound as an electron acceptor, which explains the loss of reductive dechlorination activity.
A recent study to explore the bacterial diversity in a 1,2-D-dechlorinating bioreactor derived from Saale River sediment demonstrated an apparent increase of Dehalobacter-like 16S rRNA gene sequences over an operational period of 14 months (28, 29). However, 16S rRNA gene-based approaches also detected Dehalococcoides most closely related to members of the Pinellas group, and hence, the involvement of Dehalococcoides populations in 1,2-D dechlorination in the bioreactor community cannot be excluded. Recently, De Wildeman et al. described a 1,2-dichloroethane-respiring Desulfitobacterium species that also dechlorinated 1,2-D (7). These findings suggest that reductive dechlorination of 1,2-D to propene is carried out by populations affiliated with several different bacterial genera (i.e., Dehalobacter, Desulfitobacterium, and Dehalococcoides), but among these, only Dehalococcoides populations were identified in RC and KS cultures.
Molecular characterization of KS and RC cultures showed that Dehalococcoides populations may constitute a small fraction of a community and avoid detection by some 16S rRNA gene-based approaches, like direct PCR with 16S rRNA gene targeted primer pairs. When isolated community DNA was used as the template, a Dehalococcoides population size of greater than 104 cells per ml of culture fluid was required in direct PCR. The RTm PCR approach required about 103 16S rRNA gene copies per ml in the culture fluid and indicated there were roughly 2 × 103 Dehalococcoides 16S rRNA gene copies per ml of culture fluid in KS culture. This explains the false-negative results obtained with direct PCR and further indicates that 16S rRNA gene-based approaches must be interpreted cautiously, particularly with environmental samples that have not undergone an enrichment process. In the presence of sufficient chlorinated electron acceptor or in ampicillin-amended medium, the abundance of Dehalococcoides increased, and false-negative results could be avoided.
T-RFLP has become a popular method for community profiling and tracking populations of interest (16, 25). T-RFLP, however, failed to detect a peak T-RF indicative of a Dehalococcoides population in every DNA preparation from the 1,2-D-dechlorinating enrichment cultures, despite the fact Dehalococcoides populations were present, as determined by the more sensitive nested PCR approach. Therefore, the T-RFLP approach cannot be expected to identify specific Dehalococcoides T-RFs in a mixture of 16S rRNA gene amplicons obtained from community DNA of naturally diverse sediment or aquifer samples. In silico digests of known Dehalococcoides 16S rRNA gene sequences suggested that Dehalococcoides populations belonging to the Cornell group are not detectable when a T-RFLP approach is used under certain conditions. For instance, Dehalococcoides ethenogenes strain 195 yielded a T-RF of 1,073 bases in length when digested with enzyme HhaI, a fragment too large to be resolved with T-RFLP. A missing HhaI restriction site in the 16S rRNA gene of strain 195 is consistent with the sequence difference that places strain 195 in the Cornell group of the Dehalococcoides cluster (14). Another problem arises when DNA is obtained from samples harboring complex microbial communities. For example, in silico digestion of 16S rRNA gene sequences of several Clostridium species yielded T-RFs of 514 or 518 bp with MspI and 445 bp with RsaI, which resemble T-RF sizes for known Dehalococcoides populations of the Pinellas group (i.e., T-RFs of 514 and 442 bp). Clostridia are common members of anaerobic microbial communities, and thus, clostridial T-RFs might mask or be interpreted erroneously as Dehalococcoides T-RFs, leading to a false-positive interpretation of T-RFLP profiles. HhaI digestion distinguishes clostridia (T-RFs of 223 or 233 bp) from populations of the Pinellas group, which generate 202-bp T-RFs. T-RFs of 223 or 233 bp were not found in HhaI digests of community DNA from RC and KS cultures, indicating that clostridial populations did not contribute to the Dehalococcoides signal. Yet another difficulty in interpreting T-RFLP data arises from rRNA gene copy number differences in target organisms (5). Dehalococcoides ethenogenes strain 195, strain FL2, and strain BAV1 possess a single copy of the 16S rRNA gene (http://www.tigr.org) (13). The number of 16S rRNA gene operons in Acetobacterium spp. is not yet published, but related gram-positive bacteria can possess multiple (up to 11) rRNA operons (http://rrndb.cme.msu.edu). Hence, the dominant T-RFs as well as the large number of Acetobacterium clones recovered from the clone libraries may overestimate the abundance of this bacterial group in the dechlorinating enrichment cultures. In conclusion, neither T-RFLP analysis nor the screening of clone libraries is recommended to either validate or exclude the presence of Dehalococcoides populations, unless the community DNA was extracted from highly enriched, actively dechlorinating cultures, as was done in this study.
Using 16S rRNA gene sequence analysis, we identified in the KS culture a Desulfuromonas population that is most closely related to a PCE- to cis-DCE-dechlorinating strain of Desulfuromonas michiganensis (31). The KS enrichment cultures, however, failed to dechlorinate PCE, indicating that uncultured Desulfuromonas species exist that cannot dechlorinate PCE but possess 16S rRNA genes that amplify with the primers designed to target the PCE-dechlorinating Desulfuromonas group (20). The Desulfuromonas population(s) might persist in the KS 1,2-D enrichment culture in low numbers because of the inherent ability of the family Geobacteriaceae (of which Desulfuromonas is a member) to reduce ferric iron and/or sulfur (22). While neither ferric iron nor sulfur was added to the medium, sulfide was used as a reductant. Trace amounts of oxidized sulfur compounds could explain the stable maintenance of a Desulfuromonas population in culture KS (6). The 1,2-D-dechlorinating Dehalococcoides populations implicated in 1,2-D dechlorination in cultures RC and KS share identical variable regions of the 16S rRNA gene with the chloroethene-dechlorinating isolate BAV1 (13). Neither the RC culture nor the KS culture dechlorinated vinyl chloride to ethene, and isolate BAV1 failed to dechlorinate 1,2-D (unpublished data). Since there is no firm link between 16S rRNA gene sequence and a particular dechlorinating activity among Dehalococcoides populations within the Pinellas group, 16S rRNA gene-based approaches cannot predict dechlorination activity. Supporting measurements indicative of the dechlorination process are recommended. Continued efforts focus on obtaining the 1,2-D-dechlorinating Dehalococcoides population in pure culture and identifying proteins and functional genes involved in the reductive dechlorination of 1,2-D. This would allow the design of molecular tools to complement 16S rRNA gene-based approaches and allow meaningful interpretations regarding the presence and distribution of 1,2-D-dechlorinating Dehalococcoides populations.
Acknowledgments
This research was supported by a National Science Foundation CAREER award (award number 0090496) to F.E.L. and, in part, by Regenesis and the Strategic Environmental Research and Development Program (contract DACA72-00-C-0023).
Footnotes
In memory of Olga Maltseva, a great scientist, teacher, and friend.
REFERENCES
- 1.Adrian, L., U. Szewzyk, J. Wecke, and H. Görisch. 2000. Bacterial dehalorespiration with chlorinated benzenes. Nature 408:580-583. [DOI] [PubMed] [Google Scholar]
- 2.Baena, S., M.-L. Fardeau, T. H. S. Woo, B. Ollivier, M. Labat, and B. K. C. Patel. 1999. Phylogenetic relationships of three amino-acid-utilizing anaerobes, Selenomonas acidaminovorans, ‘Selenomonas acidaminophila’ and Eubacterium acidaminophilum, as inferred from partial 16S rDNA nucleotide sequences and proposal of Termonaerovibrio acidaminovorans gen. nov., comb. nov. and Anaeromusa acidaminophila gen. nov., comb. nov. Int. J. Syst. Bacteriol. 49:969-974. [DOI] [PubMed] [Google Scholar]
- 3.Bosma, T., J. Damborsky, G. Stucki, and D. B. Janssen. 2002. Biodegradation of 1,2,3-trichloropropane through directed evolution and heterologous expression of a haloalkane dehalogenase gene. Appl. Environ. Microbiol. 68:3582-3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chastain, M., and I. Tinoco, Jr. 1991. Structural elements in RNA. Prog. Nucleic Acid Res. Mol. Biol. 41:131-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crosby, L. D., and C. S. Criddle. 2003. Understanding bias in microbial community analysis techniques due to rrn operon copy number heterogeneity. BioTechniques 34:790-794, 796, 798 passim. [DOI] [PubMed]
- 6.De Wever, H., J. R. Cole, M. R. Fettig, D. A. Hogan, and J. M. Tiedje. 2000. Reductive dehalogenation of trichloroacetic acid by Trichlorobacter thiogenes gen. nov., sp. nov. Appl. Environ. Microbiol. 66:2297-2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Wildeman, S., G. Diekert, H. Van Langenhove, and W. Verstraete. 2003. Stereoselective microbial dehalorespiration with vicinal dichlorinated alkanes. Appl. Environ. Microbiol. 69:5643-5647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Wildeman, S., A. Neumann, G. Diekert, and W. Verstraete. 2003. Growth-substrate dependent dechlorination of 1,2-dichloroethane by a homoacetogenic bacterium. Biodegradation 14:241-247. [DOI] [PubMed] [Google Scholar]
- 9.El Fantroussi, S., J. Mahillon, H. Naveau, and S. N. Agathos. 1997. Introduction of anaerobic dechlorinating bacteria into soil slurry microcosms and nested-PCR monitoring. Appl. Environ. Microbiol. 63:806-811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gossett, J. M. 1987. Measurement of Henrys law constant for C1 and C2 chlorinated hydrocarbons. Environ. Sci. Technol. 21:202-208. [Google Scholar]
- 11.Hage, J. C., D. G. Kiestra, and S. Hartmans. 2001. Co-metabolic degradation of chlorinated hydrocarbons by Pseudomonas sp. strain DCA1. Appl. Environ. Microbiol. 57:548-554. [DOI] [PubMed] [Google Scholar]
- 12.He, J., K. M. Ritalahti, M. R. Aiello, and F. E. Löffler. 2003. Complete detoxification of vinyl chloride (VC) by an anaerobic enrichment culture and identification of the reductively dechlorinating population as a Dehalococcoides species. Appl. Environ. Microbiol. 69:996-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He, J., K. M. Ritalahti, K.-L. Yang, S. S. Koenigsberg, and F. E. Löffler. 2003. Detoxification of vinyl chloride to ethene coupled to growth of an anaerobic bacterium. Nature 424:62-65. [DOI] [PubMed] [Google Scholar]
- 14.Hendrickson, E. R., J. A. Payne, R. M. Young, M. G. Starr, M. P. Perry, S. Fahnestock, D. E. Ellis, and R. C. Ebersole. 2002. Molecular analysis of Dehalococcoides 16S ribosomal DNA from chloroethene-contaminated sites throughout North America and Europe. Appl. Environ. Microbiol. 68:485-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hengstmann, U., K. J. Chin, P. H. Janssen, and W. Liesack. 1999. Comparative phylogenetic assignment of environmental sequences of genes encoding 16S rRNA and numerically abundant culturable bacteria from an anoxic rice paddy soil. Appl. Environ. Microbiol. 65:5050-5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu, W. T., T. L. Marsh, H. Cheng, and L. J. Forney. 1997. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl. Environ. Microbiol. 63:4516-4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Löffler, F. E., J. E. Champine, K. M. Ritalahti, S. J. Sprague, and J. M. Tiedje. 1997. Complete reductive dechlorination of 1,2-dichloropropane by anaerobic bacteria. Appl. Environ. Microbiol. 63:2870-2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Löffler, F. E., J. R. Cole, K. M. Ritalahti, and J. M. Tiedje. 2002. Diversity of dechlorinating bacteria, p. 53-87. In M. M. Häggblom and D. Bossert (ed.), Dehalogenation: microbial processes and environmental applications. Kluwer Academic Press, New York, N.Y.
- 19.Löffler, F. E., K. M. Ritalahti, and J. M. Tiedje. 1997. Dechlorination of chloroethenes is inhibited by 2-bromoethanesulfonate in the absence of methanogens. Appl. Environ. Microbiol. 63:4982-4985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Löffler, F. E., Q. Sun, J. Li, and J. M. Tiedje. 2000. 16S rRNA gene-based detection of tetrachloroethene (PCE)-dechlorinating Desulfuromonas and Dehalococcoides species. Appl. Environ. Microbiol. 66:1369-1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Löffler, F. E., J. M. Tiedje, and R. A. Sanford. 1999. Fraction of electrons consumed in electron acceptor reduction and hydrogen thresholds as indicators of halorespiratory physiology. Appl. Environ. Microbiol. 65:4049-4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lonergan, D. J., H. L. Jenter, J. D. Coates, E. J. P. Philips, T. M. Schmidt, and D. R. Lovley. 1996. Phylogenetic analysis of dissimilatory Fe(III)-reducing bacteria. J. Bacteriol. 178:2402-2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mabey, W. R., J. H. Smith, R. T. Podoll, H. L. Johnson, and T. Mill. 1982. Aquatic fate process data for organic priority pollutants. U.S. Environmental Protection Agency report no. 440/4-81-014. U.S. Environmental Protection Agency, Washington, D.C.
- 24.Maidak, B. L., J. R. Cole, T. G. Lilburn, C. T. Parker, Jr., P. R. Saxman, R. J. Farris, G. M. Garrity, G. J. Olsen, T. M. Schimdt, and J. M. Tiedje. 2001. The RDP-II (Ribosomal Database Project). Nucleic Acids Res. 29:173-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marsh, T. L. 1999. Terminal restriction fragment length polymorphism (T-RFLP): an emerging method for characterizing diversity among homologous populations of amplicons. Curr. Opin. Microbiol. 2:323-327. [DOI] [PubMed] [Google Scholar]
- 26.Ouattara, A. S., A. S. Traore, and J.-L. Garcia. 1992. Characterization of Anaerovibrio burkinabensis sp. nov., a lactate-fermenting bacterium isolated from rice field soils. Int. J. Syst. Bacteriol. 42:390-397. [Google Scholar]
- 27.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 28.Schlötelburg, C., C. von Wintzingerode, R. Hauck, F. von Wintzingerode, W. Hegemann, and U. B. Göbel. 2002. Microbial structure of an anaerobic bioreactor population that continuously dechlorinates 1,2-D. FEMS Microbiol. Ecol. 39:229-237. [DOI] [PubMed] [Google Scholar]
- 29.Schlötelburg, C., F. von Wintzingerode, R. Hauck, W. Hegemann, and U. B. Göbel. 2000. Bacteria of an anaerobic 1,2-dichloropropane dechlorinating mixed culture are phylogenetically related to those of other anaerobic dechlorinating consortia. Int. J. Syst. Bacteriol. 50:1505-1511. [DOI] [PubMed] [Google Scholar]
- 30.Strompl, C., B. J. Tindall, G. N. Jarvis, H. Lunsdorf, E. R. Moore, and H. Hippe. 1999. A re-evaluation of the taxonomy of the genus Anaerovibrio, with the reclassification of Anaerovibrio glycerini as Anaerosinus glycerini gen. nov., comb. nov., and Anaerovibrio burkinabensis as Anaeroarcus burkinensis [corrig.] gen. nov., comb. nov. Int. J. Syst. Bacteriol. 49:1861-1872. [DOI] [PubMed] [Google Scholar]
- 31.Sung, Y., K. M. Ritalahti, R. A. Sanford, J. W. Urbance, S. J. Flynn, J. M. Tiedje, and F. E. Löffler. 2003. Characterization of two tetrachloroethene (PCE)-reducing, acetate-oxidizing anaerobic bacteria and their description as Desulfuromonas michiganensis sp. nov. Appl. Environ. Microbiol. 69:2964-2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Swanson, P. E. 1999. Dehalogenases applied to industrial-scale biocatalysis. Curr. Opin. Biotechnol. 10:365-369. [DOI] [PubMed] [Google Scholar]
- 33.Terzenbach, D. P., and M. Blaut. 1994. Transformation of tetrachloroethylene to trichlororethylene by homoacetogenic bacteria. FEMS Microbiol. Lett. 123:213-218. [DOI] [PubMed] [Google Scholar]
- 34.Tesoriero, A. J., F. E. Löffler, and H. Liebscher. 2001. The fate and origin of fumigant-related compounds in an unconfined shallow aquifer. Environ. Sci. Technol. 35:455-461. [DOI] [PubMed] [Google Scholar]
- 35.Zhou, J., M. E. Davey, J. B. Figueras, E. Rivkina, D. Gilichinsky, and J. M. Tiedje. 1997. Phylogenetic diversity of a bacterial community determined from Siberian tundra soil. Microbiology 143:3913-3919. [DOI] [PubMed] [Google Scholar]