Abstract
Atlantic salmon (Salmo salar) were challenged with Vibrio anguillarum strains M93Sm and NB10 and empA null mutants M99 and NB12. Both wild types were virulent when administered by intraperitoneal (i.p.) injection or anal intubation. NB12 was avirulent via either route of infection. M99 virulence was attenuated when delivered by intubation, but fully virulent by i.p. injection. Northern blot analysis revealed empA expression in M93Sm and NB10 cells incubated in mucus, while incubation in Luria-Bertani broth plus 2% NaCl (LB20) induced empA expression only in NB10. Nucleotide differences between M93Sm and NB10 empA sequences were found in regions located 207 and 229 bp upstream of the empA translational start. Reverse transcription-PCR and 5′ rapid amplification of cDNA ends revealed the empA transcriptional start site 85 bp upstream of the translational start for both strains. A putative σS-dependent promoter was identified upstream of the transcriptional start in both strains. Site-directed mutagenesis was used to create rpoS mutants of M93Sm and NB10. Neither rpoS mutant exhibited protease activity. Since empA is expressed during stationary phase, the effects of conditioned medium on protease activity were examined. M99 conditioned LB20 supernatants stimulated protease activity in NB10 while allowing M93Sm to produce protease in LB20. Neither acyl homoserine lactones nor AI-2 induced protease activity. Conditioned LB20 supernatant from a V. anguillarum luxS mutant caused a more rapid induction of protease activity in wild-type cells. Our data show that expression of empA is differentially regulated in V. anguillarum strains NB10 and M93Sm and requires σS, quorum-sensing molecules, and gastrointestinal mucus.
Vibrio anguillarum is the causative agent of vibriosis in fish, bivalves, and crustaceans (3, 8, 10). Vibriosis has been a major problem for the aquaculture industry around the world. Large economic losses due to this fish pathogen are sustained by the aquaculture industry (3). Vibriosis in fish is observed as a hemorrhagic septicemia (3). Infected fish display skin discoloration and erythema around the base of the fins, vent, and mouth. Necrotic lesions are observed in the abdominal muscle. The intestinal tract and rectum become distended and filled with fluid. Mortalities within infected fish farm stocks range from 30 to 100% (3, 19, 30).
Olsson et al. (34) demonstrated that the gastrointestinal (GI) tract of fish can serve as the port of entry for V. anguillarum. Additionally, the GI tract of fish appears to be a site of colonization and amplification for pathogenic Vibrio species (36, 37). Once in the GI tract, the pathogen grows rapidly, utilizing intestinal mucus as a nutrient. Garcia et al. (15) have shown that salmon GI mucus is an excellent growth medium for V. anguillarum. Additionally, both V. anguillarum and Vibrio alginolyticus exhibit strong chemotaxis toward fish mucus (9). When growing in GI mucus, V. anguillarum specifically expresses a number of different proteins, including several outer membrane proteins (15) and EmpA protease (this study). Previously, we demonstrated that the empA metalloprotease is expressed during the stationary phase when V. anguillarum cells are incubated in mucus (13).
Quorum sensing (QS) is a term used to describe how bacteria can sense or measure their cell density. Bacteria monitor their cell density by using diffusible signal molecules termed autoinducers. This phenomenon was first studied by Hastings et al. (14, 32) in the marine symbiont Vibrio fischeri. This luminescent marine bacterium is found living in light organs of squid and fish (23, 38). Autoinducers in many gram-negative bacteria, such as V. fischeri, include acyl homoserine lactones (AHLs). These compounds, specifically 3-oxo-C6-homoserine lactone (3-oxo-C6-HSL), regulate light production (via the expression of luxICDABEGH) in V. fischeri (39). The AHL acts with the transcriptional activator LuxR to activate transcription of the AHL synthase, luxI and the luxCDABEGH genes in a positive feedback loop. Similarly, the opportunistic pathogen Pseudomonas aeruginosa regulates the production of the metalloprotease virulence factor elastase (a homologue of EmpA), using AHL-dependent QS (35, 46). Milton et al. (24, 25) have demonstrated that V. anguillarum mutants unable to produce C6-HSL and 3-OH-C6-HSL and mutants that fail to produce 3-oxo-C10-HSLs still exhibit normal production of the EmpA metalloprotease. In addition to AHLs, many gram-negative and some gram-positive organisms produce a non-AHL signal molecule called autoinducer 2 (AI-2), which has been suggested to be a furanone borate diester (11). The synthesis of AI-2 in S-adenosylmethionine metabolism requires a functional luxS gene (40). LuxS is the AI-2 synthase, and its substrate is S-ribosylhomocysteine, which is cleaved to yield homocysteine and AI-2. AI-2 has been shown to regulate by QS the expression of the luxCDABEGH in Vibrio harveyi (5). LuxS-mediated QS has also been linked to the expression of the genes in enterohemorrhagic Escherichia coli O157:H7 and enteropathogenic E. coli during attaching and effacing in the intestine (42).
It is hypothesized that the EmpA metalloprotease of V. anguillarum is important for virulence during infection of Atlantic salmon (Salmo salar). Extracellular proteases are secreted during infection in many pathogenic bacteria. The host tissue damage observed in infected fish suggests the production of proteases by V. anguillarum. The elastolytic protease of P. aeruginosa is required for tissue destruction during opportunistic infections (33). The VVP metalloprotease of V. vulnificus causes hemorrhagic damage by digesting type IV collagen after being injected intradermally into dorsal skin of guinea pigs (29). These proteases, which both result in destructive tissue damage, are highly homologous to EmpA metalloprotease of V. anguillarum.
In this study, wild-type strains (NB10 and M93Sm) and empA mutants (NB12 and M99) of V. anguillarum were tested for virulence in Atlantic salmon when introduced by intraperitoneal (i.p.) injection and by anal intubation (AIB). The two wild-type strains were chosen because they have different serotypes and we had previously examined the induction of protease activity in M93Sm (13), while Milton and her coworkers had examined empA in NB10 (26). We examined and characterized the differences in expression of empA in NB10 and M93Sm cells incubated in LB20 and mucus, using Northern blot analysis. Differences in EmpA activity in cell supernatants were also demonstrated by the azocasein protease assay. The empA genes in NB10 and M93Sm were sequenced and compared to determine and characterize the promoter and other possible upstream regulatory regions. Additionally, the effect of an rpoS mutation on EmpA expression was determined. The possibility of extracellular QS-directed expression of empA was also examined with regard to AHLs and AI-2.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
All bacterial strains and plasmids used in this report are listed in Table 1. V. anguillarum strains were routinely grown in Luria-Bertani broth plus 2% NaCl (LB20) (15, 44), supplemented with the appropriate antibiotic, on a rotary shaker at 27°C. V. harveyi strains were grown in autoinducer bioassay (AB) broth (16) on a rotary shaker at 30°C. Experimental media included LB20, marine minimal medium (3 M) (15), and nine-salt solution (NSS; a carbon-, nitrogen-, and phosphorus-free salt solution) plus 200 μg of mucus protein/ml (NSSM) (15). Gastrointestinal mucus was harvested from Atlantic salmon as previously described by Garcia et al. (15). Overnight cultures of V. anguillarum were grown in LB20 and centrifuged (9,000 × g, 10 min), and pelleted cells were washed twice with NSS (15). Washed cells were resuspended to appropriate cell densities in experimental media. Specific conditions are described in the text for each experiment. Cell densities were determined by serial dilution and plating on LB20 agar plates or by measuring optical density at 600 nm (OD600). Antibiotics were used at the following concentrations: streptomycin, 200 μg/ml; chloramphenicol, 5 μg/ml; kanamycin, 40 μg/ml; ampicillin, 100 μg/ml; and tetracycline, 200 μg/ml.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype and description | Source or reference |
|---|---|---|
| Strains | ||
| V. anguillarum | ||
| M93Sm | Spontaneous Smr mutant of M93 (serotype J-O-1) | Denkin & Nelson (13) |
| M99 | Smr Cmr; M93Sm empA mutant | This study |
| M01 | Smr Cmr; M93Sm luxS mutant | This study |
| M03 | Smr Cmr; M93Sm rpoS mutant | This study |
| NB10 | Wild type (serotype O1) | Milton et al. (26) |
| NB12 | Cmr; NB10 empA mutant | Milton et al. (26) |
| NB03 | Smr Cmr; NB10 rpoS mutant | This study |
| V. harveyi | ||
| BB120 | Wild type | Bassler et al. (43) |
| BB152 | luxMN (AI-1− AI-2+) | Bassler et al. (43) |
| BB170 | luxN::Tn5 (sensor 1− sensor 2+) | Bassler et al. (43) |
| E. coli | ||
| SM10 | thi thr leu tonA lacY supE recA RP4-2-Tc::Mu::Km (λ pir) | Milton et al. (26) |
| JB525 | Tcr; MT102(pJBA132) | Andersen et al. (2) |
| JM109L | Apr; JM109(pSB1075) | Winson et al. (47) |
| Plasmids | ||
| pNQ705-1 | Cmr; suicide vector with R6K origin | Milton et al. (26) |
| pempA | Cmr Tcr; pSUP202 with 4-kbp empA | Milton et al. (26) |
| pJBA132 | luxR-PluxI-RBSII-gfp (ASV) in pME6031 (detects C6- to C8-AHLs) | Andersen et al. (2) |
| pSB1075 | luxCDABE fused to lasRI′ in pUC18 (detects C10- to C12-AHLs) | Winson et al. (47) |
| pNQEmpA | Cmr; 440-bp empA in pNQ701-1 | This study |
| pNQLuxS | Cmr; 270-bp luxS in pNQ705-1 | This study |
| pNQRpoS | Cmr; 250-bp rpoS in pNQ705-1 | This study |
Bacterial matings.
Plasmids were introduced into V. anguillarum M93Sm from E. coli SM10 by conjugation by the procedure described by Milton et al. (27, 28). Briefly, overnight cultures of V. anguillarum M93Sm and E. coli SM10 were prepared and mixed at ratios of 1:1 or 3:1 (recipient to donor) in NSS plus 10 mM MgSO4. The cell suspension was vacuum filtered onto a 0.22-μm-pore-diameter nylon membrane, which was placed on an LB15 agar plate and allowed to incubate overnight at 27°C. Following incubation, the cells were removed from the filter by vigorous mixing in NSS plus 10 mM MgSO4. The cell suspension (100 μl) was plated on LB20 with 200-μg/ml streptomycin and 5-μg/ml chloramphenicol and allowed to incubate at 27°C until V. anguillarum colonies were observed (usually 16 to 24 h).
Site-directed mutagenesis of empA, luxS, and rpoS.
Site-directed mutagenesis was used to create gene interruptions within the structural genes of empA, luxS, and rpoS. Primers (Table 2) were generated based on the empA sequence for V. anguillarum NB10 (accession no. L02528) and M93Sm (accession no. AY428808), the luxS sequence for V. harveyi (accession no. AF120098), and Vibrio cholerae (accession no. AE004141), and the rpoS sequence for V. harveyi (accession no. AF321124), Vibrio parahaemolyticus (accession no. AF144608), and V. cholerae (accession no. AF000945). Sequences from multiple organisms were aligned, and primers were synthesized by using highly homologous regions. A 440-bp region from empA was PCR amplified with QIAGEN Taq DNA polymerase and cloned into the suicide vector pNQ705 by using SacI and XbaI restriction endonucleases to yield pNQSD8. The presence of the 440-bp empA-derived insert was confirmed by both PCR amplification and restriction analysis with SacI and XbaI. The mobilizable suicide vector containing empA was transferred into V. anguillarum by conjugation with E. coli SM10. E. coli SM10 contains the λ pir protein that is required for replication of pNQ705. Chloramphenicol-resistant colonies were selected and screened for insertion within empA. PCR and southern blot analysis were used to confirm the incorporation of pNQSD8. For PCR analysis, a primer described previously by Milton et al. (26) complementary to the pNQ705 vector was utilized (Table 2). The forward primer, SDempA-Forward (Table 2), is complementary to a region upstream of the insertion. PCR products were analyzed by electrophoresis through a 1.0% agarose gel in Tris-acetate EDTA (TAE) (4) buffer containing 0.2-μg/ml ethidium bromide. The gene was interrupted within the 440-bp region of empA, rendering the mutants resistant to chloramphenicol at 5 μg/ml. The resulting V. anguillarum empA mutant was designated as M99 (Table 1). For creating luxS and rpoS mutants, a 270-bp fragment and a 250-bp fragment, respectively, were PCR amplified separately and used in the same protocol as for the empA gene interruption.
TABLE 2.
Primers used in this study
| Primer | Sequencea |
|---|---|
| pNQ705-R | 5′-GCGTAACGGCAAAAGCACCGCCGGACATCA-3′ |
| empA-F | 5′-GCTATTCATGTACCGACGCG-3′ |
| empA-R | 5′-CGGAAGGATTTGAAAATGTCGC-3′ |
| empAUSP1 | 5′-GTTATGTGCACTATTAAATGTAC-3′ |
| empADSP1 | 5′-GACGCTCGTTTTGCACTATTTTTC-3′ |
| empAUSP2 | 5′-GATCCATTTTCAATGAGCGAATC-3′ |
| empADSP2 | 5′-GGATTGAGAAATGAAAAAAGTAC-3′ |
| empAP2-1 | 5′-GCTGGTGGTTGATTGGATGTAAG-3′ |
| empAP2-2 | 5′-GGATGTAAGTCGTCAGCGGATGC-3′ |
| empAP2-3 | 5′-GCGGATGCTAATAATAACTCTC-3′ |
| empAP2-4 | 5′-TAATAATAACTCTCTAGGATTGAG-3′ |
| empATXNST | 5′-GAATCATTCGCAGTCACGTC-3′ |
| empA-GSP1 | 5′-GCGTAAGTGCAATACTAA-3′ |
| empA-GSP2 | 5′-GGTGAAGTACTTACCACATCAGAA-3′ |
| 5′RACE-AAP | 5′-GGCCACGCGTCGACTAGTACGGGIIGGGIIGGGIIG-3′ |
| 5′RACE-AP | 5′-GGCCACGCGTCGACTAGTACAAAAAAAAAAAAAAAA-3′ |
| SDempA-F | 5′-GCTAGGAGCTCAAACCATGCAGAGGCG-3′ |
| SDempA-R | 5′-GCTAGTCTAGACCAAAGGTCATGGACG-3′ |
| SDluxS-F2 | 5′-GCTAGGAGCTCCGTATGAATGCACCRGC GG-3′ |
| SDluxS-R | 5′-GCTAGTCTAGAGGCGYACCAATCAAGCT CATG-3′ |
| SDrpoS-F | 5′-GCTAGGAGCTCCGTGGYGATGAAGCSGCWCG-3′ |
| SDrpoS-R | 5′-GCTAGTCTAGACGCAGATAAATGTTCAG CTC-3′ |
Restriction sites for SacI (GAGCTC) and XbaI (TCTAGA) are underlined, and mixed bases are in bold (R = A or G, S = G or C, W = T or A, and Y = C or T).
Fish infections.
V. anguillarum cells were prepared by growing in LB20 for 16 h on a rotary shaker at 27°C to a density of ∼2 × 109 CFU/ml. Cells were harvested by centrifugation, washed twice in NSS, resuspended in NSS, and diluted to the appropriate inoculation concentrations. Inoculum dosages were determined by serial dilution and plating on LB20 agar plates. Juvenile Atlantic salmon were infected with wild-type or mutant strains via either of two routes of administration: i.p. or AIB. In each case, fish were anesthetized in water supplemented with tricaine methane sulfonate (75 mg/liter) before challenge. Fish were inoculated i.p. or AIB with equal volumes (50 μl) of cells (ranging from ∼104 to 107 CFU/ml) in NSS or with NSS alone (control fish). Death due to vibriosis was determined by the observation of gross clinical signs and confirmed by the recovery and isolation of V. anguillarum cells that were resistant to the appropriate antibiotics from infected organs of dead fish. Observations for clinical signs of vibriosis were continued for 21 days.
Detection and quantification of protease activity.
Culture supernatants were assayed for proteolytic activity according to our previously described modification of the method by Windle and Kelleher (13, 45). Briefly, culture supernatant was incubated with azocasein (5 mg/ml) dissolved in Tris-HCl (50 mM, pH 8.0) containing 0.04% NaN3. Culture supernatant was prepared by centrifugation of 1 ml of cells at 12,000 × g (10 min, 20°C). Supernatant was removed and filtered through a 0.22-μm-pore-size cellulose-acetate filter. Filtered supernatant (100 μl) was incubated at 30°C with 100 μl of azocasein solution. An incubation time of 30 min was sufficient for assays of supernatants from cell suspensions of ≥5 × 108 cells/ml. Reactions were terminated by addition of trichloroacetic acid (TCA) (10% [wt/vol]) to a final concentration of 6.7% (wt/vol). The mixture was allowed to stand for 1 to 2 min and centrifuged (12,000 × g, 4 min) to remove unreacted azocasein, and supernatant containing azopeptides was suspended in 700 μl of 525 mM NaOH (45). Absorbance of the azopeptide supernatant was measured at 442 nm with a Pharmacia Ultrospec 4000 spectrophotometer. A blank control was prepared by boiling V. anguillarum M93Sm supernatant (100°C, 10 min). TCA was added to the blank control supernatant immediately after the addition of azocasein. The mucus used was also boiled (10 min) to destroy any inherent protease activity. Protease activity units were calculated with the following equation: 1 protease activity unit = [1,000 (OD442)/CFU] × (109) (13).
Northern blot analysis.
Samples (1 ml) of cells were centrifuged (12,000 × g, 2 min, at 4°C) and cell pellets were collected and stored at −75°C. Total RNA was isolated from V. anguillarum M93Sm and NB10 cell pellets by using the Purescript RNA isolation kit (Gentra Systems, Inc., Minneapolis, Minn.) or using the RNeasy purification kit (QIAGEN). RNA samples were treated with RQ1 (RNase-free) DNase (1 U/μl) according to the manufacturer's specifications (Promega, Madison, Wis.). RNA (5 μg) was prepared according to the method of Ausubel et al. (4) and electrophoresed at 80 V for 90 min through a 1% agarose gel containing 1.1% formaldehyde in a mixture containing 1× MOPS [0.4 M 3-(N-morpholino)propanesulfonic acid], 0.2 M sodium acetate (pH 7.0), and 0.01 M Na2EDTA (1 M Na2EDTA stock solution to pH 8.0). All buffers, reagents, and solutions were made with 0.1% diethylpyrocarbonate-treated water (Sigma). After electrophoresis, the RNA gel was rinsed in distilled water twice and then washed in 10× sodium chloride-sodium citrate (SSC; 3 M NaCl, 0.3 M sodium citrate, pH 7.0) at room temperature for 45 min. RNA was transferred to a nylon membrane (Magna Graph; Osmonics, Inc.) using Turboblotter, a rapid downward neutral transfer system according to the directions of the manufacturer (Schleicher & Schuell, Keene, N.H.). RNA was bound to the nylon membrane by using a UV cross-linker (UVC-500; Hoefer Pharmacia Biotech, Inc., San Francisco, Calif.). The blot was probed with a digoxigenin (DIG)-dUTP labeled probe. The empA gene probe was constructed by purifying the pempA plasmid from E. coli DH1 by using a Promega Wizard plus miniprep DNA purification system (Promega) according to the manufacturer's instructions. The empA gene was amplified by PCR and labeled with DIG by using a PCR-DIG probe synthesis kit (Roche). The primers used to amplify empA were empA-F and empA-R (Table 2). A pempA (Table 1) sample (60 ng) was amplified with Taq polymerase (3.5 U/100 μl; Gibco BRL Life Technologies, Bethesda, Md.) on a Perkin-Elmer GeneAmp model 9700 Thermocycler (Perkin-Elmer). Gene probes for vanM and vanI were PCR amplified and DIG labeled with a PCR-DIG probe synthesis kit (Roche) from total genomic DNA. Genomic DNA was prepared with a QIAGEN Dneasy tissue kit. Oligonucleotides were synthesized by Integrated DNA Technologies, Inc. (Coralville, Iowa). The PCR cycle conditions were 94°C for 1 min, 51°C for 2 min, and 72°C for 3 min. The reaction was run for 35 cycles and then held at 4°C until collected. The primers used for PCR amplification, derived from the empA gene sequence (26), were empA-F and empA-R.
Solvent extraction of conditioned supernatant.
A modification of the ethyl acetate extraction method described by Holden et al. (18) was used to obtain AHLs from culture supernatants. Briefly, conditioned cell culture supernatants were prepared from bacterial cultures grown (rotary shaker at 27°C, 20 h) in the experimental media described above unless otherwise noted. Cells were removed by centrifugation (7,000 × g, 10 min, 20°C). The cell supernatant was transferred to a screw-cap glass test tube and extracted twice for 30 to 60 s with acidified (100 μl of glacial acetic acid/liter) ethyl acetate (3 parts acidified ethyl acetate to 2 parts supernatant). Organic and aqueous phases were separated by centrifugation (2,000 × g, 5 min, 20°C). The organic phase was transferred to a 20-ml glass test tube and dried under nitrogen gas in a 40°C water bath. The dried samples were dissolved in 50 to 100 μl of 100% ethanol and then concentrated to a final volume of 10 μl.
AHL detection with gfp- and lux-based reporter E. coli strains.
V. anguillarum strains were grown overnight and resuspended under the appropriate experimental condition. Cell-free conditioned supernatant was prepared at indicated time points and extracted twice with acidified ethyl acetate and then dried under nitrogen. Dried material from extracts was dissolved in 100 μl of 100% ethanol. Samples were further dried and resuspended in 10 μl of 100% ethanol and then used in the gfp- or lux-based reporter assays. The gfp-based reporter strain E. coli JB525 (2) was used for detection of C6- to C8-AHLs. This reporter strain contains a gfp fusion to the rhlI promoter from P. aeruginosa PA01, allowing gfp expression in response to C6- to C8-AHLs. To detect longer-chain AHLs (C10 to C14), the lux-based reporter strain E. coli JM109 containing pSB1075 (47) was used. This second reporter strain contains the lasRI promoter region coupled to the entire lux structural operon. Fluorescence by green fluorescent protein (GFP) was measured with a spectrofluorimeter (Perkin-Elmer; LS55). Luminescence from the lux reporter was measured with a luminometer, (Turner Designs; model TD-20/20).
Preparation of cell-free conditioned medium for AI-2 assay.
To prepare cell-free conditioned medium, V. anguillarum strains were grown overnight in LB20 without antibiotics on a rotary shaker at 27°C. Cells were centrifuged at 9,000 × g for 10 min, and the cell pellets were washed twice in NSS. Cells were diluted 1:100 (∼2 × 107 to 3 × 107 CFU/ml) into LB20 or NSSM and allowed to incubate with shaking at 27°C for 10 h. V. anguillarum cells were removed by centrifugation (9,000 × g, 10 min), and the resulting supernatant was passed through a 0.22-μm-pore-size acetate filter. Samples were stored at −20°C. V. harveyi BB152 conditioned medium was prepared as described by Surette and Bassler (43).
V. harveyi luminescence bioassay for AI-2.
The assay for AI-2 production was performed as described by Surette and Bassler (43). V. harveyi BB170 was grown overnight and diluted 1:5,000 into fresh AB medium. Prepared cell-free conditioned medium was added at 10% (vol/vol) (100 μl) to the diluted sensor strain (900 μl) and allowed to incubate for 3 h with aeration at 30°C. As a control, conditioned supernatant from V. harveyi BB152 was added to the AI-2 sensor strain BB170 and luminescence was measured. Aliquots (500 μl) were taken, and luminescence was measured with a luminometer (Turner Designs; model TD-20/20).
Identification of the transcriptional start site for empA.
Total RNA was isolated from V. anguillarum cells incubated in either LB20, NSSM, 3 M, or NSS as described above for Northern blot analysis. The OneStep reverse transcription (RT)-PCR system from QIAGEN was used to approximate the empA transcriptional start. Primers used in RT-PCR are listed in Table 2. To identify the transcriptional start site, RNA was subjected to 5′ rapid amplification of cDNA ends (RACE) using the Invitrogen 5′ RACE kit. Briefly, 5 μg of RNA was used to generate specific first-strand cDNA from empA mRNA in a reverse transcriptase reaction with an empA primer (empA-GSP1). A poly(dC) or poly(dA) tail was added to the 3′ cDNA end with dCTP or dATP and terminal deoxynucleotidyl transferase. A PCR product was amplified from the tailed cDNA by using a 5′ RACE-abridged anchor primer (AAP) or a 5′ RACE anchor primer (AP) (Table 2) and a nested empA primer (empA-GSP2). The PCR product was purified with a PCR Qiaquick spin kit (QIAGEN) and sequenced. DNA sequences were aligned by using the Baylor College of Medicine search launcher multiple sequence alignment program ClustalW 1.8 (http://searchlauncher.bcm.tmc.edu).
DNA sequencing.
DNA sequencing was performed by the HHMI Biopolymer/Keck Foundation Biotechnology Resource Laboratory at Yale University (New Haven, Conn.) and by the University of Rhode Island Genomics and Sequencing Center (Kingston, R.I.). Sequencing was performed on Applied Biosystems 377 and 3100 DNA analysis instruments (Keck) and on a Beckman-Coulter CEQ 8000 (URI). Fluorescent-labeled dideoxynucleotide Big Dye terminators with Taq FS DNA polymerase (Keck) and the Dye Terminator Cycle Sequencing (DTCS) quick start kit (URI) in thermal cycling programs were used in the sequence reactions. DNA samples were mixed with the appropriate primer (Table 2) and then submitted for sequencing.
RESULTS
Role of EmpA as a virulence factor during infection of Atlantic salmon.
In order to examine the role of EmpA as a virulence factor, juvenile Atlantic salmon were infected in separate experiments by i.p. injection or by AIB with V. anguillarum wild-type strains M93Sm and NB10 or empA mutant strains M99 and NB12. The results of i.p. infections shown in Table 3 reveal that both V. anguillarum wild-type strains (M93Sm and NB10) were equally virulent. Infection with 3 × 106 to 4 × 106 CFU by the i.p. route resulted in 80% mortality by 3 to 4 days. The empA mutant strain V. anguillarum M99 exhibited virulence equal to that of the parental wild-type strain (M93Sm). In contrast, i.p. inoculation of salmon with 4 × 106 CFU of the empA mutant strain V. anguillarum NB12 resulted in no salmon mortalities over the 21-day experiment.
TABLE 3.
Virulence of V. anguillarum wild-type strains M93Sm and NB10 and empA mutants M99 and NB12 in Atlantic salmon by i.p. injectionsa
| Strain | Dose/fish (CFU) | Mortality (no. dead/no. injected) | Day of death (no. dead/no. injected) |
|---|---|---|---|
| M93Sm | 2.20 × 104 | 1/5 | 3 (1/5) |
| 3.03 × 105 | 3/5 | 2 (1/5), 3 (2/5), 4 (3/5) | |
| 2.93 × 106 | 4/5 | 1 (1/5), 2 (3/5), 3 (4/5) | |
| NB10 | 3.83 × 104 | 2/5 | 2 (1/5), 4 (2/5) |
| 3.27 × 105 | 2/5 | 3 (1/5), 5 (2/5) | |
| 3.73 × 106 | 4/5 | 3 (3/5), 4 (4/5) | |
| M99 | 2.63 × 104 | 1/5 | 3 (1/5) |
| 2.87 × 105 | 3/5 | 3 (1/5), 4 (3/5) | |
| 2.60 × 106 | 5/5 | 1 (1/5), 2 (5/5) | |
| NB12 | 4.43 × 104 | 0/5 | NAb |
| 3.87 × 105 | 0/5 | NA | |
| 4.03 × 106 | 0/5 | NA | |
| Control (NSS) | 0/15 | NA |
Fish were injected with 50 μl of washed cells suspended in NSS (experimental) or NSS alone (control) at the indicated dosage amount. The experiment continued for 21 days. Death due to vibriosis was confirmed by observing clinical signs and isolation of streptomycin-resistant V. anguillarum cells.
NA, not applicable: no death due to vibriosis during the 21-day experiment.
In a second series of experiments, salmon were infected with wild-type (M93Sm and NB10) and empA mutant strains (M99 and NB12) of V. anguillarum by AIB (Table 4). V. anguillarum wild-type strains M93Sm and NB10 administered by AIB at doses of 7.15 × 106 and 6.50 × 106 CFU, respectively, resulted in 80 to 100% mortality after 10 to 11 days. Both empA mutant strains exhibited attenuated virulence compared to their wild-type parental strains. AIB infections of salmon with strain M99 using 1.04 × 107 CFU resulted in 40% mortality to the fish. AIB infection of salmon with strain NB12 using 8.15 × 107 CFU caused no deaths in the infected salmon over the 21-day experiment. Control fish inoculated with NSS only by either the i.p. or AIB route did not show any signs of vibriosis during the 21-day experiment. These data suggest that EmpA is a virulence factor that is important during the infection of the GI tract of Atlantic salmon by V. anguillarum.
TABLE 4.
Virulence of V. anguillarum wild-type strains M93Sm and NB10 and empA mutants M99 and NB12 in Atlantic salmon by AIBa
| Strain | Dose/fish (CFU) | Mortality (no. dead/no. intubated) | Day of death (no. dead/no. intubated) |
|---|---|---|---|
| M93Sm | 9.15 × 105 | 3/5 | 5 (1/5), 10 (3/5) |
| 7.15 × 106 | 4/5 | 2 (1/5), 6 (2/5), 9 (3/5), 11 (4/5) | |
| 2.77 × 107 | 5/5 | 2 (2/5), 3 (3/5), 4 (4/5), 6 (5/5) | |
| NB10 | 1.42 × 106 | 4/5 | 3 (1/5), 5 (2/5), 10 (3/5), 11 (4/5) |
| 6.50 × 106 | 5/5 | 2 (2/5), 5 (3/5), 8 (4/5), 10 (5/5) | |
| 5.35 × 107 | 5/5 | 2 (1/5), 7 (2/5), 8 (3/5), 9 (5/5) | |
| M99 | 9.50 × 105 | 1/5 | 2 (1/5) |
| 3.47 × 106 | 1/5 | 2 (1/5) | |
| 1.04 × 107 | 2/5 | 4 (1/5), 6 (2/5) | |
| NB12 | 1.68 × 106 | 0/5 | NAb |
| 1.04 × 107 | 0/5 | NA | |
| 8.15 × 107 | 0/5 | NA | |
| Control (NSS) | 0/15 | NA |
Fish were anally intubated with 50 μl of washed cells suspended in NSS (experimental) or NSS alone (control) at the indicated dosage amount. A small amount of phenol red was added to the cell suspension to ensure there was no leakage. The experiment continued for 21 days. Death due to vibriosis was confirmed by observing clinical signs and isolation of streptomycin-resistant V. anguillarum cells.
NA, not applicable: no death due to vibriosis during the 21-day experiment.
Northern blot analysis of empA expression in LB20 and NSSM.
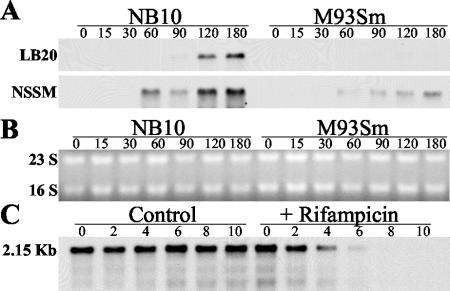
We have previously demonstrated that salmon GI mucus is a specific inducer of the EmpA metalloprotease in V. anguillarum M93Sm when cells are at high density (13). The expression of empA was examined by Northern blot analysis in M93Sm and NB10 (Fig. 1A). Since maximum protease activity was observed at high cell density (≥1 × 109 CFU/ml) (13), empA expression was examined in cells suspended at 2 × 109 CFU/ml. When cells of strain NB10 were incubated in LB20, empA mRNA was first detected 90 min after the start of incubation. Maximum levels of transcription were observed at 120 and 180 min. In contrast, no detectable empA message was detected in cells of strain M93Sm incubated in LB20 under identical conditions. When cells of either strain NB10 or M93Sm were incubated in NSSM, empA message was detected by 60 min, with maximum levels of expression observed at 180 min. Comparison of the amounts of empA transcript produced by each strain revealed that NB10 produced approximately threefold more than M93Sm at 180 min. The calculated size for the empA transcript of each strain was 2.15 kb. To determine the empA message half-life, cells were incubated in NSSM for 3 h. Rifampin was added to an aliquot of NSSM-incubated cells to block further mRNA synthesis, and samples were taken every 2 min for RNA preparation. The half-life of empA mRNA from M93Sm cells induced in mucus was 2.5 min (Fig. 1C). In both experiments, equal amounts of RNA (5 μg) were loaded into each lane. The half-life of the empA transcript in NB10 was similar to that of M93Sm (data not shown).
FIG. 1.
Northern blot analysis of empA expression in LB20 and NSSM. (A) V. anguillarum strains NB10 and M93Sm were grown overnight in LB20 to stationary phase. Cells were washed twice in NSS and resuspended under the experimental conditions at 2 × 109 CFU/ml. Cells were incubated (27°C) with shaking. Samples (1 ml) were taken at 0, 15, 30, 60, 90, 120, and 180 min. Total RNA was isolated and separated on a formaldehyde agarose gel (5 μg of RNA/sample). RNA was transferred from the gel to a nylon membrane and probed with a DIG-dUTP-labeled empA probe for 16 h at 51°C. Panel B shows the RNA gel from panel A stained with ethidium bromide to demonstrate equal loading of all samples. (C) mRNA half-life for empA was determined in M93Sm cells incubated in NSSM for 3 h. Rifampin was added to the cell suspension, and cell samples (1 ml) were taken at 0, 2, 4, 6, 8, and 10 min. Total RNA was prepared and analyzed as described above for panel A.
Protease activity in LB20 and NSSM.
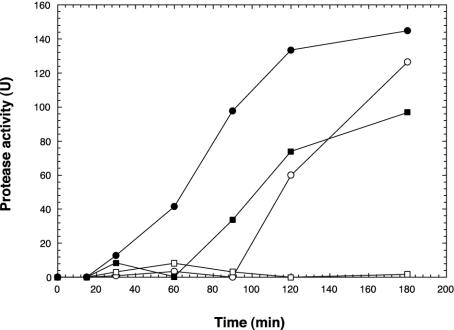
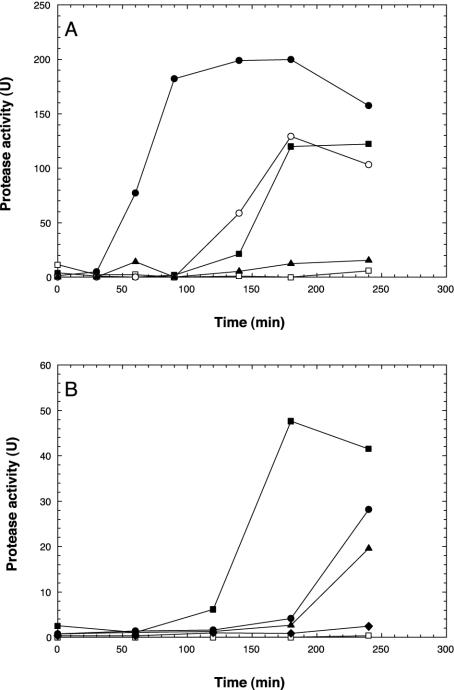
In addition to examining empA expression at the transcriptional level, cell-free supernatant was assayed with azocasein (13) to determine the amount of protease activity. Cell supernatant was prepared from cells that were used to examine empA expression in Fig. 1. When NB10 cells were incubated in LB20, significantly increased protease activity was observed at 120 min (Fig. 2). While a low level of empA message could be detected at 90 min in LB20-incubated NB10 cells (Fig. 1), empA was strongly expressed by 120 min. In contrast, protease activity was not detected in LB20-incubated M93Sm cells. When NB10 or M93Sm cells were incubated in NSSM, protease activity was first detected at 60 and 90 min, respectively. Protease activity for both wild-type cells continued to increase during the 180-min experiment, with NB10 cells exhibiting 1.5-fold-greater protease activity than M93Sm in NSSM at 180 min.
FIG. 2.
Induction of protease activity in V. anguillarum M93Sm (squares) and NB10 (circles) cells incubated in LB20 or NSSM. Cells were grown for 16 h in LB20, harvested by centrifugation (10 min at 9,000 × g, 4°C), washed twice in NSS, and resuspended at 2 × 109 CFU/ml in either LB20 (open symbols) or NSSM (solid symbols). Samples were taken at the indicated times, and the cell supernatant was assayed for protease activity.
Analysis of empA DNA sequence from M93Sm and NB10.
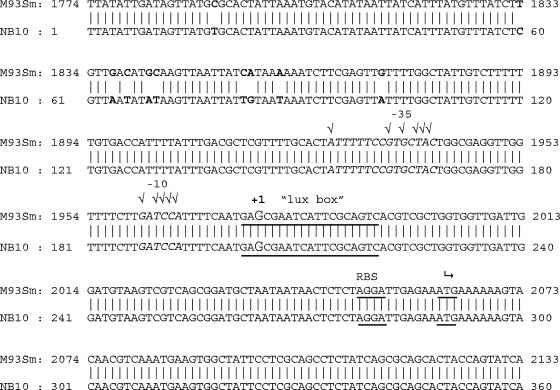
In order to determine whether upstream regulatory sequences in the empA gene could account for the differences in empA expression between strains M93Sm and NB10, the empA gene of M93Sm was cloned, sequenced (accession no. AY428808), and compared to that of NB10. The sequence alignment of empA genes from M93Sm to NB10 is shown in Fig. 3. The structural gene was >99% identical at the nucleotide level to empA in NB10. Three (out of 14) different base pair changes in the structural gene of empA resulted in a changed amino acid (data not shown). The methionines at amino acids 57 and 102 in M93Sm are a valine and leucine, respectively, in NB10. The valine at amino acid 237 in M93Sm is a methionine in NB10. However, ten base pair differences were detected between 188 and 228 bp upstream of the translational start site. There were no other differences detected between the two sequences. A putative lux box reported (26) between 69 and 87 bp upstream of the translational start in NB10 was found in the same position in M93Sm. The differences detected upstream of empA and within the structural gene were confirmed by sequencing the pempA clone from NB10.
FIG. 3.
DNA sequence alignment of the empA regulatory and 5′ regions in V. anguillarum M93Sm and NB10. The −10 and −35 promoter regions are italicized, and check marks indicate matches with consensus. The transcriptional start site is labeled +1. The ribosomal binding site is labeled RBS and is underlined. The translational start site (AtG) is denoted with a bent arrow and is underlined. The differences observed between the two sequences are in bold. The potential lux box region is underlined.
Identification of the empA transcriptional start.
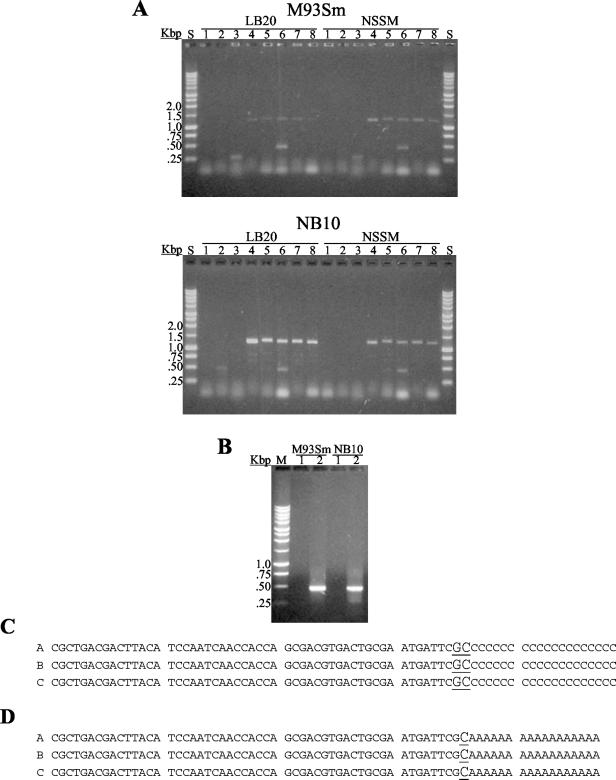
To determine the approximate transcriptional start region for empA, RT-PCR using several sense-strand primers encompassing the predicted start (Table 2) in combination with a single antisense primer (empA-R; Table 2) was initially carried out. RT-PCR products were observed only with sense strand primers designed within 79 bp upstream of the translational start for empA (Fig. 4A). These results suggested that empA transcription begins between 53 and 91 bp upstream of the ATG start translation codon. 5′ RACE was then used to determine the transcriptional start for the empA promoter. Total RNA was extracted from protease-producing cells incubated in LB20 (NB10) or in NSSM (NB10 and M93Sm) and used in the 5′ RACE determination of the transcriptional start. A 480-bp 5′ RACE product was detected by using RNA from M93Sm and NB10 cells in NSSM (Fig. 4B). The 5′ RACE products were sequenced, and all cases revealed that the empA transcriptional start is a C or G located 84 to 85 bp upstream of the translational start (Fig. 4C). To determine if transcription begins at the C or G, 5′ RACE was repeated by using dA tailing. The 5′ RACE product obtained by dA tailing also yielded a 480-bp product (data not shown) that upon sequencing revealed the transcriptional start base as a G (Fig. 4D). Analysis of the −10 and −35 sequences (Fig. 3) showed high similarity to a σS promoter (17).
FIG. 4.
Determination of empA transcriptional start by RT-PCR and 5′ RACE analysis. (A) Eight different empA sense-strand primers (lanes 1 to 8, primers empAUSP1, empADSP1, empAUSP2, empADSP2, empAP2-1, empAP2-2, empAP2-3, and empAP2-4, respectively) were used with an empA antisense strand primer (empA-R) to determine the approximate transcriptional start for empA. RNA from LB20- and NSSM-grown M93Sm (top panel) or NB10 (bottom panel) cells was prepared and used in the RT-PCRs. The RT-PCR products were visualized on a 1% agarose TAE gel containing ethidium bromide. Molecular weight standards (indicated in kilobase pairs) are in lanes S. (B) One percent agarose TAE gel showing 5′ RACE products (even-numbered lanes) from M93Sm and NB10 in NSSM. As a negative control, the tailing enzyme, terminal deoxynucleotidyl transferase, was left out of the reaction (odd-numbered lanes). A 1-kbp DNA ladder was used as the size standard (lane S). (C) DNA sequence alignments of dC-tailed and (D) dA-tailed 5′ RACE products showing the empA transcriptional start site (underlined) for NB10 in LB20 and NSSM and M93Sm under NSSM growth conditions.
Effects of conditioned LB20 cell supernatants on expression of empA.
Previously, we demonstrated that induction of EmpA activity required high cell density (13). The presence of a putative lux box suggested the possibility of cell density regulation of empA. In order to determine whether empA expression is induced by a secreted cell density-dependent factor, conditioned LB20 cell supernatant prepared from the empA mutant, M99, was added to stationary-phase LB20-grown cells of V. anguillarum strains M93Sm and NB10 (Fig. 5A). Protease activity in M93Sm cells resuspended in conditioned LB20 (at 109 CFU/ml) was induced by 140 min and reached a maximum activity by 180 min. No protease activity was observed in M93Sm cells resuspended in fresh LB20 (at 109 CFU/ml). Additionally, when NB10 cells were resuspended in conditioned LB20 (at 109 CFU/ml), EmpA protease activity was detected after 60 min of incubation. In contrast, NB10 cells suspended in fresh LB20 did not exhibit protease activity until 140 min. Furthermore, maximum levels of protease activity for NB10 cells in conditioned LB20 were about 55% greater than in NB10 cells incubated in fresh LB20. To confirm that EmpA was the protease induced, M99 was incubated in conditioned media and no activity was observed.
FIG. 5.
(A) Induction of protease activity in V. anguillarum M93Sm (squares), NB10 (circles), and M99 (triangles) by conditioned LB20 cell supernatants. V. anguillarum empA mutant strain M99 was grown for 20 h in LB20, and conditioned supernatant was prepared as described in Materials and Methods. V. anguillarum cells were grown for 16 h to 2 × 109 CFU/ml in LB20, prepared as described in the legend to Fig. 1, and resuspended in conditioned LB20 supernatant (solid symbols) or fresh LB20 (open symbols). (B) Conditioned LB20 supernatant was prepared from M99 as described and diluted with fresh LB20. V. anguillarum M93Sm was grown overnight and resuspended at 2 × 109 CFU/ml in the diluted conditioned LB20: 100% (solid squares), 87.5% (solid circles), 75% (solid triangles), 62.5% (solid diamonds), and 0% (open squares).
To demonstrate the dose-dependent nature of the inducer of protease activity in conditioned media, M93Sm cells were incubated in various concentrations of conditioned LB20 ranging from 100% to 0% (Fig. 5B). When M93Sm cells were incubated in undiluted (100%) conditioned LB20, protease activity was first observed at 120 min. However, protease activity was not detected until 180 min for cells incubated in 87.5, 75, and 62.5% conditioned LB20. Maximum EmpA activity (measured at 240 min) declined as the conditioned medium was diluted with fresh LB20. At concentrations below 62.5% conditioned LB20, protease activity was not detected (data not shown).
Production of AHLs in LB20 and mucus.
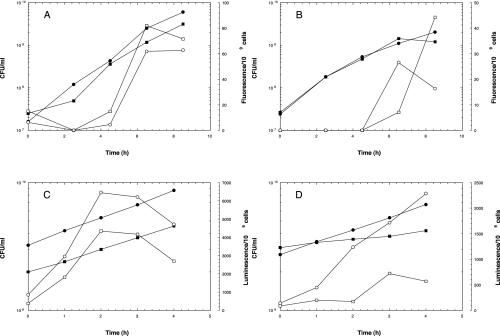
The production of AHLs in V. anguillarum M93Sm and NB10 was examined during growth in LB20 or NSSM by using gfp-based (for C6-AHLs) and lux-based (for C10-AHLs) reporter systems. Ethyl acetate extracts from cells grown in either LB20 or NSSM were added to the gfp biosensor strain, and fluorescence intensities were monitored (Fig. 6A and B, respectively). No fluorescence was detected until the cells of either ethyl acetate-extracted strain reached densities of 4 × 108 to 5 × 108 CFU/ml in LB20 (Fig. 6A) or 1 × 109 to 1.5 × 109 CFU/ml in NSSM. Since no C10-AHL production nor vanI message was detected until cells reached stationary phase (data not shown), ethyl acetate extracts from supernatants of cells incubated in either LB20 or NSSM at 2 × 109 CFU/ml were added to the lux biosensor strains to detect C10-AHL production. Production of the C10-AHL was detected within 1 h of incubation in LB20 in both M93Sm and NB10 (Fig. 6C). Maximum luminescence was detected by 2 h of incubation and declined thereafter. Additionally, M93Sm produced only ∼67% of the C10-AHL that NB10 produced in LB20. When cells were incubated in NSSM, much lower levels of C10-AHL were detected for both strains (Fig. 6D). While C10-AHL production could be detected in NB10 cells after 1 h of incubation, no significant increase in luminescence was detected in M93Sm cells until 3 h of incubation. Further, C10-AHL production in NB10 cells incubated in NSSM was >2.4-fold higher than in M93Sm cells.
FIG. 6.
AHL production measured with E. coli JB525 gfp-based (A and B) and E. coli JM109L lux-based (C and D) AHL sensor strains. Ethyl acetate extracts prepared from M93Sm (squares) and NB10 (circles) growing in LB20 (A and C) or NSSM (B and D) were added to gfp-based (which responds to C6-C8 AHLs) and lux-based (which responds to C10-C12 AHLs) sensor strains and observed for stimulation of luminescence or fluorescence, respectively, was measured. The number of CFU per milliliter (solid symbols) was determined at each time point and is indicated in addition to lux or gfp activity (open symbols).
Effects of ethyl acetate extracts containing AHLs on protease activity.
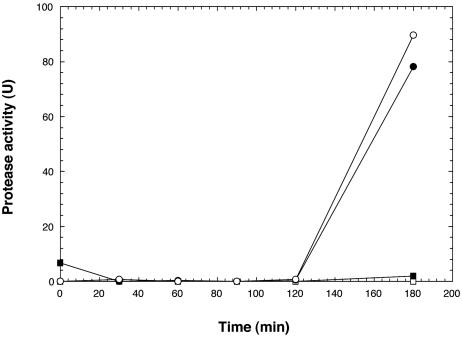
Since both M93Sm and NB10 produced AHLs when grown or incubated in LB20, conditioned supernatant from a stationary-phase culture of M99 cells was extracted for AHLs. The resulting ethyl acetate extracts, which contained levels of AHLs found in M93Sm (data not shown), were added to cells in order to determine whether the rate of EmpA induction or level of expression would be affected. No protease activity was observed when ethyl acetate extracts from either M99 conditioned LB20 or fresh LB20 was added to M93Sm cells suspended in LB20 (Fig. 7). Since protease activity is usually induced in NB10 cells in LB20 by 3 h (Fig. 2), ethyl acetate extracts from M99 conditioned LB20 were added to NB10 cells in LB20 to determine whether the rate of induction or level of expression would be affected. No increase in protease activity was observed compared to that with in incubation in ethyl acetate extract from fresh LB20. These data suggest that AHLs do not affect empA expression in either M93Sm or NB10 cells.
FIG. 7.
Induction of protease activity in V. anguillarum M93Sm and NB10 by ethyl acetate extracts from conditioned LB20 supernatants. Conditioned supernatants from the V. anguillarum empA mutant, M99, were extracted with ethyl acetate. M93Sm (squares) and NB10 (circles) cells were suspended at 2 × 109 CFU/ml in fresh LB20 containing ethyl acetate extract from either M99 (closed symbols) or fresh LB20 (open symbols). As a control, M93 and NB10 cells were suspended in ethyl acetate extract from fresh LB20 (open symbols) and observed for normal induction of protease activity.
Effects of a luxS mutation on AI-2 production and protease activity.
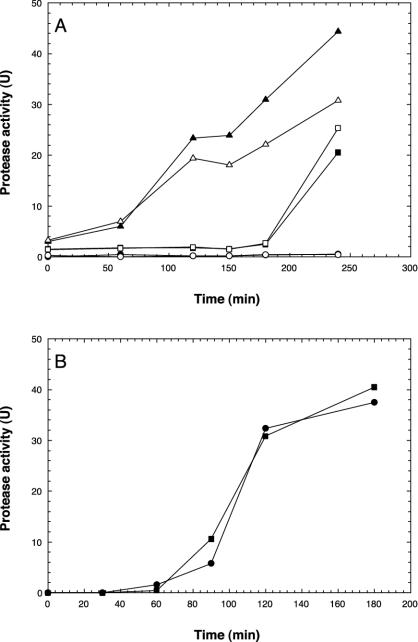
A luxS mutant, M01, of V. anguillarum M93Sm was created by site-directed mutagenesis and tested for AI-2 production. Conditioned media from M01 and M93Sm were assayed for AI-2 by using the V. harveyi BB170 sensor strain. The luxS mutant, M01, did not produce AI-2 when grown in LB20 or NSSM, whereas M93Sm did (data not shown). To determine if AI-2 had any effect on protease activity, M93Sm cells (2 × 109 CFU/ml) were resuspended in conditioned LB20 supernatant from M01 (Fig. 8A). Incubation in conditioned LB20 from M01 resulted in an earlier induction of protease activity in M93Sm compared to incubation in M99 conditioned LB20. Also, the levels of protease activity in M93Sm cells incubated in M01 conditioned LB20 for 240 min was 2.2-fold higher than the protease activity in M99 conditioned LB20. Additionally, when the luxS mutant, M01, was incubated in M01 conditioned LB20, protease activity was induced faster than when M01 was incubated in M99 conditioned LB20 (Fig. 8A). V. anguillarum M01 and M93Sm were compared for protease activity when incubated in NSSM at high cell density (2 × 109 CFU/ml). Both the luxS mutant and M93Sm exhibited the same rate of induction and amount of protease activity in mucus (Fig. 8B).
FIG. 8.
(A) Induction of protease activity in V. anguillarum by conditioned LB20 supernatants from M01 (luxS mutant). V. anguillarum M93Sm (open symbols) and M01 (closed symbols) were grown for 16 h and resuspended at 2 × 109 CFU/ml in conditioned LB20 supernatant from M01 (triangles), M99 (squares), or fresh LB20 (circles). (B) Induction of protease activity in V. anguillarum M93Sm (squares) and M01 (circles) in Atlantic salmon GI mucus. Cells were grown overnight in LB20, washed twice in NSS, and resuspended at 2 × 109 CFU/ml in 200 μg of mucus protein/ml of NSS (NSSM).
Effects of an rpoS mutation on protease activity in LB20 and NSSM.
Since the −10 and −35 promoter regions for empA exhibited a strong similarity to the consensus sequence for a σS-regulated promoter, we searched for rpoS in V. anguillarum. Cells were grown, prepared, and assayed for protease activity as described in the legend to Fig. 2. PCR primers were designed based on the rpoS sequences from V. cholerae, V. harveyi, and V. parahaemolyticus (Table 2) and used to amplify a 250-bp product from V. anguillarum genomic DNA. Sequencing revealed 82% identity to rpoS of V. cholerae and V. parahaemolyticus and 83% identity to rpoS of V. harveyi. The two rpoS mutants (M03 and NB03) created by site-directed mutagenesis were derived from the wild-type strains M93Sm and NB10, respectively. These mutants did not exhibit any protease activity when incubated in NSSM at 2 × 109 CFU/ml for 0 or 4 h. There was no protease activity at 0 h for M93Sm and NB10, whereas at 4 h, the levels of protease activity were 96.9 U for M93Sm and 133.4 U for NB10. No protease activity was observed in NB03 cells when incubated in LB20 (data not shown). These data strongly suggest that the regulation of empA metalloprotease in LB20 and NSSM is controlled by σS.
DISCUSSION
We have previously demonstrated that empA metalloprotease is strongly induced when cells at high density are incubated in Atlantic salmon GI mucus (13). These findings suggested a role for EmpA during colonization and amplification of the bacteria while in the salmon GI tract. In this study, we present data that demonstrate that empA is important for virulence by V. anguillarum in Atlantic salmon. Wild-type strains NB10 and M93Sm of V. anguillarum kill 60 to 80% of salmon when administered by i.p. injection or by AIB at doses of ∼1 × 106 total CFU/fish. The empA mutant strain, NB12 (derived from NB10), is completely avirulent in fish by either i.p. injection or AIB. However, the empA mutant M99 (derived from M93Sm) exhibits wild-type virulence when i.p. injected, but is attenuated in virulence when administered by AIB. While these data show that empA is required for pathogenesis by V. anguillarum NB10 in Atlantic salmon, differences in virulence observed for M99 when administered by i.p. injection and AIB suggest a role for empA during colonization and amplification within the fish intestine. It should be emphasized that the empA mutations in NB12 and M99 are nearly identical, with insertions within the same 200- to 300-bp region in the first third of the coding sequence. While Milton et al. (26) demonstrated that the empA mutant, NB12, exhibited somewhat attenuated virulence when introduced into rainbow trout by i.p. injection or immersion (20- and 50-fold, respectively), it should be noted that the host species used in the two studies are different. Atlantic salmon show much greater resistance to V. anguillarum infection than rainbow trout. In Atlantic salmon, the 50% lethal dose (LD50) of i.p.-injected V. anguillarum is ∼2 × 105 CFU, while in rainbow trout, the LD50 is 100 CFU.
The observation that the empA mutants M99 and NB12 exhibit differential virulence suggested that empA might be differentially regulated in the parental wild-type strains M93Sm and NB10. This was confirmed: M93Sm expresses empA only when at high cell density when incubated in GI mucus, while NB10 expressed empA when at high cell density in all growth media tested (NSSM, LB20, or 3 M). Further, both Northern blot analysis and protease activity assays show that empA is expressed at higher levels in NB10. Examination of the empA structural gene in M93Sm and NB10 sequences reveals 99.2% identity at the nucleotide level with three amino acid differences (amino acids 57, 102, and 237). The three amino acid differences are conserved and are located in the leader sequence for EmpA, not within the mature protein. Therefore, differences in protease activity in M93Sm and NB10 are not linked to the activity of EmpA. Further, the 187 nucleotides upstream of the start translation are identical. Additionally, 5′ RACE analysis shows that the transcriptional start site is identical in both strains under all conditions of empA expression. Both strains regulate transcription by identical σS-dependent promoters. This raises the question of why these two strains display differential regulation of empA. Since vanT (the V. harveyi luxR homolog) mutants do not produce protease (12; this study), it can be hypothesized that the regulation of empA involves VanT binding to a lux box. However, identical putative lux box sequences are found in each strain, which makes it unlikely that differences in empA expression are due to differences in binding efficiencies of VanT. We hypothesize that differences in empA expression may result from differences in nucleotide sequences of M93Sm and NB10 beginning 100 bp upstream of the promoter site.
Toxin-coregulated pilus (tcpA) is differentially expressed in classical and El Tor strains of V. cholerae (22, 31). Classical V. cholerae exhibits tcpA expression under many growth conditions, similar to V. anguillarum NB10 empA expression under multiple growth conditions and starvation at high density (data not shown). In contrast, tcpA expression in El Tor strains of V. cholerae occurs only under bicarbonate-containing (e.g., AKI media) growth conditions. In a similar manner, V. anguillarum M93Sm only produces protease when cells are incubated at high density in mucus. The nucleotide differences in potential upstream empA regulatory sequences of M93Sm and NB10 are very similar to the nucleotide differences observed upstream of the tcpA promoter in classical and El Tor strains of V. cholerae. In V. cholerae, there is a single nucleotide difference (A versus G) between classical and El Tor strains in the tcpA regulatory region located 65 to 66 bp upstream of the transcriptional start. This difference (A versus G) between NB10 and M93Sm sequences was detected 104 bp upstream of the transcriptional start. In V. cholerae, when the nucleotide difference is changed from A to G and G to A for classical and El Tor strains, respectively, the opposite phenotype of tcpA expression is observed (22). Additional base pair differences found 60 bp upstream of the initial difference are conserved between the two strains of V. cholerae and V. anguillarum. Further studies will be necessary to prove that these nucleotide differences upstream of empA account for the expression patterns observed in the two wild-type strains.
The contribution of metalloproteases to virulence in P. aeruginosa has been studied with the rat chronic lung infection model (49). The elastase mutant was less virulent than the parental strain in the rat lung model. The lasB gene of P. aeruginosa encodes the elastase metalloendopeptidase (7), which is highly homologous to empA metalloprotease of V. anguillarum. Comparison of the amino acid sequences of LasB elastase to EmpA by BLASTP (1) reveals that the proteins exhibit 48% identity and 65% similarity. Expression of lasB is cell density dependent and involves QS using AHLs (35). Additionally, Zhu et al. (50) suggested that expression of the hemagglutinin protease (encoded by hapA) in V. cholerae is positively regulated by QS signals. They showed that hapR, the positive regulator of hapA, is negatively regulated by luxO. They also suggest that luxO is negatively regulated by QS signals. Thus, at high cell density, AHLs and/or AI-2 would repress luxO and allow the induction of hapR, which would activate hapA transcription. In contrast, we show that in V. anguillarum neither AHLs nor AI-2 induces protease activity. We have previously shown that empA expression is cell density dependent (13). V. anguillarum produces three AHLs: 3-oxo-C10-AHL, C6-AHL, and 3-OH-C6-AHL (24, 25). The production of C6-AHL and 3-OH-C6-AHL was detected in M93Sm and NB10 when the cells reached late log phase in LB20 and NSSM. The lux-based reporter for detection of long-chain AHLs showed production of 3-oxo-C10-AHL by both strains in LB20 and NSSM when the cells reached stationary phase and increased for 3 h. In this study, we demonstrated that ethyl acetate extracts containing AHLs produced by V. anguillarum had no effect on protease production. Our result correlated with the findings of Milton et al. (24, 25). They cloned and made null mutations in the V. anguillarum AHL synthase genes vanI and vanM. The vanI and vanM mutants still exhibited empA protease activity.
While ethyl acetate extracts containing AHLs did not activate empA expression, NB10 and M93Sm cells incubated in conditioned cell LB20 supernatant from a stationary-phase culture of V. anguillarum M99 exhibited earlier and greater expression of protease activity. We tested LB20 supernatants of V. anguillarum luxS mutants and found no AI-2 activity when using the V. harveyi BB170 strain. Conditioned LB20 supernatant from the luxS null mutant (M01) allowed for a more rapid induction of protease activity in both wild-type and luxS mutant cells compared to incubation in either M99 conditioned LB20 or fresh LB20 (Fig. 8). One possibility is that AI-2 represses the expression of empA metalloprotease. Another possibility is the buildup of intermediates in the pathway for synthesis of AI-2 (48) in the luxS mutant (M01) somehow causes an earlier induction of protease activity when the conditioned supernatant is added to cells. Microarray analysis of an E. coli O157:H7 luxS mutant revealed over 400 genes up- or down-regulated in comparison to the parental strain (41). However only two observable phenotypes were restored when in vitro-synthesized AI-2 was added to luxS mutant cells (42). This suggested the possibility of a third unidentified signal, AI-3, which is synthesized by LuxS (42). Recently vanT of V. anguillarum, the luxR homologue of V. harveyi that regulates the lux operon, has been cloned and sequenced (12). A vanT mutant of V. anguillarum did not exhibit protease activity. Although this gene has been demonstrated to control empA, the autoinducer that may regulate the activity of this transcriptional regulator is not an AHL or AI-2 and has not been identified. In V. cholerae, the LysR homolog, AphB, regulates differential activation of the tcpPH promoter in classical and El Tor biotypes (22). Since the nucleotide differences between M93Sm and NB10 correspond to that observed in the tcpPH regulatory region, it could be hypothesized that empA of V. anguillarum is regulated by a similar mechanism. Finally, the requirement of stationary-phase conditions for protease activity and the sequence of the promoter region of empA suggested that empA expression is dependent upon σS encoded by rpoS. This was shown to be correct when EmpA expression was abolished in each of the rpoS mutant strains constructed. The expression of the empA homologues vvpE in V. vulnificus and hapA in V. cholerae has also been shown to be dependent upon rpoS (6, 20, 21). In summary, our data demonstrate that empA is regulated by at least three factors: (i) σS-dependent expression during stationary phase, (ii) gastrointestinal mucus as an inducer, and (iii) QS.
Acknowledgments
This research was supported by USDA NRICGP grant 2002-35204-4811 awarded to D.R.N.
We thank Bonnie Bassler of Princeton University, Princeton, N.J., for the V. harveyi strains; David Kirke of the University of Nottingham, University Park, Nottingham, United Kingdom, for pSB1075; and Michael Givskov and Jens Bo Andersen of the Technical University of Denmark, Lyngby, for the E. coli AHL reporter strains.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersen, J. B., A. Heydorn, M. Hentzer, L. Eberl, O. Geisenberger, B. B. Christensen, S. Molin, and M. Givskov. 2001. gfp-based N-acyl homoserine-lactone sensor systems for detection of bacterial communication. Appl. Environ. Microbiol. 67:575-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Austin, B., and D. A. Austin. 1993. Bacterial fish pathogens: diseases in farmed and wild fish, 2nd ed. Ellis Horwood, Ltd., Chichester, United Kingdom.
- 4.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1987. Current protocols in molecular biology. Wiley, New York, N.Y.
- 5.Bassler, B. L., M. Wright, and M. R. Silverman. 1994. Multiple signalling systems controlling expression of luminescence in Vibrio harveyi: sequence and function of genes encoding a second sensory pathway. Mol. Microbiol. 13:273-286. [DOI] [PubMed] [Google Scholar]
- 6.Benitez, J. A., A. J. Silva, and R. A. Finkelstein. 2001. Environmental signals controlling production of hemagglutinin/protease in Vibrio cholerae. Infect. Immun. 69:6549-6553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bever, R. A., and B. H. Iglewski. 1988. Molecular characterization and nucleotide sequence of the Pseudomonas aeruginosa elastase structural gene. J. Bacteriol. 170:4309-4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bolinches, J., A. E. Toranzo, A. Silva, and J. L. Barja. 1986. Vibriosis as the main causative factor of heavy mortalities in the oyster culture industry in northwestern Spain. Bull. Eur. Assoc. Fish Pathol. 6:1-4. [Google Scholar]
- 9.Bordas, M. A., M. C. Balebona, J. M. Rodriguez-Maroto, J. J. Borrego, and M. A. Moriñigo. 1998. Chemotaxis of pathogenic Vibrio strains towards mucus surfaces of gilt-head sea bream (Sparus aurata L.). Appl. Environ. Microbiol. 64:1573-1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bowser, P. R., R. Rosemark, and C. R. Reiner. 1981. A preliminary report of vibriosis in cultured American lobsters, Homarus americanus. J. Invertebr. Pathol. 37:80-85. [DOI] [PubMed] [Google Scholar]
- 11.Chen, X., S. Schauder, N. Potier, A. Van Dorsselaer, I. Pelczer, B. L. Bassler, and F. M. Hughson. 2002. Structural identification of a bacterial quorum-sensing signal containing boron. Nature 415:545-549. [DOI] [PubMed] [Google Scholar]
- 12.Croxatto, A., V. J. Chalker, J. Lauritz, J. Jass, A. Hardman, P. Williams, M. Cámara, and D. L. Milton. 2002. VanT, a homologue of Vibrio harveyi LuxR, regulates serine, metalloprotease, pigment, and biofilm production in Vibrio anguillarum. J. Bacteriol. 184:1617-1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Denkin, S. M., and D. R. Nelson. 1999. Induction of protease activity in Vibrio anguillarum by gastrointestinal mucus. Appl. Environ. Microbiol. 65:3555-3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eberhard, A., A. L. Burlingame, C. Eberhard, G. L. Kenyon, K. H. Nealson, and N. J. Oppenheimer. 1981. Structural identification of autoinducer of Photobacterium fischeri luciferase. Biochemistry 20:2444-2449. [DOI] [PubMed] [Google Scholar]
- 15.Garcia, T., K. Otto, S. Kjelleberg, and D. R. Nelson. 1997. Growth of Vibrio anguillarum in salmon intestinal mucus. Appl. Environ. Microbiol. 63:1034-1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greenberg, E. P., J. W. Hastings, and S. Ulitzur. 1979. Induction of luciferase synthesis in Beneckea harveyi by other marine bacteria. Arch. Microbiol. 120:87-91. [Google Scholar]
- 17.Hengge-Aronis, R. 1993. The role of rpoS in early stationary-phase gene regulation in Escherichia coli K12, p. 171-200. In S. Kjelleberg (ed.), Starvation in bacteria. Plenum Press, New York, N.Y.
- 18.Holden, M. T., S. Ram Chhabra, R. de Nys, P. Stead, N. J. Bainton, P. J. Hill, M. Manefield, N. Kumar, M. Labatte, D. England, S. Rice, M. Givskov, G. P. Salmond, G. S. Stewart, B. W. Bycroft, S. Kjelleberg, and P. Williams. 1999. Quorum-sensing cross talk: isolation and chemical characterization of cyclic dipeptides from Pseudomonas aeruginosa and other gram-negative bacteria. Mol. Microbiol. 33:1254-1266. [DOI] [PubMed] [Google Scholar]
- 19.Horne, M. T., R. H. Richards, R. J. Roberts, and P. C. Smith. 1977. Peracute vibriosis in juvenile turbot Scophthalmus maximus. J. Fish Pathol. 11:355-361. [Google Scholar]
- 20.Hülsmann, A., T. M. Rosche, I.-S. Kong, H. M. Hassan, D. M. Beam, and J. D. Oliver. 2003. RpoS-dependent stress response and exoenzyme production in Vibrio vulnificus. Appl. Environ. Microbiol. 69:6114-6120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jeong, H. S., M. H. Lee, K. H. Lee, S. J. Park, and S. H. Choi. 2003. SmcR and CRP coactivate Vibrio vulnificus vvpE encoding elastase through RpoS-dependent promoter in synergistic manner. J. Biol. Chem. 278:45072-45081. [DOI] [PubMed] [Google Scholar]
- 22.Kovacikova, G., and K. Skorupski. 2000. Differential activation of the tcpPH promoter by AphB determines biotype specificity of virulence gene expression in Vibrio cholerae. J. Bacteriol. 182:3228-3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McFall-Ngai, M. J., and E. G. Ruby. 2000. Developmental biology in marine invertebrate symbioses. Curr. Opin. Microbiol. 3:603-607. [DOI] [PubMed] [Google Scholar]
- 24.Milton, D. L., V. J. Chalker, D. Kirke, A. Hardman, M. Camara, and P. Williams. 2001. The LuxM homologue VanM from Vibrio anguillarum directs the synthesis of N-(3-hydroxyhexanoyl)homoserine lactone and N-hexanoylhomoserine lactone. J. Bacteriol. 183:3537-3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Milton, D. L., A. Hardman, M. Camara, S. R. Chhabra, B. W. Bycroft, G. S. A. B. Stewart, and P. Williams. 1997. Quorum sensing in Vibrio anguillarum: characterization of the vanI/vanR locus and identification of the autoinducer N-(3-oxodecanoyl)-l-homoserine lactone. J. Bacteriol. 179:3004-3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Milton, D. L., A. Norqvist, and H. Wolf-Watz. 1992. Cloning of a metalloprotease gene involved in the virulence mechanism of Vibrio anguillarum. J. Bacteriol. 174:7235-7244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Milton, D. L., A. Norqvist, and H. Wolf-Watz. 1995. Sequence of a novel virulence-mediating gene, virC, from Vibrio anguillarum. Gene 164:95-100. [DOI] [PubMed] [Google Scholar]
- 28.Milton, D. L., R. O'Toole, P. Hörstedt, and H. Wolf-Watz. 1996. Flagellin A is essential for the virulence of Vibrio anguillarum. J. Bacteriol. 178:1310-1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miyoshi, S.-I., H. Nakazawa, K. Kawata, K.-I. Tomochika, K. Tobe, and S. Shinoda. 1998. Characterization of the hemorrhagic reaction caused by Vibrio vulnificus metalloprotease, a member of the thermolysin family. Infect. Immun. 66:4851-4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Munn, C. B. 1977. Vibriosis in fish and its control. Fish Manag. 8:11-15. [Google Scholar]
- 31.Murley, Y. M., P. A. Carroll, K. Skorupski, R. K. Taylor, and S. B. Calderwood. 1999. Differential transcription of the tcpPH operon confers biotype-specific control of the Vibrio cholerae ToxR virulence regulon. Infect. Immun. 67:5117-5123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nealson, K. H., T. Platt, and J. W. Hastings. 1970. Cellular control of the synthesis and activity of the bacterial luminescent system. J. Bacteriol. 104:313-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nicas, T. I., and B. H. Iglewski. 1985. The contribution of exoproducts to virulence of Pseudomonas aeruginosa. Can. J. Microbiol. 31:387-392. [DOI] [PubMed] [Google Scholar]
- 34.Olsson, J. C., A. Jobörn, A. Westerdahl, L. Blomberg, S. Kjelleberg, and P. L. Conway. 1996. Is the turbot, Sophthalmus maximus (L.), intestine a portal of entry for the fish pathogen Vibrio anguillarum? J. Fish Dis. 19:225-234. [Google Scholar]
- 35.Passador, L., J. M. Cook, M. J. Gambello, L. Rust, and B. H. Iglewski. 1993. Expression of Pseudomonas aeruginosa virulence genes requires cell-to-cell communication. Science 260:1127-1130. [DOI] [PubMed] [Google Scholar]
- 36.Ransom, D. P. 1978. Bacteriologic, immunologic and pathologic studies of Vibrio sp. pathogenic to salmonids. Ph.D. thesis. Oregon State University, Corvallis.
- 37.Ransom, D. P., C. N. Lannan, J. S. Rohovecm, and J. L. Fryer. 1984. Comparison of histopathology caused by Vibrio anguillarum and Vibrio ordalii in three species of Pacific salmon. J. Fish Dis. 7:107-115. [Google Scholar]
- 38.Ruby, E. G., and K. H. Nealson. 1976. Symbiotic association of Photobacterium fischeri with the marine luminous fish Monocentris japonica: a model of symbiosis based on bacterial studies. Biol. Bull. 151:574-586. [DOI] [PubMed] [Google Scholar]
- 39.Schaefer, A. L., D. L. Val, B. L. Hanzelka, J. E. Cronan, Jr., and E. P. Greenberg. 1996. Generation of cell-to-cell signals in quorum sensing: acyl homoserine lactone synthase activity of a purified Vibrio fischeri LuxI protein. Proc. Natl. Acad. Sci. USA 93:9505-9509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schauder, S., K. Shokat, M. G. Surette, and B. L. Bassler. 2001. The LuxS family of bacterial autoinducers: biosynthesis of a novel quorum-sensing signal molecule. Mol. Microbiol. 41:463-476. [DOI] [PubMed] [Google Scholar]
- 41.Sperandio, V., A. G. Torres, J. A. Girón, and J. B. Kaper. 2001. Quorum sensing is a global regulatory mechanism in enterohemorrhagic Escherichia coli O157:H7. J. Bacteriol. 183:5187-5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sperandio, V., A. G. Torres, B. Jarvis, J. P. Nataro, and J. B. Kaper. 2003. Bacteria-host communication: the language of hormones. Proc. Natl. Acad. Sci. USA 100:8951-8956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Surette, M. G., and B. L. Bassler. 1998. Quorum sensing in Escherichia coli and Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 95:7046-7050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vaantanen, P. 1976. Microbiological studies in coastal waters of the northern Baltic Sea. I. Distribution and abundance of bacteria and yeasts in the Tvarminne area. Walter Ander Nottbeck Found. Sci. Rep. 1:1-58. [Google Scholar]
- 45.Windle, H. J. P., and D. Kelleher. 1997. Identification and characterization of a metalloprotease activity from Helicobacter pylori. Infect. Immun. 65:3132-3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Winson, M. K., M. Camara, A. Latifi, M. Foglino, S. R. Chhabra, M. Daykin, M. Bally, V. Chapon, G. P. Salmond, B. W. Bycroft et al. 1995. Multiple N-acyl-L-homoserine lactone signal molecules regulate production of virulence determinants and secondary metabolites in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 92:9427-9431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Winson, M. K., S. Swift, L. Fish, J. P. Throup, F. Jorgensen, S. R. Chhabra, B. W. Bycroft, P. Williams, and G. S. Stewart. 1998. Construction and analysis of luxCDABE-based plasmid sensors for investigating N-acyl homoserine lactone-mediated quorum sensing. FEMS Microbiol. Lett. 163:185-192. [DOI] [PubMed] [Google Scholar]
- 48.Winzer, K., K. R. Hardie, N. Burgess, N. Doherty, D. Kirke, M. T. Holden, R. Linforth, K. A. Cornell, A. J. Taylor, P. J. Hill, and P. Williams. 2002. LuxS: its role in central metabolism and the in vitro synthesis of 4-hydroxy-5-methyl-3(2H)-furanone. Microbiology 148:909-922. [DOI] [PubMed] [Google Scholar]
- 49.Woods, D. E., S. J. Cryz, R. L. Friedman, and B. H. Iglewski. 1982. Contribution of toxin A and elastase to virulence of Pseudomonas aeruginosa in chronic lung infections of rats. Infect. Immun. 36:1223-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhu, J., M. B. Miller, R. E. Vance, M. Dziejman, B. L. Bassler, and J. J. Mekalanos. 2002. Quorum-sensing regulators control virulence gene expression in Vibrio cholerae. Proc. Natl. Acad. Sci. USA 99:3129-3134. (First published 19 February 2002; 10.1073/pnas.052694299.) [DOI] [PMC free article] [PubMed] [Google Scholar]