Abstract
In this study, a molecular-beacon-based real-time reverse transcription (RT)-PCR assay was developed to detect the presence of hepatitis A virus (HAV) in environmental samples. A 125-bp, highly conserved 5′ noncoding region of HAV was targeted. The sensitivity of the real-time RT-PCR assay was tested with 10-fold dilutions of viral RNA, and a detection limit of 1 PFU was obtained. The specificity of the assay was demonstrated by testing with other environmental pathogens and indicator microorganisms, and only HAV was positively identified. When combined with immunomagnetic separation, the real-time RT-PCR assay successfully detected as few as 20 PFU in seeded groundwater samples. Because of its simplicity and specificity, this assay has broad applications for the rapid detection of HAV in contaminated foods or water.
Hepatitis A virus (HAV) is an important cause of acute hepatitis worldwide that can lead to severe illness or even death. It is transmitted by the fecal-oral route through the consumption of contaminated foods or water. Traditional methods for HAV detection based on cell culture propagation are often difficult to perform and can take more than 1 week before any visual cytopathic effects are observed (2). Molecular methods such as reverse transcription (RT)-PCR and integrated cell culture-PCR (6, 11, 15) have largely superseded the cell culture methods, offering improved sensitivity and specificity. However, positive detection still relies on visual detection of an appropriately sized DNA band, followed by specific hybridization with a radiolabeled DNA probe, which can take up to 15 h and is not amenable to automation. Recent developments in the real-time RT-PCR technique have engendered wider acceptance of the PCR assay, as it eliminates the need for gel electrophoresis and improves the speed, sensitivity, and reproducibility of detection (13).
Molecular beacons (MBs) are single-stranded fluorescence probes that form a stem-loop structure. In the presence of a target sequence, the MB undergoes a spontaneous conformational reorganization that forces the stem hybrid to dissociate and causes fluorescence to occur. Since unhybridized MBs do not have to be separated, they can be included in PCRs, permitting the progress of the reaction to be followed in real time (21).
Pretreatments of environmental samples before RT-PCR are often necessary to reduce reaction volumes and to remove PCR inhibitors naturally present in the samples (1, 10, 18). Immunomagnetic separation (IMS) is a simple and powerful tool for quick and effective separation and isolation of bacteria and viruses from environmental water samples (4, 5, 7, 12). IMS depends on isolation of the antigen from the sample with either monoclonal or polyclonal antibodies coupled to magnetic beads. In this paper, an MB-based real-time RT-PCR assay for HAV was developed. We demonstrate that the reported assay is highly sensitive and specific for HAV and, when combined with IMS, allows detection of HAV in seeded ground water samples.
Selection of the target region for RT-PCR.
The primers and MB employed in this work were designed on the basis of an alignment of the sequences of 15 different HAV strains obtained from the GenBank database. Although most of the published RT-PCR assays for HAV targeted the VP1 and VP2 regions (7, 9, 14), our alignment results indicated that the 5′ noncoding region (5′-NCR) is more conserved and it was chosen as the target region. As shown in Table 1, primers KH1 (380-397) (5′-ATCTTCCACAAGGGGTAG-3′) and KH2 (487-504) (5′-CGGCGTTGAATGGTTTTT-3′) were designed to amplify a 125-bp region of the 5′-NCR. MB 5′-FAM-CTTGCGGGATAGGGTAACAGCGGCGGCGCAAG-DABCYL-3′ (21) was designed to be perfectly complementary to a 20-bp region of the amplicon (Midland Certified Reagent Co., Midland, Tex.).
TABLE 1.
Alignment of the genomic sequences of different HAV strains in the working region of the PCR primers in the 5′ NCR
| Human HAV strain | Accession no. | Upstream primer KH1 region (380-397) sequence (5′-3′)a | Downstream primer KH2 region (488-504) sequence (3′-5′) |
|---|---|---|---|
| HHAV-HM175 | M14707 | ATCTTCCACAAGGGGTAG | AAAACCATTCAACGCCG |
| HHAV-HM175 | M16632 | ------------------ | ----------------- |
| HHAV-HM175 (HPA18F) | M59808 | ------------------ | ----------------- |
| HHAV-HM175 (HPA24A) | M59810 | ------------------ | ----------------- |
| HHAV-HM175 (HPA43C) | M59809 | ------------------ | ----------------- |
| HHAV-GBM | X75216 | ------------------ | ----------------- |
| HHAV-FG | X83302 | ------------------ | ----------------- |
| HHAV-AH2 | AB020565 | ------------------ | ----------------- |
| HHAV-FH1 | AB020567 | ------------------ | ----------------- |
| HHAV-FH2 | AB020568 | ------------------ | ----------------- |
| HHAV-FH3 | AB020569 | ------------------ | ----------------- |
| HHAV-HAF-203 | AF268396 | ------------------ | ----------------- |
| HHAV-LA | K02990 | ------------------ | ----------------- |
| HHAV-LU38 | AF357222 | ------------------ | ----------------- |
| HHAV-LY6 | AF485328 | ------------------ | ----------------- |
| HHAV-AH1 | AB020564 | -----T-----A------ | ----------------- |
| HHAV-AH3 | AB020566 | -----------AA----C | ----------------- |
| HHAV-MBB | M20273 | -----------A------ | ----------------- |
| HHAV-SLF88 | AY03286 | -----------A------ | ------T---------- |
Dashes represent nucleotides identical to those in the sequence at the top.
Real-time RT-PCR assay.
The ability of the real-time RT-PCR assay to detect HAV was investigated with cytopathic HAV strain HM175, which was obtained from the American Type Culture Collection (ATCC), Manassas, Va. Fetal rhesus monkey kidney (FRhK-4) cells were used for virus propagation (2), and viral loads were determined by the standard plaque assay (17). Total viral RNA was extracted by the phenol-chloroform method (16).
All RT-PCRs were performed with a GeneAmp Gold RNA PCR Core Kit (Applied Biosystems, Foster City, Calif.) as described by the manufacturer. An iQ icycler (Bio-Rad, Hercules, Calif.) was used for all real-time RT-PCRs. In the RT step, deoxyribonucleoside triphosphates and primer KH2 were used at final concentrations of 250 and 0.4 μM, respectively. For the PCR step, 0.4 μM KH1 and 0.5 μM MB were used in a final volume of 50 μl. PCRs were performed with 50 cycles of melting at 95°C for 1 min, annealing at 56°C for 1 min, and extension at 72°C for 1 min. Fluorescence measurements were recorded at each annealing step. The threshold cycle (Ct) of each amplification reaction was calculated on the basis of the first PCR cycle at which the fluorescence was 10-fold higher than the standard deviation of the mean baseline emission.
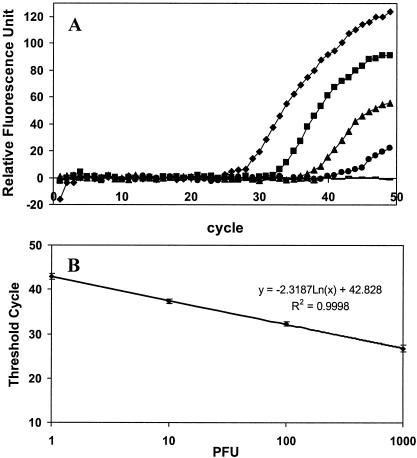
Serial dilutions of viral RNA were evaluated to ascertain the sensitivity of the assay. A 125-bp fragment was correctly amplified by the primers, and the amplicon was detected by the MB as indicated by a significant increase in fluorescence (Fig. 1A). As little as 1 PFU per reaction was detected. Although a viral load of less than 1 PFU was sometimes detected, the frequency was too low to confirm positive detection. The reproducibility of the assay was evaluated by comparing the Ct values obtained from five independent sets of real-time RT-PCR assays with less than 2% variability observed. A linear standard curve of 1,000 to 1 PFU per reaction was obtained (Fig. 1B).
FIG. 1.
Sensitivity of the real-time RT-PCR assay. (A) Detection of serial dilutions of viral RNA at 1,000 (⧫), 100 (▪), 10 (▴), and 1 (•) PFU per reaction was done. A negative control (−) containing only water was used for comparison. (B) Standard curve generated by plotting the Ct value versus the number of PFU. The data represent the results of five independent experiments.
Specificity of the real-time RT-PCR assay.
The selectivity of the MB-based RT-PCR relies on the selected sequence of the primer set and the probe moiety of the beacon, both of which were based on the 5′-NCR of HAV strain HM-175. Experiments were conducted to determine the specificity of the assay for other potential pathogens and indicator microorganisms found in contaminated foods or water (Table 2). None of these species produced significant fluorescence that could be detected by the real-time RT-PCR assay, demonstrating the specificity of the assay. The ability of the assay to detect HAV strains other than HM-175 was confirmed by the observation of a positive response with a clinical isolate (GA76) obtained from the Centers for Disease Control and Prevention.
TABLE 2.
Pathogens tested to determine the specificity of the assay
| Agent | Strain | Detection | Sourcea |
|---|---|---|---|
| HAV | HM175 | + | ATCC |
| HAV | GA76 | + | CDC |
| Coxsackievirus B1 (ATCC VR-28) | Conn-5 | − | ATCC |
| Coxsackievirus B3 (ATCC VR-30) | Nancy | − | ATCC |
| Coxsackievirus B6 (ATCC VR-155) | Schmitt | − | ATCC |
| Echovirus 11 (ATCC VR-41) | Gregory | − | ATCC |
| Echovirus 17 (ATCC VR-1058) | CHHE-29 | − | ATCC |
| Echovirus 19 (ATCC VR-1060) | Bruke | − | ATCC |
| Human paraechovirus type 1 | Valencia | − | CSD |
| Adenovirus type 2 | Sewage isolate | − | CSD |
| Adenovirus type 15 | Sewage isolate | − | CSD |
| Poliovirus 1 | LscAb | − | ATCC |
| Rotavirus | Wa | − | ATCC |
| MS2 (ATCC 15597-B1) | − | ATCC | |
| φX174 (ATCC 13706-B1) | − | ATCC |
CDC, Hepatitis Branch, National Center for Infectious Diseases, Centers for Disease Control and Prevention, Atlanta, Ga. CSD, County Sanitation Districts of Los Angeles County, Whittier, Calif.
Combined IMS-MB-RT-PCR assay for detection of HAV in seeded groundwater.
The utility of the real-time RT-PCR assay for environmental samples was tested with seeded groundwater samples obtained from southern California. Sample collection and processing were done by standard methods. Briefly, 1,000 liters of groundwater was collected by using an electropositive MDS filter (AMF CUNO, Meriden, Conn.). The filter cartridge was then flushed with 1 liter of 1.5% beef extract V containing 0.05 M glycine (pH 7.5; Becton Dickinson, Sparks, Md.). The eluate was concentrated by adjusting the pH from 7.4 to 3.5 with 0.10 M HCl and centrifugation for 15 min at 4,000 × g. The resulting pellet was resuspended in 20 ml of 0.15 M Na2HPO4 buffer, and the pH was adjusted to 7.4 with 0.10 M NaOH (19). The concentrate was analyzed for the presence of HAV by molecular methods and cell culture and found to be negative.
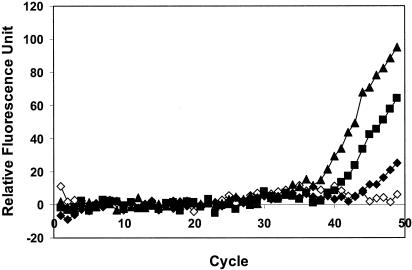
When different dilutions of HAV were added to the groundwater concentrates, no visible increase in fluorescence was observed in the real-time RT-PCR assay, indicating severe interference from the inhibitors present in the samples. Although many pretreatment methods are available for the removal of inhibitors, IMS is particularly attractive because of the increased potential for detecting intact and infective viruses. To recover HAV with IMS, magnetic beads coated with a human polyclonal HAV antibody (Gamimune N; Bayer Co., Elkhart, Ind.) were added to groundwater concentrates containing different dilutions of HAV. Beads were recovered with a strong magnetic particle separator stand (Novagen Inc., Madison, Wis.) after 2 h of incubation at room temperature. Viral RNA was released by heating at 99°C for 5 min in 13 μl of RT-PCR buffer and subjected to RT-PCR assay. A strong positive signal was obtained for all of the dilutions, with as few as 20 PFU detected (Fig. 2). These results demonstrated the potential of the combined IMS-real-time RT-PCR assay for rapid and quantitative detection of HAV in contaminated water and foods.
FIG. 2.
Detection of HAV in seeded groundwater samples by a combined IMS-MB-RT-PCR assay. Results are shown for 2,000 (▴), 200 (▪), and 20 (⧫) PFU and distilled water (◊) as a control.
Conclusion.
Up to now, only one real-time RT-PCR assay, based on TaqMan technology, has been reported for HAV detection (3). However, the use of MB has several advantages over this technology, including improved specificity and an increased signal-to-noise ratio (20). Our findings demonstrate that the reported MB-based RT-PCR assay is sensitive and specific for the detection of HAV, and as little as 1 PFU was detected. These detection limits are similar to those reported with the TaqMan RT-PCR assay and a recent report based on detection by the nucleic acid sequence-based amplification method (8).
The presence of inhibitory compounds in environmental water represents a major problem that necessitates the concentration of large volumes (hundreds of liters) of water and the removal of concentrated inhibitors. This was observed in the present study by the inability to detect viral RNA directly extracted from groundwater concentrates. By using IMS to separate HAV from the inhibitory substances, a detectable signal was obtained for as few as 20 PFU. These observations further confirm the potential of the combined IMS-MB-RT-PCR assay as a simple tool for rapid detection and quantification of HAV. Since the principle of IMS is based on antigen capture with surface epitopes of HAV, a positive detection result could potentially be correlated with the presence of intact and infectious HAV in a sample. This possibility is under investigation.
Acknowledgments
This work was funded in part by a grant from the UC Water Resources Center. Khaled H. Abd El Galil was supported by the Egyptian Ministry of High Education.
REFERENCES
- 1.Abbaszadegan, M., M. S. Huber, C. P. Gerba, and I. L. Pepper. 1993. Detection of enteroviruses in ground water with the polymerase chain reaction. Appl. Environ. Microbiol. 59:1318-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bidawid, S., J. M. Farber, and S. A. Sattar. 2000. Contamination of foods by food handlers: experiments on hepatitis A virus transfer to food and its interruption. Appl. Environ. Microbiol. 66:2759-2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Costa-Mattioli, M., S. Monpoeho, E. Nicand, M. H. Aleman, S. Billaudel, and V. Ferré. 2002. Quantification and duration of viraemia during hepatitis A infection as determined by real-time RT-PCR. J. Viral Hepatitis 9:101-106. [DOI] [PubMed] [Google Scholar]
- 4.Deng, M. Y., S. P. Day, and D. O. Cliver. 1994. Detection of hepatitis A virus in environmental samples by antigen capture PCR. Appl. Environ. Microbiol. 60:1927-1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grinde, B., T. O. Jonassen, and H. Ushijima. 1995. Sensitive detection of group A rotaviruses by immunomagnetic separation and reverse transcription polymerase chain reaction. J. Virol. Methods 55:327-338. [DOI] [PubMed] [Google Scholar]
- 6.Hurst, C. J., G. R. Knudsen, M. J. McInerney, L. D. Stetzenbach, and M. V. Walter. 2001. Manual of environmental microbiology. ASM Press, Washington, D.C.
- 7.Jansen, R. W., G. Siegl, and S. M. Lemon. 1990. Molecular epidemiology of human hepatitis A virus defined by an antigen-capture polymerase chain reaction. Proc. Natl. Acad. Sci. USA 87:2867-2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jean, J., B. Blais, A. Darveau, and I. Fliss. 2001. Detection of hepatitis A virus by the nucleic acid sequence-based amplification technique and comparison with reverse transcription-PCR. Appl. Environ. Microbiol. 67:5593-5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jothikumar, N., R. Paulmurugan, P. Padmanabhan, R. B. Sundari, S. Kamatchiammal, and K. S. Rao. 2000. Duplex RT-PCR for simultaneous detection of hepatitis A and hepatitis E virus isolated from drinking water samples. J. Environ. Monit. 2:587-590. [DOI] [PubMed] [Google Scholar]
- 10.Kreader, C. A. 1995. Design and evaluation of bacteroids DNA probes for the specific detection of human fecal pollution. Appl. Environ. Microbiol. 61:1171-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Legeay, O., Y. Caudrelier, C. Cordevant, L. Rigottier-Gois, and M. Lange. 2000. Simplified procedure for detection of enteric pathogenic viruses in shellfish by RT-PCR. J. Virol. Methods 90:1-14. [DOI] [PubMed] [Google Scholar]
- 12.López-Sabater, E. I., M. Y. Deng, and D. O. Cliver. 1997. Magnetic immunoseparation PCR assay (MIPA) for detection of hepatitis A virus (HAV) in American oyster (Crassostrea virginica). Lett. Appl. Microbiol. 24:101-104. [DOI] [PubMed] [Google Scholar]
- 13.Mackay, I. M., K. E. Andren, and A. Nitsche. 2002. Real-time PCR in virology. Nucleic Acids Res. 30:1292-1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Monceyron, C., and B. Grinde. 1994. Detection of hepatitis A virus in clinical and environmental samples by immunomagnetic separation and PCR. J. Virol. Methods 46:157-166. [DOI] [PubMed] [Google Scholar]
- 15.Reynolds, K. A., C. P. Gerba, M. Abbaszadegan, and I. L. Pepper. 2001. ICC/PCR detection of enteroviruses and hepatitis A virus in environmental samples. Can. J. Microbiol. 47:153-157. [PubMed] [Google Scholar]
- 16.Sambrook J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 17.Sattar, S. A., V. S. Springthorpe, Y. Karim, and P. Loro. 1989. Chemical disinfection of nonporous inanimate surfaces experimentally contaminated with four human pathogenic viruses. Epidemiol. Infect. 102:493-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schwab, K. J., R. De Leon, and M. D. Sobsey. 1993. Development of PCR methods for enteric virus detection in water. Water Sci. Tech. 27:211-218. [Google Scholar]
- 19.Sobsey, M. D., and S. J. Glass. 1980. Poliovirus concentration from tap waters with electropositive absorbent filters. Appl. Environ. Microbiol. 40:201-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Täpp, I., L. Malmberg, E. Rennel, M. Wik, and A. C. Syvänen. 2000. Homogeneous scoring of single-nucleotide polymorphisms: comparison of the 5′-nuclease Taqman assay and molecular beacon probes. BioTechniques 28:732-738. [DOI] [PubMed] [Google Scholar]
- 21.Tyagi, S., and F. R. Kramer. 1996. Molecular beacons: probes that fluoresce upon hybridization. Nat. Biotechnol. 14:303-308. [DOI] [PubMed] [Google Scholar]