Abstract
Yarrowia lipolytica contains five acyl-coenzyme A oxidases (Aox), encoded by the POX1 to POX5 genes, that catalyze the limiting step of peroxisomal β-oxidation. In this study, we analyzed morphological changes of Y. lipolytica growing in an oleic acid medium and the effect of POX deletions on lipid accumulation. Protrusions involved in the uptake of lipid droplets (LDs) from the medium were seen in electron micrographs of the surfaces of wild-type cells grown on oleic acid. The number of protrusions and surface-bound LDs increased during growth, but the sizes of the LDs decreased. The sizes of intracellular lipid bodies (LBs) and their composition depended on the POX genotype. Only a few, small, intracellular LBs were observed in the mutant expressing only Aox4p (Δpox2 Δpox3 Δpox5), but strains expressing either Aox3p or both Aox3p and Aox4p had the same number of LBs as did the wild type. In contrast, strains expressing either Aox2p or both Aox2p and Aox4p formed fewer, but larger, LBs than did the wild type. The size of the LBs increased proportionately with the amount of triacylglycerols in the LBs of the mutants. In summary, Aox2p expression regulates the size of cellular triacylglycerol pools and the size and number of LBs in which these fatty acids accumulate.
The yeast Yarrowia lipolytica can grow on alkanes or fatty acids as the sole carbon source (for a review, see reference 5). Alkanes must be converted to fatty acids before they can serve as substrates for different metabolic pathways. In mammalian cells, the degradation of fatty acids occurs through β-oxidation in mitochondria and peroxisomes. In yeasts, the enzymes in this pathway are present only in peroxisomes. Fatty acid degradation is a multiple-step process requiring four different enzymatic activities. The first step is catalyzed by an acyl-coenzyme A (CoA) oxidase (Aoxp). Saccharomyces cerevisiae contains only one Aox (28), but Y. lipolytica contains five Aox isoenzymes with different substrate specificities and different activity levels (Fig. 1) (27, 28). Aox3p is specific for short-chain acyl-CoAs (17), Aox2p preferentially oxidizes long-chain acyl-CoAs (18), and the Aox4p and Aox5p activities do not appear to be sensitive to the length of the aliphatic chain of the acyl-CoA (29). Previous studies revealed that the mutants MTLY36 (Δpox2 Δpox3 Δpox5) and MTLY37 (Δpox2 Δpox3 Δpox4 Δpox5) either had a growth defect or did not grow at all, respectively, when cultivated on oleic acid minimal medium (28).
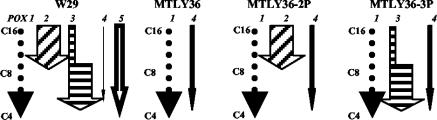
FIG. 1.
Schematic representation of acyl-CoA oxidase genotypes and activities in Y. lipolytica strains W29, MTLY36, MTLY36-2P, and MTLY36-3P. The wild-type strain W29 contains five POX genes (POX1 to POX5) coding for the acyl-CoA oxidases Aox1p to Aox5p. The mutant strain MTL36 expresses only Aox1p (inactive) and Aox4. In MTL36-2P, we reintroduced the POX2 gene, and in MTLY36-3P, we reintroduced the POX3 gene. The POX genotype is indicated at the top and the Aox activity is indicated by an arrow; the length indicates the chain-length specificity (C16, C16-CoA; C8, C8-CoA; C4, C4-CoA) and the width represents the activity relative to that in the wild-type strain.
Fatty acids can also be utilized as storage molecules when they are incorporated into triacylglycerols. All eukaryotic organisms and some gram-positive bacteria store triacylglycerols in intracellular compartments that are variously termed lipid particles, lipid droplets (LDs), lipid bodies (LBs), oil bodies, oleosomes, or spherosomes (in plants). The structure of these lipid-rich compartments is similar in all cell types and is rather simple: LBs consist of a hydrophobic core formed from neutral lipids, mainly triacylglycerols and/or steryl esters, which is surrounded by a phospholipid monolayer with a few embedded proteins (for a review, see reference 31). LBs were first considered to serve only as an energy source and/or a source of fatty acids and sterols needed for membrane biogenesis. More recently, however, the participation of LBs in the formation of specific lipophilic components, e.g., steroid hormones or prostaglandins, was demonstrated (6, 11, 12, 30). Several hypotheses to explain the mechanism of LB formation have been proposed. LB biogenesis probably results from budding of the endoplasmic reticulum (ER) (21). Enzymes in the ER synthesize neutral lipids that are deposited between the two leaflets of the ER membrane to form a pre-LB. When this structure reaches a certain size, it buds off to form a mature LB (31).
Our objective in this study was to define the effects of the acyl-CoA oxidases of Y. lipolytica on (i) cellular growth and morphology and (ii) the lipid content and fatty acid composition of the LDs. We hypothesized that protrusions on the surfaces of cells grown in the presence of fatty acids play a role in the uptake of these compounds from the medium. We found that the Aox composition affects the lipid content and fatty acid composition of the LBs, either positively or negatively. Particularly, a mutant expressing Aox2p had an increased lipid content, which was accumulated in LBs, mainly as triglycerides.
MATERIALS AND METHODS
Experimental rationale.
Fatty acid adsorption, consumption, and intracellular accumulation were evaluated in a set of mutants lacking certain POX genes encoding Aox proteins. Aox2p or Aox3p was reintroduced into the mutants. After precultures were grown in a glucose-containing medium, the cells were cultivated on an oleic acid medium to determine the role of the POX gene products in fatty acid metabolism.
Yeast strains, growth, and culture conditions.
The Y. lipolytica strains used for this study were derived from the wild-type Y. lipolytica strain W29 (ATCC 20460) or from mutants that contained disruptions of one or more of the POX genes (Table 1). Escherichia coli DH5α was used for gene manipulations. The media and growth conditions used for Y. lipolytica were described by Barth and Gaillardin (5) and those for E. coli were described by Sambrook et al. (26). We introduced a uracil auxotrophy into strain MTLY37 by transformation, using a 1.5-kb PCR fragment carrying the ura3-41 allele, followed by the selection of transformants on YNB-5-fluoroorotic acid medium (9), which gave rise to strain MTLY40. Rich medium (YPD), minimal glucose medium (YNB), and minimal medium with Casamino Acids (YNBcas) were prepared as described previously (28). Minimal oleic acid medium (YNBO) contained 1.7 g of yeast nitrogen base (without amino acids and ammonium sulfate [YNBww]) (Difco, Paris, France)/liter, 5 g of NH4Cl/liter, 1 g of yeast extract/liter, 50 g of oleic acid (Merck, Fontenay-sous-Bois Cedex, France)/liter, and 50 mM phosphate buffer (pH 6.8). Oleic acid was emulsified by sonication in the presence of 0.2 g of Tween 40/liter (19). Uracil (100 mg/liter) was added as required. Cells grown to the exponential-growth phase on YPD were used as a preculture for inoculation of the respective fresh medium to an optical density at 600 nm (OD600) of 0.5.
TABLE 1.
Y. lipolytica strains used for this study
| Strain | Strain no. | Genotype | Auxotrophy | Reference |
|---|---|---|---|---|
| W29 | JMY399 | Mata | None | 5 |
| MTLY35 | JMY154 | Δpox2 Δpox5 pox3::URA3 | None | 28 |
| MTLY36 | JMY155 | Δpox2 Δpox3 Δpox5 | Ura− | 28 |
| MTLY37 | JMY172 | Δpox2 Δpox3 Δpox5 pox4::URA3 | None | 28 |
| MTLY40 | JMY755 | Δpox2 Δpox3 Δpox4 Δpox5 | Ura− | This study |
| MTLY36-2P | JMY798 | Δpox2 Δpox3 Δpox5 POX2-URA3 | None | This study |
| MTLY36-3P | JMY805 | Δpox2 Δpox3 Δpox5 POX3-URA3 | None | This study |
| MTLY40-2P | JMY794 | Δpox2 Δpox3 Δpox4 Δpox5 POX2-URA3 | None | This study |
| MTLY40-3P | JMY802 | Δpox2 Δpox3 Δpox4 Δpox5 POX3-URA3 | None | This study |
To measure cell growth, we centrifuged the cultures at 10,000 × g for 10 min at 20°C and washed the cell pellet twice with equal volumes of solution S (9 g of NaCl/liter) or solution SB (9 g of NaCl/liter plus 5 g of bovine serum albumin [BSA]/liter). Biomass production was determined by measuring the OD600 and estimating the cell dry weight after drying at 80°C for 24 h.
Construction of pYEG1-POX2 and pYEG1-POX3 plasmids.
The POX2 and POX3 genes were amplified by PCR with oligonucleotide primer pairs POX2-ATG/POX2-STOP and POX3-ATG/POX3-STOP, respectively. The sequences of the primers were as follows: POX2-ATG, 5′-TCCGCCTAGGCACAATGAACCCCAACAACACTGGCACCATTG-3′; POX2-STOP, 5′-CAGGCCCGCGGGGCCCTATTCCTCATCAAGCTCGCAAATGTC-3′; POX3-ATG, 5′-GATCCGCCTAGGCACAATGATCTCCCCCAACCTCACAGCT-3′; and POX3-STOP, 5′-GAGGCCCGCGGGGCCCTATTCCTCGTCCAGCACGCAAATG-3′. The PCR fragments were cleaved with BlnI and SfiI and inserted into the plasmid pYEG1 (22), giving rise to the plasmids pYEG1-POX2 and pYEG1-POX3, respectively. These plasmids allow the expression of the POX2 and POX3 genes under the control of the POX2 promoter. The POX2 and POX3 promoters have similar strengths (22). Both expression cassettes were transformed into MTLY36 and MTLY40 by a lithium acetate transformation technique (5). Prior to the transformation of yeast cells, the plasmids were digested with NotI to free the expression cassette, and Ura+ transformants were selected on YNBcas (22).
Electron microscopy.
Cells were prefixed by the addition of glutaraldehyde (final concentration, 2%) to a culture that had been growing for 1 h at 28°C with shaking at 250 rpm. The cells were harvested by centrifugation for 10 min at 10,000 × g at 20°C, resuspended in 50 mM phosphate buffer (pH 6.8) containing 3% glutaraldehyde to an OD600 of 10, and fixed for 24 h at room temperature. For specific lipid staining, the cells were postfixed with 2% OsO4 in 200 mM imidazole buffer (pH 7.5) for 1 h (3). After being washed with 100 mM imidazole buffer (pH 7.5), the cells were dehydrated in a graded acetone series in the following order: 30, 40, 70, and 100% (vol/vol).
Samples for cryo-scanning electron microscopy (SEM) were spotted onto HTTP 0.4-μm-pore-size filters (Millipore Corp., St. Quentin Yvelines, France), mounted, and cryofixed with an Oxford System Cryotrans CT 1500 instrument. The samples were then coated with gold, frozen under liquid nitrogen, and vacuum dried at −160°C. For inspection of the samples, a Phillips SM525 M microscope (Phillips Electron Optics, Eindhoven, The Netherlands) and a CCD ISIS 200 camera (Megaview III; Eloïse, Roissy CDG, France) were used. Picture exposition was performed with a soft imaging analysis system (Eloïse).
For transmission electron microscopy (TEM), the cells were embedded in medium LR white resin and 50-nm-thick sections were cut with an Ultracut E ultramicrotome (Leica, Rueil Malmaison, France). Sections were mounted on 200-mesh grids and examined with a Phillips 420 microscope (Phillips Electron Optics).
Light microscopy.
Ten milliliters of a growing yeast culture was prefixed by the addition of 1.34 ml of a formaldehyde stock solution (50 mM potassium phosphate buffer [pH 6.8], 0.5 mM MgCl2, 4.8% formaldehyde) and then incubated for 1 h at 28°C with shaking at 250 rpm. The prefixed cells were harvested, resuspended to an OD600 of 2.5 in the formaldehyde stock solution, and incubated for 5 h at room temperature. The cells were washed twice with 50 mM potassium phosphate buffer (pH 6.8) and stored in 100 mM potassium phosphate buffer (pH 7.5) at an OD600 of 2.5 at 4°C until light microscopy observation. For the visualization of LBs, Nile red (1-mg/ml solution in acetone) (Molecular Bioprobe, Montluçon, France) was added to the cell suspension (1:10 [vol/vol]) and incubated for 1 h at room temperature. The cells were harvested, washed twice with distilled water, and resuspended in 50 mM potassium phosphate buffer (pH 6.8) to an OD600 of 2.5. Microscopy was performed with an Olympus BX 51 light microscope (Micro Mecanique, Evry, France) with a 100× oil immersion objective. To record pictures, we used Photometrics CoolSNAP software (Micro Mecanique).
Isolation and characterization of LBs.
LBs were isolated from yeast cells grown to the early stationary phase. In brief, Yarrowia cells were grown on oleic acid medium to the early stationary phase (20 h after transfer into YNBO medium), harvested, washed three times with 5 g of BSA/liter and once with H2O, and converted to spheroplasts (8, 15). Spheroplasts were washed twice with 1.2 M sorbitol in 20 mM potassium phosphate buffer (pH 7.4) and homogenized in a lysis buffer containing 10 mM morpholineethanesulfonic acid (MES)-Tris (pH 6.9), 12% Ficoll 400, 0.2 mM EDTA, and 1 mM phenylmethylsulfonyl fluoride to a final concentration of 0.2 g of cell wet weight/ml. The homogenate was overlaid with lysis buffer and centrifuged for 1 h at 100,000 × g at 4°C in an SW-28 swinging bucket rotor (Kontron). The floating layer, containing LBs, was collected from the top of the gradient and resuspended in lysis buffer. The LBs were further purified by three additional sequential floating steps in (i) 10 mM MES-Tris (pH 6.9), 8% Ficoll 400, 0.2 mM EDTA, and 1 mM phenylmethylsulfonyl fluoride; (ii) 10 mM MES-Tris (pH 6.9), 0.25 M sorbitol, and 0.2 mM EDTA; and (iii) 10 mM MES-Tris (pH 6.9), 0.15 M sorbitol, and 0.2 mM EDTA, with centrifugation at 100,000 × g for 30 min at 4°C after each step. The floating layer from the top of the last gradient contained LBs.
Lipid determination.
The lipid composition of the medium (ExLip) and the composition of lipids present on the cell surface (SurfLip), in total cells (InLip), and in LBs (LBLip) were determined as follows. For ExLip, yeast cultures were centrifuged for 10 min at 10,000 × g at 20°C, and the supernatants were collected for lipid analysis. Harvested cells were washed twice with either aqueous BSA (5 g/liter) (Sigma-Aldrich, St. Louis, Mo.) or hexane (20), giving rise to fractions containing SurfLip. InLip was extracted (10) from cells washed with SB buffer (9 g of NaCl/liter plus 5 g of BSA/liter), and LBLip was extracted from isolated LBs. Heptadecanoic acid was added as an internal standard.
Thin-layer chromatographic (TLC) analysis of the InLip was performed on silica gel 60 F254 plates (Merck, VWR, Fontenay sous Bois, France) with hexane-acetic acid-diethyl ether (40:1:10 [vol/vol/vol]) as the developing solvent. For the quantification of lipids, standard lipids (5 or 10 μg) were separated on the same plate. Lipids were visualized with phosphomolybdenic acid, the plates were scanned at a 300-dpi resolution, and the images were stored as TIFF files. The amounts of lipids were determined by analyses of image files with Image Quant (version 4.2a) software (Molecular Dynamics; Amersham Biosciences Europe GmbH, Orsay, France).
To analyze the neutral lipid composition of isolated LBs (LBLip), we applied extracts to silica gel 60 plates with the aid of a sample applicator (Linomat IV; CAMAG, Muttenz, Switzerland) and then developed and analyzed chromatograms as previously described (4).
For the analysis of fatty acid species, fatty acids were converted to methyl esters by methanolysis with BF3-methanol (14%), as described previously (4), and were analyzed by using an SP-2380 (Varian, Les Ulis, France) gas chromatograph equipped with a capillary column (30 m by 0.25 mm, 0.2-μm-thick film, poly 90% biscyanopropyl-10% cyanopropylphenyl siloxane, with He as a carrier gas at a linear flow rate of 4.3 ml/min). The temperature program used for the analysis of fatty acid species was as follows: injection temperature, 220°C; flame ionization detector temperature, 250°C; and increase from 120 to 210°C at a rate of 3°C/min. Fatty acid species were identified and quantified by comparisons to fatty acid methyl ester standards (Laboratoire de Biochimie INRA, Rennes, France).
Protein analysis.
Proteins were quantified by the method of Lowry et al. (16), with BSA used as a standard. Prior to protein analysis of the LB fraction, the samples were delipidated. Lipids were extracted with 2 volumes of diethyl ether, the organic phase was discarded, and residual diethyl ether was removed under a stream of nitrogen. Proteins were precipitated from the extracted aqueous phase with trichloroacetic acid (final concentration, 10%) and were solubilized in 0.1% sodium dodecyl sulfate in 0.1 M NaOH. Cross-contamination of the LB fraction with peroxisomes was tested by Western blotting (13) using antisera raised against Aox3p (28).
RESULTS
Growth of Y. lipolytica mutants with multiple deletions of Aox proteins.
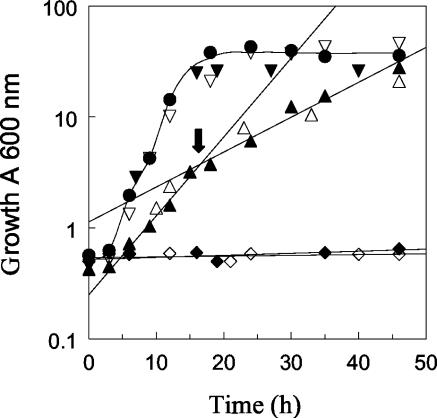
To determine if the growth defect of the triple MTLY36 (Δpox2 Δpox3 Δpox5) and quadruple MTLY37 (Δpox2 Δpox3 Δpox4 Δpox5) deletion mutants could be rescued by the expression of either Aox2p or Aox3p, we reintroduced either POX2 or POX3 into the mutants. Transformation with both genes fully restored the growth defect of the triple and quadruple deletion mutants (Fig. 2), indicating that the expression of either Aox2p or Aox3p is sufficient for growth on an oleic acid medium. Furthermore, diauxic growth was observed for the triple mutant MTLY35 and its Ura− derivative MTLY36 (Fig. 2, arrow), with the first μmax being 0.50 ± 0.02 h−1 and the second being 0.25 ± 0.02 h−1, which may reflect differences in fatty acid utilization.
FIG. 2.
Growth of wild-type W29 and mutant strains with altered POX genotypes in oleic acid medium (YNBO). The growth of the wild-type and mutant strains listed in Table 1 was monitored over time. Symbols: closed circles, W29; closed triangles, MTLY35; open triangles, MTLY36; open inverted triangles, MTLY36-2P; closed inverted triangles, MTLY36-3P; open diamonds, MTLY37; and closed diamonds, MTLY40. The arrow indicates the diauxic growth shift. The results are mean values from three independent experiments. The standard deviations were <10% of the values of the points.
Cell surface and intracellular structure of Y. lipolytica grown on oleic acid.
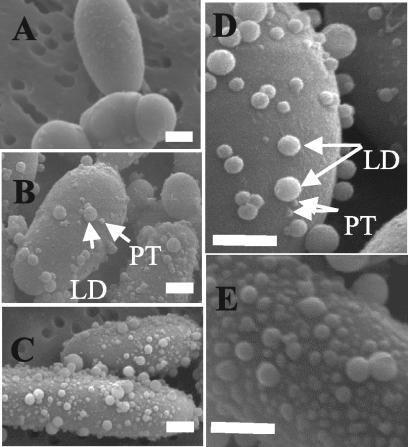
Wild-type W29 cells were grown in YPD medium to the exponential growth phase and then transferred to a minimal medium containing oleic acid, decane (C10), or glucose as the carbon source. After the shift to the fresh medium, cell morphologies were compared by cryo-SEM. Glucose-grown cells had smooth surfaces (Fig. 3A), while cells grown on fatty acids had small protrusions scattered across their surfaces (Fig. 3B). The number of protrusions increased with the time of incubation with oleic acid (Fig. 3C). LDs were also observed on the cell surfaces, and their number increased with the number of protrusions. The LD sizes ranged from 20 to 500 nm in diameter and decreased during growth on oleic acid. Two hours after induction, few protrusions or LDs were seen (Fig. 3D). The number of protrusions increased 18 h after induction (Fig. 3E).
FIG. 3.
SEM of Y. lipolytica cells grown in YNBO medium. Glucose-grown cells were transferred into oleic acid medium, and samples were withdrawn and fixed at various time points. SEM micrographs of exponentially growing cells in glucose (A) and of cells grown in YNBO medium at 2 h (B) and 18 h (C) revealed that, as an adaptive response for the utilization of hydrophobic substrates, protrusions (PT) appeared on the yeast cell surfaces. LDs were visible on the cell surfaces. The numbers of both protrusions and LDs increased between 2 and 18 h. (D and E) Enlargement of cell surfaces at 2 and 18 h, respectively. Bar = 1 μm.
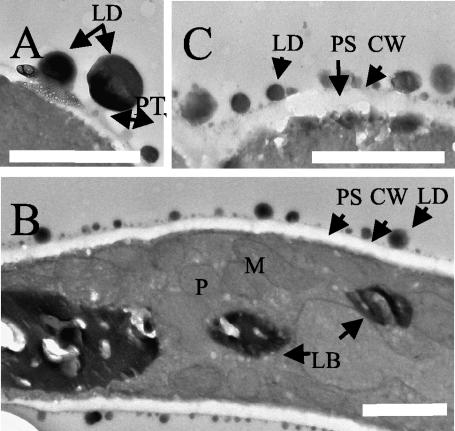
The numbers and sizes of protrusions and LDs were also seen by TEM. The LDs on the cell surface appeared as spherical black structures after osmium staining (Fig. 4). After a 2-h shift to oleic acid medium, the number of LDs per micrometer at the cell surface in TEM pictures was 4.0 ± 0.5, with an average diameter of 310 ± 18 nm. After 40 h, the number of LDs increased to 7.3 ± 0.4/μm of cell surface, whereas the average diameter decreased to 51 ± 30 nm. LDs were attached to protrusions (Fig. 4), which are labeled less with osmium (gray) than are the LDs (black). Differences in the thickness of the cell wall and the width of the periplasmic space were observed for cells grown on glucose and oleic acid, as was previously described for Y. lipolytica grown on crude oil (14). The periplasmic space width increased from approximately 80 nm (at 2 h) (Fig. 4A) to approximately 150 nm (at 40 h) (Fig. 4C) after transfer to an oleic acid-containing medium, and the cell wall thickness decreased from 40 to 25 nm.
FIG. 4.
TEM of Y. lipolytica cells grown in YNBO medium. Glucose-grown cells were transferred into oleic acid medium, and samples were withdrawn and fixed at various time intervals after the transfer. TEM micrographs revealed lipids (black staining) as LDs on the cell surface or as an intracellular accumulation of LBs. The cells were fixed at 2 h (A), 18 h (B), and 40 h (C). LDs were bound at the tops of protrusions (PT) (A and B). Bar = 1 μm. Abbreviations: CW, cell wall; M, mitochondria; P, peroxisome; and PS, periplasmic space.
Differences in the intracellular structure were also observed when cells were shifted to oleic acid medium. The number of peroxisomes increased (data not shown), and lipids accumulated as LBs in the cytoplasm (Fig. 4B). Cells grown on oleic acid were fixed with formaldehyde at different growth phases and were stained with either fluorescent lipids or fluorescent dyes specifically directed against neutral lipids. Small LBs were observed 6 h after the transfer of cells to YNBO medium, but only a few small LBs were observed in glucose-grown cells. The number of LBs decreased during growth on YNBO medium, but their size increased. Thus, LBs filled a large part of the cell 40 h after induction (data not shown).
Lipid accumulation in POX mutants during growth on oleic acid medium.
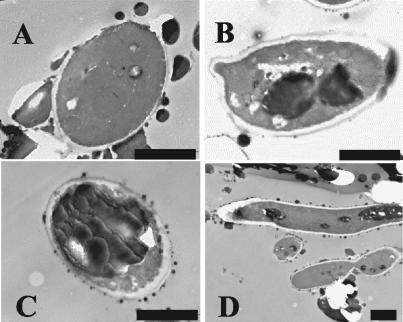
MTLY36, MTLY40, and their derivatives were transformed with a plasmid encoding either Aox2p or Aox3p, grown on YNBO medium, and stained with Nile red or observed by TEM. The number of LBs in all strains expressing Aox3p was similar to their number in wild-type cells. More LBs were observed in strains expressing Aox2p, but in the triple mutant MTLY35, a smaller number of small LBs was present (data not shown). The identity of the observed structures as LBs and differences in the level of lipid accumulation by the POX mutants were confirmed by TEM (Fig. 5). Few, small LBs were observed in W29 cells 2 h after transfer to an oleic acid-containing medium (Fig. 5A). After 40 h of growth in oleic acid-containing medium, the presence of one to three large and a few small LBs was observed in the wild-type strain (Fig. 5B). In strains expressing Aox2p, one large LB filled almost the entire cell 40 h after induction (Fig. 5C). MTLY35 accumulated smaller LBs than the wild type did (Fig. 5D).
FIG. 5.
Lipid accumulation in LBs of wild-type strain W29 and of mutants with altered POX genotypes. Strains W29 (A and B), MTLY36-2P (C), and MTLY35 (D) were grown in YNBO medium in baffled Erlenmeyer flasks at 28°C with shaking at 250 rpm. At different time intervals, samples were harvested, fixed, and prepared for TEM. (A) Strain W29 at 2 h; (B to D) Strains W29 (B), MTLY36-2P (C), and MTLY35 (D) grown on LB for 40 h. Bar = 2 μm.
Effect of POX mutations on lipid content of Y. lipolytica during growth on oleic acid.
The ExLip in the growth medium, SurfLip, and InLip of strains W29, MTLY35, and MTLY36-2P were analyzed by TLC and gas-liquid chromatography. ExLip and SurfLip were similar in composition for all three strains when the strains were grown on oleic acid, indicating that no accumulation of fatty acid intermediates occurred in the medium.
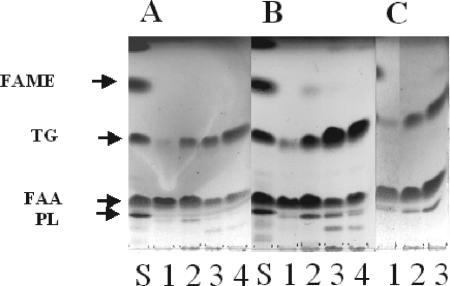
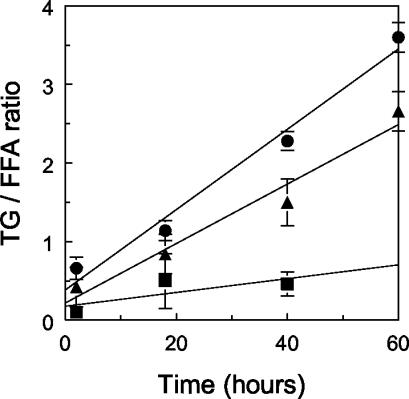
There were differences in InLip that depended on the POX genotype. We observed primarily free fatty acids (FFAs) after 2 h of growth in oleic acid-containing medium (Fig. 6A and B, lanes 1). Over time, the amount of FFAs decreased and the amount of triacylglycerols increased (Fig. 6A and B, lanes 4). At 18 h, the level of triacylglycerols that had accumulated in strain MTLY35 was lower than that in strain W29, and strain MTLY36-2P had a higher level of triacylglycerols than strain W29. The TLC plates were scanned, and the triacylglycerol/FFA ratios were calculated (Fig. 7). MTLY35 accumulated low levels of lipids, mainly as FFAs, while MTLY36-2P accumulated more lipids than the wild type, mainly as triacylglycerols. Thus, MTLY35 does not accumulate lipids, the wild type accumulates both FFAs and triacylglycerols, and MTLY36-2P accumulates a higher level of lipids, primarily triacylglycerols.
FIG. 6.
Cellular lipid analysis in wild-type strain W29 and in mutants with altered POX genotypes. Strains W29, MTLY36-2P, and MTLY35 were grown in YNBO medium, pH 6.8, at 28°C. The total cellular lipids were extracted and analyzed on TLC plates at different times of growth. A time course analysis for W29 (A) and MTLY36-2P (B) at 2 h (lanes 1), 18 h (lanes 2), 40 h (lanes 3), and 60 h (lanes 4) and the cellular lipid composition for strains MTLY35 (lane 1), W29 (lane 2), and MTLY36-2P (lane 3) at 18 h (C) are shown. The lipid standard (S) was composed of cholesterol (20%), cholesteryl oleate (20%) (PL), oleic acid (20%) (FFA), methyl oleate (20%) (FAME), and triolein (20%) (TG).
FIG. 7.
Triglyceride/FFA ratio (TG/FFA ratio) over time for wild-type strain W29 (triangles) and the mutants MTLY35 (squares) and MTLY36-2P (circles). The results are means of four different TLC analyses. Error bars indicate standard deviations.
LB content.
LBs were isolated after 20 h of growth on YNBO medium (Table 2). Strain MTLY36-2P accumulated more neutral lipids (triacylglycerols, ergosterol, and steryl esters) in this compartment than the wild type did, but only small amounts of neutral lipids were present in strain MTLY35. Introducing the POX2 gene into the triple deletion mutant restored triacylglycerol accumulation in the LBs and resulted in a higher accumulation of triacylglycerols than in the wild type.
TABLE 2.
Lipid content of isolated LBs from Y. lipolytica wild-type strain W29 and POX-altered mutants MTL36-2P and MTLY35a
| Strain | Amt of lipid (μg/μg of protein)
|
||||
|---|---|---|---|---|---|
| Ergosterol (E) | Steryl esters (SE) | Triacylglycerol (TG) | Total neutral lipid | TG/lipid (E + SE) | |
| W29 | 0.06 | 0.17 | 13 | 14 | 56 |
| MTLY36-2P | 0.04 | 0.05 | 16 | 17 | 178 |
| MTLY35 | 0.03 | 0.01 | 0.39 | 0.43 | 10 |
LBs were isolated from YNBO medium-grown cells after 20 h of growth and were analyzed by TLC.
Fatty acid profile of lipid bodies.
The contents of unsaturated fatty acid species were similar in the wild type and strain MTLY36-2P, but the amount of saturated fatty acids was slightly, but significantly, smaller in strain MTLY35 (Table 3). The higher amount of C14:1 in MTLY35 than in the wild type reflects the inability of the mutant to degrade short-chain fatty acids.
TABLE 3.
Fatty acid profiles of isolated LBs from Y. lipolytica wild-type strain W29 and POX-altered mutants MTLY36-2P and MTLY35a
| Fatty acid | % of total fatty acids in strain:
|
||
|---|---|---|---|
| W29 | MTLY36-2P | MTLY35 | |
| C14:0 | 0.6 | NDb | NDb |
| C14:1 | 1.3 | 4.2 | 5.7 |
| C16:0 | 5.5 | 5.2 | 3.4 |
| C16:1 | 16 | 12 | 11 |
| C18:0 | 1.1 | 0.9 | NDb |
| C18:1 | 66 | 70 | 72 |
| C18:2 | 8.7 | 7.3 | 7.3 |
| C20:1 | 0.7 | 1.1 | 1.5 |
| Unsaturated/saturated fatty acid ratio | 13 | 15 | 28 |
| C14:1/C16:1/C18:1 | 2/24/100 | 6/17/100 | 8/15/100 |
LBs were isolated from YNBO medium-grown cells at 20 h. Fatty acids in the LBs were converted to methyl esters and analyzed by gas-liquid chromatography.
ND, not detected.
DISCUSSION
Y. lipolytica can use fatty acids and alkanes as carbon sources. This yeast secretes emulsifiers (liposan) (7), modifies its cell surface hydrophobicity (2), stores lipids in specifically developed organelles, and uses a special set of enzymes to store and use alkanes or lipids as carbon sources. The β-oxidation pathway, which is heavily involved in the utilization of these substrates, is a common target for modifying the ability of Y. lipolytica to use lipids and alkanes as carbon sources. Thus, we tested Y. lipolytica strains with different POX genotypes to determine the effect of the major acyl-CoA oxidases on lipid accumulation.
The expression of either Aox2p or Aox3p, encoded by POX2 and POX3, respectively, in Δpox2 Δpox3 Δpox5 or Δpox2 Δpox3 Δpox4 Δpox5 mutant backgrounds restored wild-type growth even though these enzymes differ in substrate specificity, i.e., Aox2p has a preference for long-chain fatty acids while Aox3p prefers short-chain fatty acids as a substrate (17, 18). One explanation for this observation is that the ratio of unsaturated to saturated fatty acids changes when Aox2p is expressed. This difference may lead to changes in the physical properties of the membranes and may alter growth properties.
Structural changes on the surfaces of cells grown on oleic acid result in the formation of protrusions that enable the yeast to take up the hydrophobic compounds from the medium. The number of protrusions on the cell surface during growth in a fatty acid-containing medium is accompanied by an increase in the number of LDs adhering to these structures. The size of the LDs decreases with time. These observations are consistent with a hypothesis that the protrusions are important for the uptake of neutral lipids by the cell. In addition to forming protrusions on the cell surface, cells grown in fatty acids also have an increased periplasmic space (Fig. 4), as previously described by Kim et al. (14), and a decreased cell wall thickness. These structural alterations also could facilitate the uptake of fatty acids.
Some changes in intracellular structures result from the absence or presence of certain acyl-CoA oxidases. The triple mutant MTLY35, which is defective in the three major Aox proteins, Aox2p, Aox3p, and Aox5p, forms few and relatively small intracellular LBs, raising the question of why this strain does not accumulate fatty acids as triacylglycerols in a large LB. One hypothesis is that this strain cannot efficiently degrade fatty acids. The small volume of LBs in this mutant could result from a low level of ATP, which is formed by β-oxidation in this cell but is preferentially used in pathways other than lipid storage. Alternatively, the lack of Aox proteins could downregulate triacylglycerol synthesis. Finally, a regulatory mechanism could prevent lipid storage in the form of triacylglycerols in LBs if long-chain fatty acids cannot be used as an energy source due to the absence of active Aox proteins. This last hypothesis is supported by our observation that, in contrast to the triple mutant MTLY35, MTLY36-2P, which expresses Aox2p, contains a large LB (Fig. 5C) and stores larger amounts of neutral lipids, mainly triacylglycerols, than does the wild type.
In conclusion, the accumulation of neutral lipids is strongly influenced by the presence of certain Aox proteins. Genetic engineering can therefore be used to improve the ability of Y. lipolytica to store and utilize lipids. A modification of the POX genotype could be a useful alternative to the modification of growth conditions and medium composition (1, 23-25) that was previously used to improve intracellular lipid accumulation in Y. lipolytica, making strain MTLY36-2P a good candidate for single-cell oil production.
Acknowledgments
We thank F. Jaunet and J. Laperonnie (Service de Microscopie Électronique, INRA, Versailles, France) for sample preparation and microscopic observations and Daniel Catheline (Laboratoire de Biochimie INRA, Rennes, France) for providing fatty acid methyl ester standards. E. Roux thanks Günther Daum for allowing her to work in his lab.
E. Roux thanks FEBS for providing a summer fellowship. K. Mlíčková was the recipient of a Marie Curie training fellowship during her stay at the INRA Grignon Yeasts of Technological Interest (YETI) training site. This work was financially supported by the Fonds zur Förderung der Wissenschaftlichen Forschung in Österreich (project 14468 to G.D. and project T113 to K.A.).
REFERENCES
- 1.Aggelis, G., and M. Komaitis. 1999. Enhancement of single cell oil production by Yarrowia lipolytica growing in the presence of Teucrium polium L. aqueous extract. Biotechnol. Lett. 21:747-749. [Google Scholar]
- 2.Aguedo, M., Y. Waché, V. Mazoyer, A. Sequeira-Le Grand, and J.-M. Belin. 2003. Increased electron donor and electron acceptor characters enhance the adhesion between oil droplets and cells of Yarrowia lipolytica as evaluated by a new cytometric assay. J. Agric. Food Chem. 7:3007-3011. [DOI] [PubMed] [Google Scholar]
- 3.Angermuller, S., and D. H. Fahimi. 1982. Imidazole-buffered osmium tetroxide: an excellent stain for visualization of lipids in transmission electron microscopy. Histochem. J. 14:823-835. [DOI] [PubMed] [Google Scholar]
- 4.Athenstaedt, K., D. Zweytick, A. Jandrositz, S. D. Kohlwein, and G. Daum. 1999. Identification and characterization of major lipid particle proteins of the yeast Saccharomyces cerevisiae. J. Bacteriol. 181:6441-6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barth, G., and C. Gaillardin. 1996. Yarrowia lipolytica, p. 313-388. In W. K. Wolf (ed.), Nonconventional yeasts in biotechnology, vol. 1. Springer-Verlag, Berlin, Germany. [Google Scholar]
- 6.Bisgaier, C. L., R. Chanderbhan, R. W. Hinds, and G. V. Vahouny. 1985. Adrenal cholesterol esters as substrate source for steroidogenesis. J. Steroid Biochem. 23:967-974. [DOI] [PubMed] [Google Scholar]
- 7.Cirigliano, M. C., and G. M. Carman. 1984. Isolation of a bioemulsifier from Candida lipolytica. Appl. Environ. Microbiol. 48:747-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daum, G., P. C. Böhni, and G. Schatz. 1982. Import of proteins into mitochondria. Cytochrome b2 and cytochrome c peroxidase are located in the intermembrane space of yeast mitochondria. J. Biol. Chem. 10:13028-13033. [PubMed] [Google Scholar]
- 9.Fickers, P., J.-M. Nicaud, J. Destain, and P. Thonart. 2003. Overproduction of lipase by Yarrowia lipolytica mutants. Appl. Microbiol. Biotechnol. 63:136-142. [DOI] [PubMed] [Google Scholar]
- 10.Folch, J., M. Lees, and G. H. Sloane-Stanley. 1957. A simple method for the isolation and purification of total lipids from animal tissues. J. Biol. Chem. 226:497-509. [PubMed] [Google Scholar]
- 11.Freeman, D. A., and M. Ascoli. 1982. Studies on the source of cholesterol used for steroid biosynthesis in cultured Leydig tumor cells. J. Biol. Chem. 10:14231-14238. [PubMed] [Google Scholar]
- 12.Gwynne, J. T., and J. F. Strauss. 1982. The role of lipoproteins in steroidogenesis and cholesterol metabolism in steroidogenic glands. Endocr. Rev. 3:299-329. [DOI] [PubMed] [Google Scholar]
- 13.Haid, A., and M. Suissa. 1983. Immunochemical identification of membrane proteins after sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Methods Enzymol. 96:192-205. [DOI] [PubMed] [Google Scholar]
- 14.Kim, T. H., O. Young-Sook, and S. J. Kim. 2000. The possible involvement of the cell surface in aliphatic hydrocarbon utilization by an oil degrading yeast, Yarrowia lipolytica 180. J. Microbiol. Biotechnol. 10:333-337. [Google Scholar]
- 15.Leber, R., E. Zinser, G. Zellnig, F. Paltauf, and G. Daum. 1994. Characterization of lipid particles of the yeast Saccharomyces cerevisiae. Yeast 10:1421-1428. [DOI] [PubMed] [Google Scholar]
- 16.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the Folin-phenol reagents. J. Biol. Chem. 193:265-275. [PubMed] [Google Scholar]
- 17.Luo, Y., H. Wang, K. V. Gopalan, D. K. Srivastava, J.-M. Nicaud, and T. Chardot. 2000. Purification and characterization of the recombinant form of acyl CoA oxidase 3 from the yeast Yarrowia lipolytica. Arch. Biochem. Biophys. 384:1-8. [DOI] [PubMed] [Google Scholar]
- 18.Luo, Y., J.-M. Nicaud, P. van Veldhoven, and T. Chardot. 2002. The acyl-CoA oxidases from the yeast Y. lipolytica: characterization of Aox2p. Arch. Biochem. Biophys. 403:32-38. [DOI] [PubMed] [Google Scholar]
- 19.Mauersberger, S., H. Wang, C. Gaillardin, G. Barth, and J.-M. Nicaud. 2001. Insertional mutagenesis in the n-alkane-assimilating yeast Yarrowia lipolytica: generation of tagged mutations in genes involved in hydrophobic substrate utilization. J. Bacteriol. 183:5102-5109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Montet, D., R. Ratomahenina, P. Galzy, M. Pina, and J. Graille. 1985. A study of the influence of the growth media on the fatty acid composition in Candida lipolytica diddens and lodder. Biotechnol. Lett. 7:733-736. [Google Scholar]
- 21.Murphy, D. J. 2001. The biogenesis and functions of lipid bodies in animals, plants and microorganisms. Prog. Lipid Res. 40:325-438. [DOI] [PubMed] [Google Scholar]
- 22.Nicaud, J.-M., C. Madzak, P. van den Broek, G. Gysler, P. Duboc, P. Niederberger, and C. Gaillardin. 2002. Protein expression and secretion in the yeast Yarrowia lipolytica. FEMS Yeast Res. 2:371-379. [DOI] [PubMed] [Google Scholar]
- 23.Papanikolaou, S., I. Chevalot, M. Komaitis, G. Aggelis, and I. Marc. 2001. Kinetic profile of the cellular lipid composition in an oleaginous Yarrowia lipolytica capable of producing a cocoa-butter substitute from industrial fats. Antonie Leeuwenhoek 80:215-224. [DOI] [PubMed] [Google Scholar]
- 24.Papanikolaou, S., and G. Aggelis. 2002. Lipid production by Yarrowia lipolytica growing on industrial glycerol in a single-stage continuous culture. Bioresource Technol. 82:43-49. [DOI] [PubMed] [Google Scholar]
- 25.Papanikolaou, S., I. Chevalot, M. Komaitis, I. Marc, and G. Aggelis. 2002. Single cell oil production by Yarrowia lipolytica growing on an industrial derivative of animal fat in batch cultures. Appl. Microbiol. Biotechnol. 58:308-312. [DOI] [PubMed] [Google Scholar]
- 26.Sambrook, J., T. Maniatis, and E. F. Fritsch. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 27.Wang, H., A. Le Clainche, M.-T. Le Dall, Y. Waché, Y. Pagot, J.-M. Belin, C. Gaillardin, and J.-M. Nicaud. 1998. Cloning and characterization of the peroxisomal acyl CoA oxidase ACO3 gene from the alkane-utilizing yeast Yarrowia lipolytica. Yeast 15:1373-1386. [DOI] [PubMed] [Google Scholar]
- 28.Wang, H., M.-T. Le Dall, Y. Waché, P. Laroche, J.-M. Belin, C. Gaillardin, and J.-M. Nicaud. 1999. Evaluation of acyl coenzyme A oxidase isozyme function in the n-alkane-assimilating yeast Yarrowia lipolytica. J. Bacteriol. 181:5140-5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang, H., M.-T. Le Dall, Y. Waché, P. Laroche, J.-M. Belin, C. Gaillardin, and J.-M. Nicaud. 1999. Cloning, sequencing, and characterization of five genes coding for acyl-CoA oxidase isozymes in the yeast Yarrowia lipolytica. Cell. Biochem. Biophys. 31:165-174. [DOI] [PubMed] [Google Scholar]
- 30.Weller, P. F., and A. M. Dvorac. 1994. Lipid bodies: intracellular sites for eicosanoid formation. Allergy Clin. Immunol. 94:1152-1156. [DOI] [PubMed] [Google Scholar]
- 31.Zweytick, D., K. Athenstaedt, and G. Daum. 2000. Intracellular lipid particles of eukaryotic cells. Biochim. Biophys. Acta 18:101-120. [DOI] [PubMed] [Google Scholar]