Background: Conflicting claims exist concerning the occurrence of the glyoxylate cycle in cyanobacteria.
Results: The genes for isocitrate lyase and malate synthase were identified in Chlorogleopsis fritschii PCC 9212 and the purified enzymes were characterized.
Conclusion: C. fritschii has a functional glyoxylate cycle and can grow in the dark on acetate.
Significance: These results clarify the occurrence of the glyoxylate cycle in cyanobacteria.
Keywords: acetyl coenzyme A (acetyl-CoA), cyanobacteria, metabolism, photosynthesis, tricarboxylic acid cycle (TCA cycle) (Krebs cycle), TCA cycle, glyoxylate cycle, isocitrate lyase, malate synthase, poly-3-hydroxybutyrate
Abstract
Cyanobacteria are important photoautotrophic bacteria with extensive but variable metabolic capacities. The existence of the glyoxylate cycle, a variant of the TCA cycle, is still poorly documented in cyanobacteria. Previous studies reported the activities of isocitrate lyase and malate synthase, the key enzymes of the glyoxylate cycle in some cyanobacteria, but other studies concluded that these enzymes are missing. In this study the genes encoding isocitrate lyase and malate synthase from Chlorogloeopsis fritschii PCC 9212 were identified, and the recombinant enzymes were biochemically characterized. Consistent with the presence of the enzymes of the glyoxylate cycle, C. fritschii could assimilate acetate under both light and dark growth conditions. Transcript abundances for isocitrate lyase and malate synthase increased, and C. fritschii grew faster, when the growth medium was supplemented with acetate. Adding acetate to the growth medium also increased the yield of poly-3-hydroxybutyrate. When the genes encoding isocitrate lyase and malate synthase were expressed in Synechococcus sp. PCC 7002, the acetate assimilation capacity of the resulting strain was greater than that of wild type. Database searches showed that the genes for the glyoxylate cycle exist in only a few other cyanobacteria, all of which are able to fix nitrogen. This study demonstrates that the glyoxylate cycle exists in a few cyanobacteria, and that this pathway plays an important role in the assimilation of acetate for growth in one of those organisms. The glyoxylate cycle might play a role in coordinating carbon and nitrogen metabolism under conditions of nitrogen fixation.
Introduction
Under natural growth conditions, all bacteria continually face changing nutrient availability, and consequently they must strategically adapt their metabolic capabilities in response to such changes. In addition to utilizing various storage compounds, the capacity to take up and use dissolved carboxylic acids, such as acetate, lactate, pyruvate, and succinate, from the surrounding environment is important for sustainable growth under many conditions (1–3). The ability to assimilate organic carbons also exists in some autotrophic bacteria, including cyanobacteria, even though most are able to synthesize all essential precursor metabolites from CO2 (4). The assimilation of dissolved carboxylic acids by heterotrophic bacteria has been known and studied for decades (5, 6). Acetate is one of the most common and important carbon sources for many bacteria, and acetate is frequently used as a carbon source by eukaryotic microalgae (7, 8). Once acetate is transported into the cytosol, it is first converted by acetyl-CoA synthetase to acetyl coenzyme A (acetyl-CoA),2 which can then be used by the tricarboxylic acid (TCA) cycle or the glyoxylate cycle to produce other important precursor metabolites, such as 2-oxoglutarate and oxaloacetate (9, 10).
Sir Hans Adolf Krebs, who also established the urea/ornithine cycle as well as the TCA cycle, discovered the glyoxylate cycle (11, 12). The glyoxylate cycle is usually described as a modified TCA cycle, because it shares the activities of malate dehydrogenase, citrate synthase, and aconitase with the TCA cycle (Fig. 1). However, the difference lies in the two key enzymes that are used in the glyoxylate cycle but which are not used in the TCA cycle, namely isocitrate lyase (AceA) and malate synthase (AceB), which convert isocitrate and acetyl-CoA into succinate and malate (Fig. 1). In more detail, isocitrate is split into succinate and glyoxylate by isocitrate lyase, after which glyoxylate and acetyl-CoA are condensed to form malate with the release of CoA by malate synthase. Malate is further converted to oxaloacetate by malate dehydrogenase to continue the cycle, and succinate is released as the net product. Overall, the net reaction of the glyoxylate cycle, which can be used to produce precursors for amino acid or carbohydrate biosynthesis, allows cells to convert two acetyl-CoA units into succinate and avoid the CO2-releasing steps of the TCA cycle. Thus, the glyoxylate cycle enables cells to utilize C2 units (i.e. acetyl-CoA) more efficiently for biomass production. These C2 units can be derived from ethanol or acetate as the sole carbon source, and collectively these reactions are usually correlated with the ability of bacteria to assimilate acetate (13).
FIGURE 1.
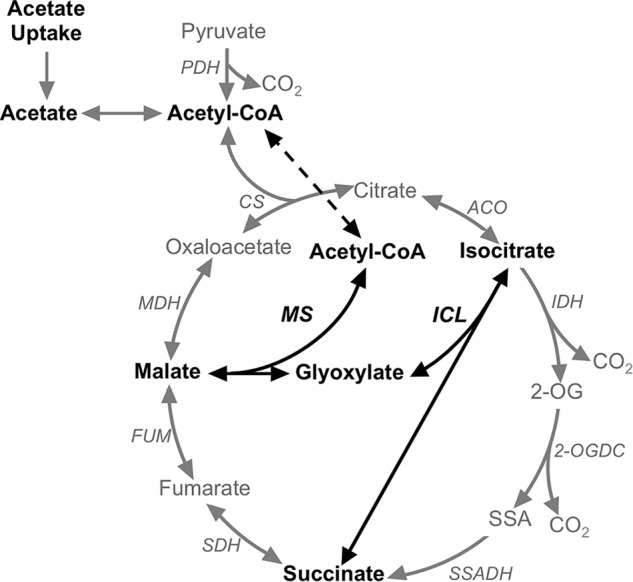
Scheme showing the glyoxylate and TCA cycles in some cyanobacteria. Abbreviations used were: 2-OG, 2-oxoglutarate; 2-OGDC, 2-oxoglutarate decarboxylase; ACO, aconitase; CS, citrate synthase; FUM, fumarase; ICL, isocitrate lyase; IDH, isocitrate dehydrogenase; MDH, malate dehydrogenase; MS, malate synthase; PDH, pyruvate dehydrogenase; SDH, succinic acid dehydrogenase; SSA, succinic semialdehyde; SSADH, succinic semialdehyde dehydrogenase. The heavy arrows show the two reactions specific for the glyoxylate cycle.
The glyoxylate cycle has been found in many chlorophototrophic bacteria (14, 15). Isocitrate lyase and malate synthase are found in all chlorophototrophic members of the Chloroflexi (e.g. Chloroflexus spp., Oscillochloris trichoides, and Roseiflexus spp.). By using the glyoxylate cycle, all of these organisms are able to photoassimilate acetate, and some can even grow heterotrophically on acetate (16, 17). In addition, the glyoxylate cycle occurs in most purple sulfur bacteria, which can also photoassimilate acetate. However, no genes encoding these enzymes have yet been identified in most purple non-sulfur bacteria (15). Heliobacteria, green sulfur bacteria, and Chloracidobacterium thermophilum lack isocitrate lyase and malate synthase, and thus the glyoxylate cycle is absent in these bacteria. Interesting, heliobacteria and green sulfur bacteria use a different acetate assimilation mechanism, the carboxylation of acetyl-CoA by pyruvate synthase, and thus these bacteria are thus able to assimilate both acetate and CO2 at the same time (18, 19).
Cyanobacteria are a large group of oxygenic chlorophototrophic bacteria with highly diverse metabolic capabilities, but the occurrence of the glyoxylate cycle in these organisms has remained controversial (20). Although it has been reported that isocitrate lyase and/or malate synthase activities were detected in some cyanobacteria (21, 22), and that some cyanobacteria were able to assimilate acetate (23, 24), a recent study in Synechocystis sp. PCC 6803 failed to detect the enzymes of the glyoxylate cycle (25). However, a recent genome sequencing study reported that two Cyanothece spp. (strains PCC 7424 and PCC 7822) have an operon encoding the isocitrate lyase and malate synthase (26). However, this study did not demonstrate acetate utilization or the enzyme activities of the genes in question. Database searches showed that similar operons were also found in the genomes of two Chlorogloeopsis sp., strains, PCC 6912 and PCC 9212. Consistent with the presence of these two genes and thus the glyoxylate cycle in the Chlorogloeopsis spp., one of the organisms had been reported to assimilate acetate under both light and dark conditions (23).
Further confusing the properties of the TCA and glyoxylate cycle enzymes in cyanobacteria, no gene encoding fumarase was initially identified in the annotation of the genome of Synechococcus sp. PCC 7002, although a fumarase was annotated in the genome of Synechocystis sp. PCC 6803. BLASTP searches showed that, among all the gene products in Synechococcus sp. PCC 7002, the product of the open reading frame of SYNPCC7002_A2041 had the highest sequence identity (43%) to the fumarase (slr0018) from Synechocystis sp. PCC 6803. Although it had initially been misannotated as aspartate ammonia-lyase, it thus seemed likely that this gene encodes fumarase.
In this study, we describe the biochemical validation of the predicted fumarase (SYNPCC7002_A2041) from Synechococcus sp. PCC 7002, as well as for two genes in Chlorogloeopsis fritschii PCC 9212 that encode the key enzymes, isocitrate lyase and malate synthase, of the glyoxylate cycle. We show that C. fritschii PCC 9212 can take up acetate under both light and dark conditions, and that the organism grows faster when acetate is supplied in the medium. Whole cell transcription profiling showed that the mRNA levels of these two genes increased when cells were grown with acetate. Furthermore, C. fritschii PCC 9212 cells accumulated much higher poly-3-hydroxybutyrate (PHB) levels when cells were supplied with acetate. This observation suggested that the extra carbon supplied as acetate was mainly stored as PHB. Additionally, when the genes for isocitrate lyase and malate synthase were overexpressed in Synechococcus sp. PCC 7002, this cyanobacterium exhibited an enhanced capacity for acetate uptake, confirming that the glyoxylate cycle can play an important role in acetate utilization even for an organism that normally lacks this capability. Overall, this study validates the existence of the glyxoxylate cycle in cyanobacteria but demonstrates that only a small number of cyanobacteria actually have this cycle. Our studies show that the glyoxylate cycle is not a common or prominent feature of cyanobacterial metabolism, but it may nevertheless be important for acetate utilization in those few organisms that have the enzymes of this pathway.
Experimental Procedures
Strains and Growth Conditions
C. fritschii PCC 9212 was obtained from the Pasteur Culture Collection (PCC) and routinely grown in medium BG-11 at 26 °C (27). To emphasize the effects of acetate supplementation, cells were grown under constant irradiance of 50 μmol photons m−2 s−1, which was provided by cool white fluorescent tubes, and cultures were sparged with 1% (v/v) CO2 in air (standard growth conditions). Low CO2 growth conditions were achieved by bubbling cultures with air while keeping all other growth conditions the same. When required, the growth medium was supplemented with 10 mm sodium acetate. The wild-type strain of Synechococcus sp. PCC 7002 as well as a strain overexpressing the genes of the glyoxylate cycle (strain Synechococcus 7002-glyox) were grown in liquid A+ medium under standard conditions for this organism (28): cells were grown at an irradiance of 250 μmol photons m−2 s−1 provided by cool white fluorescent lights, at 38 °C and cultures were sparged with 1% (v/v) CO2 in air. Low irradiance or low CO2 growth conditions were produced by growing cells under 50 μmol photons m−2 s−1 or by sparging cultures with air under otherwise standard conditions. When required for experiments with Synechococcus sp. PCC 7002, 10 mm sodium acetate was added to the A+ medium.
Acetate Concentration Measurement in Growth Medium
The concentration of acetate in the medium at different growth stages was determined by high-performance liquid chromatography (HPLC). In detail, aliquots (0.5 ml) of cell culture were removed from the growth medium at different growth stages. After centrifugation, the supernatant was filtered through 0.2-μm sterile syringe filter (VWR, Philadelphia, PA). A 20-μl aliquot of the filtered solution was loaded directly onto a Shimadzu LC-20AB HPLC system equipped with 210-nm UV detector SPD-20A. Different components in the medium were separated on a Supelcogel C610H column (Supelco, Bellefonte, PA), using 4 mm H2SO4 as the mobile phase. The flow rate was 0.5 ml min−1 and the chromatography was performed at 30 °C. Acetate concentrations were calculated on the basis of peak area using a standard curve generated from known concentrations of sodium acetate.
Cloning, Protein Purification, and Protein Identification
Open reading frames SYNPCC7002_A2041, encoding the putative fumarase of Synechococcus sp. PCC 7002, UYEDRAFT_02681, encoding the putative isocitrate lyase and UYEDRAFT_02682, encoding the putative malate synthase of C. fritschii PCC 9212 were amplified by polymerase chain reaction (PCR) with Phusion DNA polymerase (New England Biolabs, Ipswich, MA) and separately cloned into plasmid pAQ1Ex-PcpcBA (29). Primer set ICLF-ICLR was used to amplify UYEDRAFT_02681, primer set MSF-MSR was used to amplify UYEDRAFT_02682, and primer set FUMF-FUMR was used to amplify SYNPCC7002_A2041 (Table 1). An N-terminal His10 tag was introduced into isocitrate lyase and fumarase to facilitate subsequent protein purification. Initial attempts to add a His10 tag to the N terminus of malate synthase were not successful, and subsequently, a His6 tag was successfully added to the C terminus. The resulting plasmids pAQ1Ex-PcpcBA::A2041, pAQ1Ex-PcpcBA::U02681, and pAQ1Ex-PcpcBA::U02682 were verified by DNA sequencing and transformed into Escherichia coli strain DH5-α. Cells were grown overnight in 1 liter of Luria-Bertani (LB) medium containing 50 μg ml−1 gentamycin, harvested by centrifugation at 4 °C at 5,000 × g, and washed once with 50 mm Tris-HCl buffer, pH 8.0. Cells were disrupted by three passages through a chilled French pressure cell operated at 138 MPa. Soluble lysates were obtained by centrifugation at 20,000 × g for 30 min and loaded onto a Ni2+-NTA affinity resin (Goldbio, St. Louis, MO), which was pre-equilibrated with 10 mm imidazole in 50 mm Tris-HCl, pH 8.0, and washed with 30 mm imidazole in 50 mm Tris-HCl, pH 8.0, 300 mm NaCl. Proteins were eluted stepwise with 50, 100, 150, 200, and 250 mm imidazole in 50 mm Tris-HCl, pH 8.0, 300 mm NaCl. Fractions containing the recombinant proteins were monitored by polyacrylamide gel electrophoresis in the presence of sodium dodecyl sulfate (SDS-PAGE) and concentrated by ultrafiltration using Centriprep columns (Millipore, Billerica, MA). Purified proteins were further analyzed by SDS-PAGE and immunoblotting with commercial antibodies (Rockland, Limerick, PA) to the poly-His tags. Proteins were also positively identified by tryptic peptide mass fingerprinting as previously described (20).
TABLE 1.
Oligonucleotide primers used for PCR in this study
| Name | Sequence (5′ to 3′) |
|---|---|
| MSF | TTGCGTCCATGGCAAAAGGCTTTTCCTCTG |
| MSR | GTGGTGCTCGAGAAAACTGACAAGATGACG |
| ICLF | GCGCGGCATATGAGTAAATCTACTTTTGAA |
| ICLR | TAGTATGGATCCCTACTCATCATCCATGCG |
| FUMF | CAGTCAAAACATATGAGCGCAGAT |
| FUMR | CGCCTTGGATCCTCCCGATTA ATT |
Enzymatic Assays
Fumarase activity was assayed by separately measuring the reversible interconversion of malate into fumarate, as catalyzed by the recombinant enzyme. For the conversion of malate to fumarate, the reaction mixture (0.2 ml) contained 10 mm malate, 50 mm K-phosphate, pH 7.0, and 50 μg of purified SynPCC7002_A2041. The mixture was incubated at room temperature for 1 h, and an aliquot (20 μl) of the reaction mixture was injected into the HPLC for analysis. For the conversion of fumarate to malate, the reaction mixture (0.2 ml) contained 2.5 mm fumarate, 50 mm K-phosphate, pH 7.0, and 50 μg of purified SynPCC7002_A2041. The assay mixture was incubated at room temperature for 1 h, and an aliquot (20 μl) of the reaction mixture was injected into the HPLC for analysis. Control experiments were performed in a similar manner but without the addition of the purified enzyme.
For enzyme assays with isocitrate lyase, the reaction mixture (0.2 ml) contained 2 mm isocitrate, 50 mm K-phosphate, pH 7.8, 2 mm MgCl2, and 50 μg of purified UYEDRAFT_02681. The mixture was incubated at room temperature for 1 h, and then an aliquot (20 μl) of the reaction mixture was injected into the HPLC for analysis. The condensation of succinate and glyoxylate to isocitrate by isocitrate lyase was also assayed. The reaction mixture (0.2 ml) contained 1 mm glyoxylate, 1 mm succinate, 50 mm K-phosphate, pH 7.8, 1 mm MgCl2, and 50 μg of purified UYEDRAFT_02681. The mixture was incubated at room temperature for 1 h, and then an aliquot (20 μl) of the reaction mixture was injected into the HPLC for analysis. Control experiments were performed similarly but without the addition of the purified enzyme.
For enzyme assays with malate synthase, the reaction mixture (0.2 ml) contained 2 mm acetyl-CoA, 2 mm glyoxylate, 2 mm MgCl2, 50 mm K-phosphate, pH 7.8, and 50 μg of purified UYEDRAFT_02682. The mixture was incubated at room temperature for 1 h, and an aliquot (20 μl) of the reaction mixture was injected into the HPLC for analysis. Control experiments were performed similarly but without the addition of the purified enzyme. The elution times and concentrations of substrates and products were determined by comparison of results obtained from analyses of individual compounds.
Overexpression of Glyoxylate Cycle Genes in Synechococcus sp. PCC 7002
Open reading frames UYEDRAFT_02681 (isocitrate lyase) and UYEDRAFT_02682 (malate synthase) form an apparent operon in C. fritschii PCC 9212, and the entire operon was amplified by PCR and inserted into the pAQ1-based expression system (29) using primer sets MSF and ICLR (Table 1). The resulting plasmid was verified by DNA sequencing and transformed into wild type Synechococcus sp. PCC 7002 as previously described (30). The presence of the desired genes in strain Synechococcus 7002-glyox was confirmed by PCR using primer set MSF and ICLR (Table 1).
PHB Extraction and Quantification
Quantification of PHB was performed as previously described (31, 32). Briefly, parallel liquid cultures (20 ml) of C. fritschii PCC 9212 were grown to different growth stages and at selected times, the cells were harvested by centrifugation for 10 min at 10,000 × g. The pellets were washed once with double-deionized water (20 ml). The resulting cell pellets were lyophilized to obtain dried cells. The dried cells and PHB standards (Sigma) were placed into glass tubes with sealed rubber caps. Chloroform (1 ml) and acidified methanol (15% v/v H2SO4) (1 ml) were added to each sample. The samples were heated in a 97 °C water bath for 3 h to convert the PHB into 3-hydroxybutyrate methyl ester. After methanolysis, double-deionized water (1 ml) was added to each sample. Following phase separation, the bottom chloroform phase (2 μl) was extracted and loaded directly onto a GC/MS for analysis as previously described (31). The concentrations and inferred cellular contents of PHB were calculated on the basis of a standard curve generated with known concentrations of PHB (Sigma).
Transcription Profiling
Global transcriptome profiling was performed by RNA-seq as previously described (33). The C. fritschii PCC 9212 was fully adapted to acetate growth conditions by serially subculturing cells three times in liquid BG-11 medium containing 10 mm acetate. The control strain was similarly grown three times on medium without acetate, and each culture was harvested at A750 nm = 1. Total RNA was then extracted from these two strains, and rRNA depletion was performed as described (33). The construction of cDNA libraries and Illumina sequencing (50 nucleotides, single read) were performed in the Genomic Core Facility at The Pennsylvania State University. Mapping against the C. fritschii PCC 9212 genome was performed using the BWA software package, allowing a maximal 4 mismatches per read. The resulting alignment files were further analyzed with self-developed scripts to extract relative expression levels for each gene. The RNA sequencing data have been deposited in the NCBI Sequence Read Archive under accession number SRP052045.
Results
Enzyme Characterization
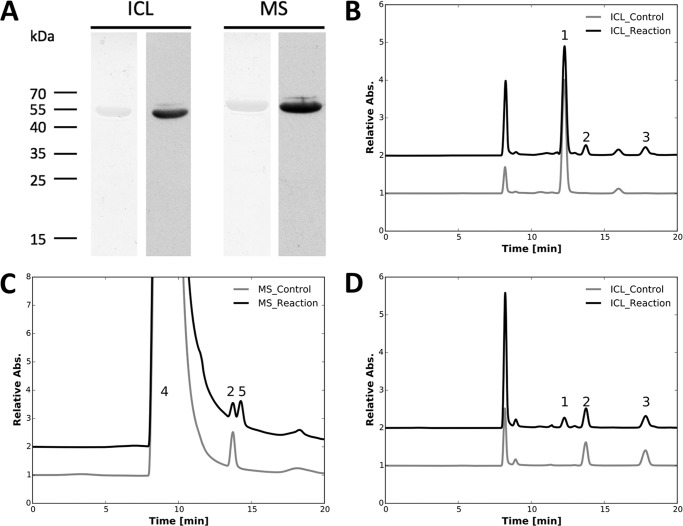
Fig. 2A shows the results of SDS-PAGE and immunoblotting analysis of the purified recombinant product of open reading frame (ORF) SYNPCC7002_A2041. The purified protein had an apparent molecular weight of 50,000 and cross-reacted with commercial antibodies to the poly-His tag. Moreover, tryptic peptide mass fingerprinting showed that the major protein present in the preparation was the product of ORF SYNPCC7002_A2041 (data not shown). When the product of SynPCC7002_A2041 was incubated with 10 mm malate at room temperature, 2.1 mm fumarate was formed and 2.2 mm malate was consumed (Fig. 2B). When the product of SynPCC7002_A2041 was incubated with 2.5 mm fumarate at room temperature, 1.9 mm malate was formed and 2.1 mm fumarate was consumed (Fig. 2C). These results confirm that SynPCC7002_A2041 encodes fumarase, which catalyzes the reversible interconversion of malate and fumarate. Together with the recently identified 2-oxoglutarate decarboxylase and succinic semialdehyde dehydrogenase, these three enzymes closed the three gaps in the TCA cycle of Synechococcus sp. PCC 7002 and demonstrate that the TCA cycle is complete in most cyanobacteria (20).
FIGURE 2.
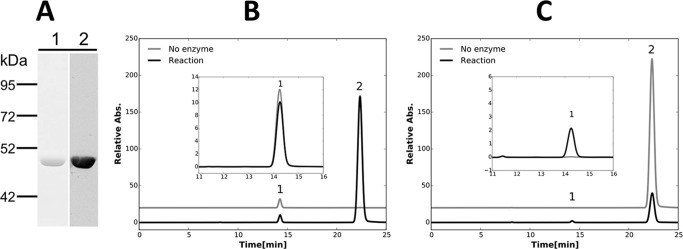
Characterization of purified recombinant fumarase (SYNPCC7002_A2041). A, SDS-PAGE (lane 1) and immunoblotting analysis with a commercial antibody to the poly-His6 tag (lane 2) of the purified fumarase. B, HPLC analysis showing that malate (peak 1) was converted to fumarate (peak 2) by fumarase (SYNPCC7002_A2041). Specifically, when the product of SynPCC7002_A2041 was incubated with 10 mm malate at room temperature, 2.1 mm fumarate was formed and 2.2 mm malate was consumed. C, HPLC analysis showing the formation of malate (peak 1) from fumarate (peak 2) catalyzed by the purified fumarase (SYNPCC7002_A2041). When the product of SynPCC7002_A2041 was incubated with 2.5 mm fumarate at room temperature, 1.9 mm malate was formed and 2.1 mm fumarate was consumed (C). The differences in the peak areas for identical amounts of fumarate and malate are due to the very different molar extinction coefficients of these two compounds at 210 nm. Insets represent the enlarged parts of the elution curves from 11 to 16 min to illustrate the changes observed more clearly. Other details of the assay conditions are described under ”Experimental Procedures.“
The isocitrate lyase (ORF UYEDRAFT_02681) of C. fritschii PCC 9212 was successfully expressed and purified from E. coli as an N-terminally poly-His-tagged protein. The purified protein had an apparent molecular weight of 52,000 on SDS-PAGE and was positively immunoreactive with commercial antibodies to the poly-His tag (Fig. 3A). The purified protein was conclusively identified by tryptic peptide mass fingerprinting (data not shown). As mentioned under “Experimental Procedures,” the malate synthase (ORF UYEDRAFT_02682) of C. fritschii PCC 9212 could not be overproduced in E. coli when the protein was produced with an N-terminal His tag, possibly due to protein misfolding. However, moving the poly-His tag to the C terminus resulted in the production of active, recombinant malate synthase. The recombinant protein had a molecular weight of 64,000 on SDS-PAGE and was positively immunoreactive with commercial antibodies to the poly-His tag (Fig. 3A). The identity of the protein was further confirmed by tryptic peptide mass fingerprinting (data not shown).
FIGURE 3.
Characterizations of purified recombinant isocitrate lyase (UYEDRAFT_02681) and malate synthase (UYEDRAFT_02682). A, SDS-PAGE and immunoblotting analysis with a commercial antibody to the poly-His6 tag for isocitrate lyase (ICL) and malate synthase (MS). B, HPLC analysis showing that isocitrate (peak 1) was converted to glyoxylate (peak 2) and succinate (peak 3) by the purified isocitrate lyase (UYEDRAFT_02681). When the protein product of UYEDRAFT_02681 was incubated with 2 mm isocitrate, 0.3 mm isocitrate was consumed and 0.25 mm succinate and 0.27 mm glyoxylate were produced. C, HPLC analysis showing the formation of malate (peak 5) from glyoxylate (peak 2) and acetyl-CoA (peak 4) catalyzed by purified malate synthase (UYEDRAFT_02682). Specifically, when the protein product from ORF UYEDRAFT_02682 was incubated with 2 mm glyoxylate and 2 mm acetyl-CoA, 1.2 mm glyoxylate and 1.1 mm acetyl-CoA were consumed, and 0.9 mm malate was produced. D, HPLC analysis showing production of isocitrate (peak 1) from glyoxylate (peak 2) and succinate (peak 3) catalyzed by the purified isocitrate lyase. Specifically, 0.15 mm isocitrate was produced, and 0.19 mm glyoxlyate and 0.15 mm succinate were consumed, when 1 mm succinate and 1 mm glyoxylate were incubated with the product of UYEDRAFT_02681. The large differences in the peak area for identical amounts of acetyl-CoA and glyoxylate are due to the different molar extinction coefficients of these two compounds at 210 nm. Detailed assay conditions are described under ”Experimental Procedures.“
To establish that the isocitrate lyase and malate synthase had the anticipated enzymatic activities, assays were performed to characterize the enzymes. When the protein product of UYEDRAFT_02681 was incubated with 2 mm isocitrate, 0.3 mm isocitrate was consumed and 0.25 mm succinate and 0.27 mm glyoxylate were produced (Fig. 3B). Demonstrating that this reaction is reversible, 0.15 mm isocitrate was produced, and 0.19 mm glyoxlyate and 0.15 mm succinate were consumed, when 1 mm succinate and 1 mm glyoxylate were incubated with the product of UYEDRAFT_02681 (Fig. 3D). When the protein product from ORF UYEDRAFT_02682 was incubated with 2 mm glyoxylate and 2 mm acetyl-CoA, 1.2 mm glyoxylate and 1.1 mm acetyl-CoA were consumed, and 0.9 mm malate was produced (Fig. 3C). These biochemical results established that UYEDRAFT_02681 encodes isocitrate lyase and that UYEDRAFT_02682 encodes malate synthase. Acting together, these two enzymes can catalyze the conversion of isocitrate and acetyl-CoA into malate and succinate (data not shown). This results in the incorporation of C2 units into metabolic intermediates of key precursor metabolites of the central metabolism. These biochemical assays also confirm that the glyoxylate cycle is present and probably active in C. fritschii PCC 9212.
Growth of C. fritschii PCC 9212 with and without Acetate
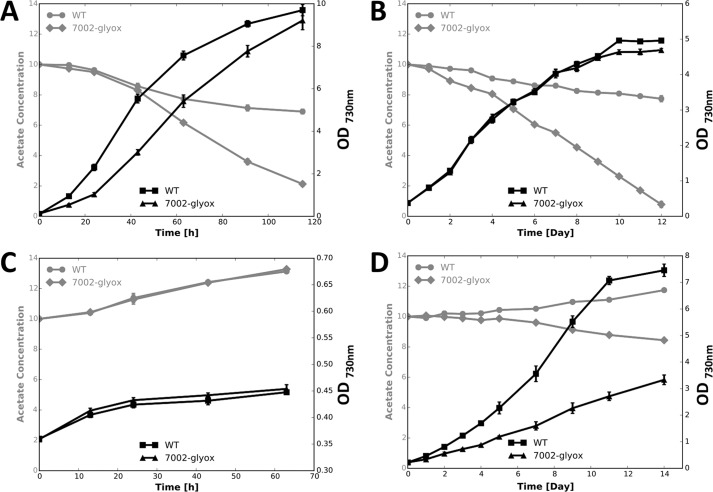
Because the glyoxylate cycle is generally believed to be involved in the acetate assimilation and metabolism, we tested whether C. fritschii PCC 9212 could assimilate acetate under different growth conditions. As described under “Experimental Procedures,” under standard growth conditions C. fritschii PCC 9212 grew faster when the medium was supplemented with 10 mm acetate (Fig. 4A). In agreement with the faster growth rate, acetate was consumed from the medium, and all of the acetate was consumed by the end of the cultivation period (Fig. 4A). As expected, C. fritschii PCC 9212 grew more slowly when cultures were sparged with air (Fig. 4B). Cells again grew faster when acetate was added to the growth medium but the magnitude of the stimulation was similar to that observed for cultures sparged with air containing 1% (v/v) CO2. This result shows that acetate can stimulate growth but certainly is not able to supplant CO2 fixation as the major route of carbon acquisition during growth under these conditions. C. fritschii PCC 9212 was able to grow very slowly in the dark when the medium contained acetate, but no growth was observed in the dark when acetate was eliminated from the medium (Fig. 4C).
FIGURE 4.
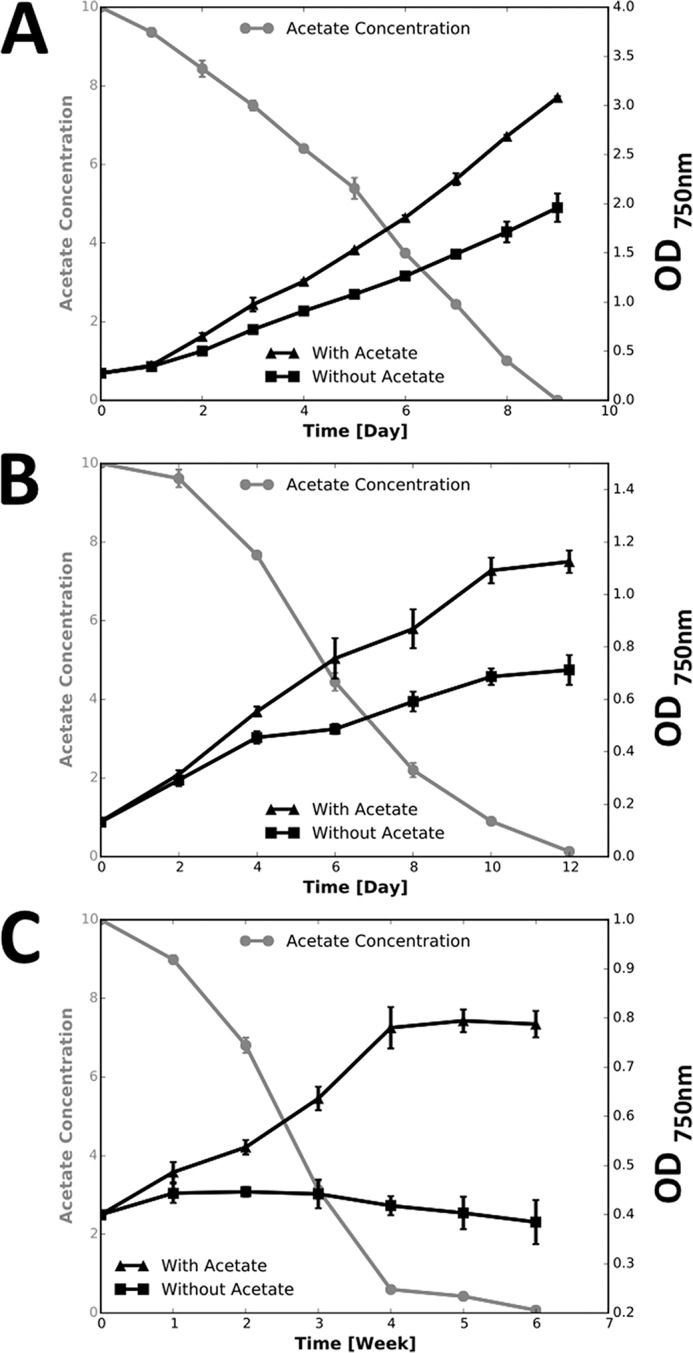
Acetate assimilation and growth curves for C. fritschii PCC 9212. Black lines indicate the cell density and gray lines indicate the acetate concentrations in the medium at different times during the batch growth cycle. A, C. fritschii PCC 9212 growing under standard conditions. B, C. fritschii PCC 9212 growing under low CO2 conditions (cultures were sparged with air). C, C. fritschii PCC 9212 grown under dark conditions (1% CO2 in air). The data shown are averages of three biological replicates, and the error bars show the standard deviation. Other details concerning the growth conditions are described under ”Experimental Procedures.“
Growth of Synechococcus sp. PCC 7002 with and without Acetate
We have not yet developed the ability to perform gene knock-out experiments to test the function of glyoxylate cycle in C. fritschii PCC 9212. Thus, we decided to study the function of the glyoxylate cycle and acetate utilization in the model cyanobacterium, Synechococcus sp. PCC 7002, which lacks the glyoxylate cycle. The aceBA operon encoding the two glyoxylate cycle genes of C. fritschii PCC 9212 was introduced into the pAQ1Ex expression plasmid system (29), which was subsequently transformed into Synechococcus sp. PCC 7002. The presence of the plasmid and the incorporation of the aceBA genes into Synechococcus sp. PCC 7002 was verified by PCR amplification of the aceBA operon and was further confirmed by sequencing the amplicon (Fig. 5). When the wild type and the strain carrying the aceBA genes, hereafter denoted as Synechococcus 7002-glyox, were grown under standard conditions, Synechococcus 7002-glyox had a slower growth rate but a faster acetate assimilation rate compared with WT (Fig. 6A). This indicated that the enzymes of the glyoxylate cycle were active in the recombinant strain and supported acetate assimilation. The slower growth rate may have been due to the overexpression of these two genes and the additional energy and nutrient resources required to synthesize the two foreign proteins. Furthermore, when the two strains were grown under low irradiance conditions, they had very similar growth rates and an even larger difference in acetate uptake was observed (Fig. 6B). This suggested that acetate was possibly more important in supplying energy for growth when light was limiting. However, no acetate uptake occurred under dark or low-CO2 conditions for WT cells (Fig. 6, C and D). WT cells exhibited net acetate excretion under these conditions (Fig. 6, C and D), and the same was observed for Synechococcus 7002-glyox under dark conditions. Acetate assimilation was still observed under low-CO2 conditions in strain Synechococcus 7002-glyox (Fig. 6D), although the assimilation rate was much slower compared with the rates observed under standard or low-light conditions. A previous study had also shown that in Aphanocapsa sp. PCC 6308 and Synechococcus elongatus PCC 6301, the CO2 concentration was crucial for acetate uptake and the acetate uptake rate was reduced by almost 50% in the absence of CO2 (4). These observations confirm that acetate assimilation, the glyoxylate cycle, and CO2 fixation are closely related metabolic processes that may possibly be coordinately regulated under different growth conditions.
FIGURE 5.
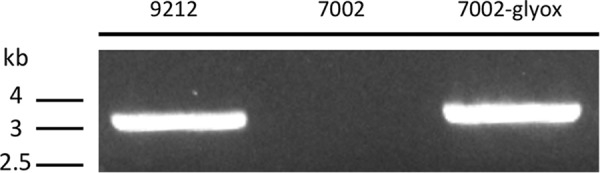
Verification of the presence of glyoxylate cycle genes (aceBA) in Synechococcus sp. PCC 7002 strain 7002-glyox by PCR. The template DNA was derived from wild-type C. fritschii PCC 9212 (lane 9212), from wild-type Synechococcus sp. PCC 7002 (lane 7002), and from the recombinant strain 7002-glyox (lane 7002-glyox), which has the aceBA genes from C. fritschii PCC 9212 inserted in plasmid pAQ1-Ex as described under ”Experimental Procedures.“
FIGURE 6.
Acetate assimilation and growth analysis of Synechococcus sp. PCC 7002. Black lines indicate cell density and gray lines indicate the acetate concentrations in the medium at different growth stages. A, Synechococcus sp. PCC 7002 grown under standard conditions; B, Synechococcus sp. PCC 7002 growing under low light conditions; C, Synechococcus sp. PCC 7002 growing under dark conditions; D, Synechococcus sp. PCC 7002 growing under low CO2 conditions. WT, wild type Synechococcus sp. PCC 7002; 7002-glyox, Synechococcus sp. PCC 7002 strain with aceBA genes of C. fritschii PCC 9212 expressed from pAQ1. The data shown are averages of three biological replicates, and the error bars show the S.D.
Gene Neighborhood Analysis of the Glyoxylate Cycle Genes
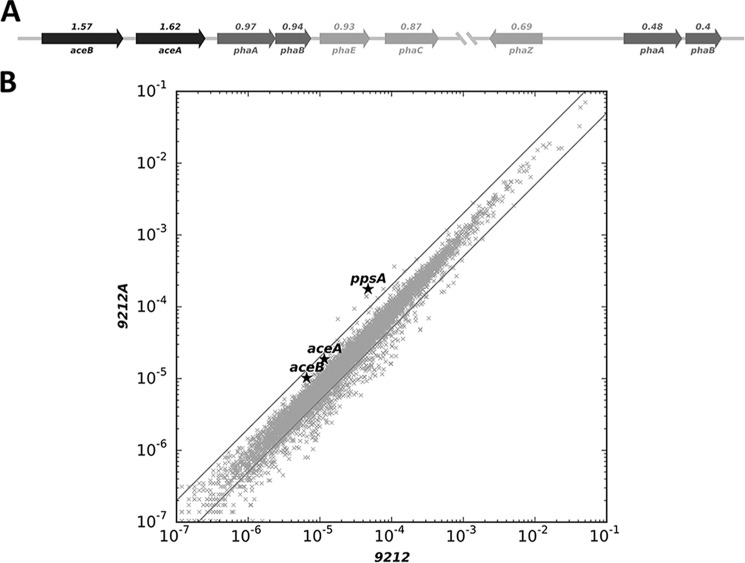
To study the possible relationships between the glyoxylate cycle and other metabolic pathways, BLASTP analysis and gene neighborhoods surrounding the aceBA operon were also investigated. As mentioned before, the aceBA genes, encoding isocitrate lyase and malate synthase, respectively, are located in an apparent operon in C. fritschii PCC 9212 (Fig. 7A). BLASTP analysis showed that these two genes also occur in C. fritschii PCC 6912, Cyanothece sp. strains PCC 7822 and 7424, Pleurocapsa minor PCC 7327, Fischerella sp. PCC 9605, Cyanobacterium PCC 7702, Mastigocoleus testarum, and Tolypothrix bouteillei. Interestingly, many of these cyanobacteria (C. fritschii PCC 9212, C. fritschii PCC 6912, P. minor PCC 7327, Fischerella sp. PCC 9605, M. testarum) are capable of growth in far-red light and exhibit the far-red light photoacclimation (FaRLiP) response (33, 34). Furthermore, all of these strains are able to fix nitrogen; this suggests that the glyoxylate cycle may serve as an additional control point for balancing the carbon and nitrogen metabolism of these cyanobacteria.
FIGURE 7.
Relative transcript abundances for mRNAs in C. fritschii PCC 9212 grown with or without acetate. A, gene neighborhood around the glyoxylate cycle genes. Numbers above each gene indicate the fold-difference of mRNA abundance in cells grown with acetate compared with cells grown without acetate. B, scatter plot showing the relative abundance of all the mRNAs under growth conditions without acetate (9212) or with acetate (9212A). Gray lines indicate a 2-fold increase or 50% decrease in mRNA level. aceA, isocitrate lyase; aceB, malate synthase; phaA, acetyl-CoA acetyltransferase; phaB, acetoacetyl-CoA reductase; phaE, poly(R)-hydroxyalkanoic acid synthase, class III, PhaE subunit; phaC, poly(R)-hydroxyalkanoic acid synthase, class III, PhaC subunit; phaZ, poly(3-hydroxybutyrate) depolymerase; ppsA, phosphoenolpyruvate synthase.
Further examination of the genes near the aceBA operon in C. fritschii PCC 9212 indicates that there is also an apparent operon of PHB-related genes (phaABEC) located downstream (Fig. 7A). Additionally, a poly-(3-hydroxybutyrate) depolymerase gene (phaZ) as well as paralogous copies of acetyl-CoA acetyltransferase (phaA) and acetoacetyl-CoA reductase (phaB) are located further downstream in the same gene neighborhood. Considering that the glyoxylate cycle and PHB metabolic pathway both use the important metabolite acetyl-CoA, and considering that all of these genes are colocalized in the genome, it is highly likely that these two pathways interact closely with each other in carbon metabolism. Synechococcus sp. PCC 7002 does not fix nitrogen, lacks the glyoxylate cycle genes, and lacks enzymes for production and mobilization of PHB.
Global Transcription Profiling of C. fritschii PCC 9212
To investigate whether other metabolic pathways in addition to the glyoxylate cycle are involved in acetate assimilation and utilization, global transcription profiling was performed for C. fritschii PCC 9212 cells grown in the presence and absence of acetate. The results showed that transcripts for the isocitrate lyase and malate synthase genes increased ∼1.6-fold in the presence of acetate, and further indicated that cells expressed these genes at relatively high levels even when acetate was not present in the medium. However, transcript levels for the genes involved in PHB metabolism (phaABEC) had similar abundance levels in cells grown with or without acetate. The different expression pattern for the aceBA and phaABEC operons suggested that the PHB metabolism genes and the glyoxylate cycle genes were probably expressed from different promoters. It should be noted that the C. fritschii PCC 9212 genome contains two copies of phaA and phaB; transcript levels for the second copies actually decreased about 2-fold when acetate was added to the growth medium. This could indicate that the distal phaAB genes might be involved in PHB degradation/utilization. Transcript levels for phosphoenolpyruvate synthase (ppsA) increased about 4-fold in the presence of acetate, which suggests that cells increase carbon flux toward glycolysis in the presence of acetate (Fig. 7B). A similar response was reported in E. coli cells grown in the presence of acetate (35).
Acetate Increases the Production of PHB
Because the genes for the glyoxylate cycle and for PHB metabolism were co-localized, we determined whether the PHB content of C. fritschii PCC 9212 cells would be affected by the addition of acetate. PHB accumulation was first tested in C. fritschii PCC 9212, and our results showed that PHB accumulated and represented about ∼5% of the cell dry weight under standard growth conditions (Fig. 8). When acetate was supplied to the medium under the same conditions, the PHB content increased to ∼15% of total cell dry weight (Fig. 8, inset). These results indicated that the assimilated acetate that was metabolized by the glyoxylate cycle might mainly be stored in the form of PHB. Interestingly, genes for PHB metabolism are present in many cyanobacteria that lack the glyoxylate cycle (e.g. Synechocystis sp. PCC 6803, Leptolyngbya sp. strain JSC-1). Genes for PHB metabolism are also found in other strains capable of performing FaRLiP (e.g. P. minor PCC 7327, Fischerella sp. PCC 9605) (34), suggesting that PHB biosynthesis could serve as a major carbon storage mechanism and the synthesized PHB might be used during cellular adaptation to different growth environments, such as far-red light conditions. In agreement with this hypothesis, the relative transcript abundances for genes of PHB metabolism increased ∼7-fold in Leptolyngbya sp. strain JSC-1 when cells were shifted to far-red light conditions (33). Along with this change, transcript abundances for numerous carbon transporter genes also increased. This indicated that the cells were probably using both internal carbon stores as well as extracellular carbon sources from the environment to provide the energy and nutrients needed for cell growth. These resources could be limiting when cells are remodeling their photosynthetic apparatus to use far-red light (33). Thus, together with the PHB biogenesis and degradation pathways to balance the acetyl-CoA concentrations inside cells, the glyoxylate cycle may provide an efficient way to use storage carbon sources under conditions when energy or carbon supply is limited. These results, together with the fact that mRNA levels of many photosynthesis related genes were also regulated in the presence of acetate, suggests that the glyoxylate cycle, acetate assimilation, and PHB metabolism are all important for the massive metabolic and physiological changes during the shift of growth condition to far-red light in FaRLiP strains.
FIGURE 8.
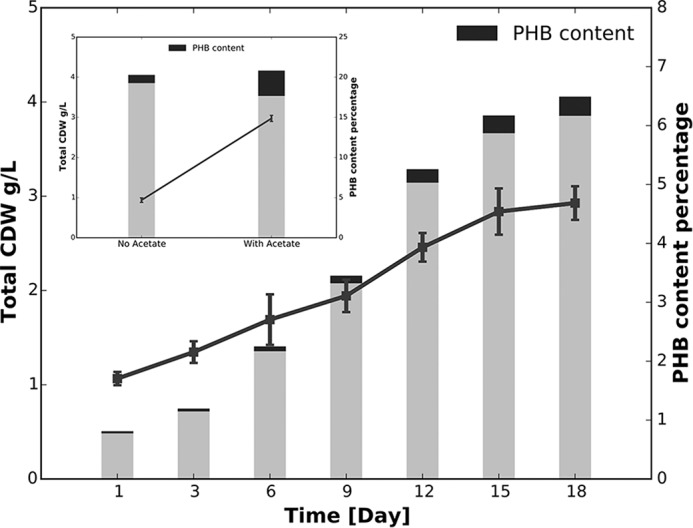
Accumulation of PHB in C. fritschii PCC 9212. PHB contents of C. fritschii PCC 9212 were monitored as a function of batch growth under standard conditions for C. fritschii PCC 9212. The bars indicate the total cell dry weight (CDW), and the black portions of the bars show the PHB content at different growth stages. This information is also plotted to emphasize the kinetics of PHB production. In the absence of added acetate, PHB accounted for ∼5% of total CDW. The inset shows that CDW changed only slightly when 10 mm acetate was added to the medium, but PHB accumulated to a much higher level, ∼15% of total CDW. The data shown are averages of three biological replicates, and the error bars show the S.D.
Discussion
The glyoxylate cycle and the TCA cycle can both be used to metabolize acetate (i.e. acetyl-CoA), and their reactions provide essential precursor metabolites (e.g. 2-oxogluatarate, oxaloacetate and sometimes succinate) and reducing power (e.g. NADH) for cells. These two cycles share many enzymes and intermediates (Fig. 1), which makes them intrinsically interconnected. However, by using two specific enzymes, isocitrate lyase and malate synthase, the glyoxylate cycle is able to bypass the CO2 releasing, oxidative steps of the TCA cycle (isocitrate dehydrogenase, and 2-oxoglutarate dehydrogenase or 2-oxoglutarate decarboxylase (20)). As a result, the glyoxylate cycle can more efficiently assimilate carbon from acetyl-CoA for biomass production, which could be derived from assimilated acetate, ethanol, or the degradation of fatty acids or poly-3-hydroxybutyrate. The net product of the glyoxylate cycle is succinate, which can be used to replenish TCA cycle intermediates or to generate metabolites for gluconeogenesis and other biosynthetic processes. Thus, the glyoxylate cycle provides an effective route for growth on fatty acids and C2 compounds such as acetate and ethanol.
Since its discovery, the glyoxylate cycle has been identified and studied in many different organisms, including bacteria, archaea, protists, plants, and fungi (13, 36, 37). Although isocitrate lyase and malate synthase activities were reportedly detected in birds and amphibians (38), no genes for isocitrate lyase have been identified in animals. The nematode, Caenorhabditis elegans, and the protest, Euglena gracilis, have a single, fused gene encoding a bifunctional enzyme (39, 40). In Chlamydomonas reinhardtii, the glyoxylate cycle was shown to be essential for dark growth on acetate, and for efficient growth in the light when acetate is supplied (41). However, it should be noted that acetate is ineffective as a growth substrate and is even toxic for some marine algae (42). In addition to allowing the growth of bacteria on C2 compounds, together with the β-oxidation of fatty acids, the glyoxylate cycle is also important in providing carbohydrates and biosynthetic precursors during the early stage of seedling establishment for plants (36, 43). It was reported that the β-oxidation pathway and glyoxylate cycle enzymes were induced in senescing leaves, possibly used for the breakdown of membrane lipids and gluconeogenesis (44).
Despite the fact that the glyoxylate cycle is essential for the assimilation and metabolism of acetate, there are still a number of acetate-using microorganisms that lack one or both of the enzymes involved in the glyoxylate cycle. More recently, some other metabolic pathways that can be used for acetate assimilation have been identified. The glyoxylate cycle is not found in green sulfur bacteria and heliobacteria, and these bacteria instead use pyruvate synthase for acetate assimilation (45). This enzyme requires ferredoxin to supply the necessary reducing power for pyruvate synthesis and thus primarily occurs in anaerobic bacteria. CO2 is also required for the growth of heliobacteria when acetate is supplied as the only organic carbon source (46). Another metabolic pathway that has been demonstrated to be involved in acetate assimilation is the ethylmalonyl-CoA pathway. This pathway is responsible for the production of glyoxylate, which can be further converted to phosphoenolpyruvate via the serine cycle pathway (47). The ethylmalonyl-CoA pathway is found in Rhodobacter sphaeroides, in which glyoxylate is condensed with acetyl-CoA to produce malyl-CoA and further hydrolyzed to malate and CoA (48). A third acetate-assimilation pathway, the methylaspartate cycle, was recently described in Haloarcula marismortui (49). This new pathway also results in the net synthesis of succinate but requires nearly three times as many steps as the glyoxylate cycle to generate oxaloacetate from citrate (49). Furthermore, in the methylaspartate cycle, isocitrate is first decarboxylated to 2-oxoglutarate, which is then converted to glutamate, and thus nitrogen metabolism is also linked to acetate assimilation in this cycle (50). Why certain microorganisms use very complex strategies, such as the ethylmalonyl-CoA pathway and the methylaspartate cycle, for acetate assimilation rather than the simple glyoxylate cycle remains unclear and requires further study.
Although acetate assimilation has been studied in many microorganisms, the assimilation of acetate in cyanobacteria was poorly understood and confusing. Previous studies had suggested that some cyanobacteria could use acetate as the sole carbon and energy source (22). However, the pathway(s) that were used to assimilate acetate in cyanobacteria remained unclear. As mentioned, pyruvate synthase is highly sensitive to oxygen and thus cannot function in oxygenic cyanobacteria when they grow in the light (however, pyruvate:ferredoxin oxidoreductase is used to decarboxylate pyruvate oxidatively during fermentation in the dark (51)). The ethylmalonyl-CoA pathway and the methylaspartate cycle have not yet been shown to occur in cyanobacteria. Some previous studies reported that the enzymatic activities of the glyoxylate cycle could be detected in cyanobacteria (21), and thus the glyoxylate cycle has been included in some recent FBA models for Synechocystis sp. PCC 6803 to investigate the possible roles of this cycle in cyanobacteria (25, 52). However, the genes encoding isocitrate lyase and malate synthase are not present in Synechocystis sp. PCC 6803 nor are they present in the genomes of most other cyanobacteria. Furthermore, these enzymatic activities were not identified when more refined and sensitive methods were employed with Synechocystis sp. PCC 6803 (25). Consistent with the absence of the detected enzyme activities, recent isotopic tracing studies also indicated that the glyoxylate cycle is not functional and that the glyoxylate cycle may mainly be used for glycine synthesis (53).
By identifying and characterizing the isocitrate lyase and malate synthase from C. fritschii PCC 9212, our results clearly demonstrate that the glyoxylate cycle does exist in a few cyanobacterial strains and that it plays an important role in acetate assimilation in one of those organisms. We also demonstrated that the ability to assimilate acetate could be significantly improved by introducing the aceBA genes to Synechococcus sp. PCC 7002, which normally lacks the glyoxylate cycle. However, the absence of these two genes, and thus the glyoxylate cycle, in most cyanobacteria implies that the few organisms with this pathway probably obtained the genes recently by lateral gene transfer. In addition to the intracellular metabolism of acetate, one interesting question would be to identify a potential acetate transporter (assuming that there is one). It has been reported that the yjcG gene is responsible for acetate transportation in E. coli, and another transport system for acetate may also exist (54). However, homologs of the yjcG gene have not been identified in cyanobacteria, and a different type of transport system might be used. Under dark aerobic conditions, microalgae use a monocarboxylic/proton transporter protein, which is a member of the Major Facilitator Superfamily, to transport acetate across the membrane (55, 56). Our transcription profiling results showed that transcript levels for several putative transporter genes increased modestly when acetate was supplied to the medium (supplemental Table S1), and further studies with these transporters might provide clues that could answer this question definitively.
Considering the importance of the glyoxylate cycle in acetate assimilation and its intrinsic link with the TCA cycle, the operation of glyoxylate cycle must be regulated properly to accommodate changes in the chemical environments of cells. Indeed, in algae the enzyme activities of isocitrate lyase have been found to increase under many different growth conditions when acetate is supplied, and the glyoxylate cycle is operated interactively with the TCA cycle and the oxidative pentose phosphate pathway (10, 56, 57). Our results showed that transcript levels for malate synthase and isocitrate lyase increased only slightly when acetate was being actively metabolized. Previous studies reported that the enzyme activities for malate synthase and isocitrate lyase did not increase when acetate was supplied to the medium, indicating that there might be regulation at other levels (23). Consistent with this hypothesis, purified isocitrate lyase from C. reinhardtii was shown to be inactivated by glutathionylation and reactivated by glutaredoxin, which implies that the glyoxylate cycle may be actively regulated under specific environmental conditions (58). However, the functional significance of these post-translational modifications in response to different growth conditions, as well as the possible regulation and interactions between the glyoxylate cycle and many other metabolic pathways (e.g. the TCA cycle, the PHB metabolism) in cyanobacteria, are not yet well understood and will require further detailed investigation.
Supplementary Material
Acknowledgments
We thank Dr. Douglas Warui for assistance with the GC/MS analysis of PHB metabolites. We also thank Candace Price, Daniel Hannon, and Dr. Craig Praul for assistance with the preparation and sequencing of the cDNA libraries for transcription analyses (RNA-seq).
This work was supported by National Science Foundation Grant MCB-1021725 (to D. A. B.) and Air Force Office of Scientific Research MURI Grants FA9550-05-1-0365 and FA9550-11-1-0148 (to D. A. B.).

This article contains supplemental Table S1.
- acetyl-CoA
- acetyl coenzyme A
- FaRLiP
- far-red light photoacclimation
- ICL
- isocitrate lyase
- PCC
- Pasteur Culture Collection
- PHB
- poly-3-hydroxybutyrate
- TCA
- tricarboxylic acid.
References
- 1. Kreth J., Lengeler J. W., Jahreis K. (2013) Characterization of pyruvate uptake in Escherichia coli K-12. PLoS One 8, e67125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Pinchuk G. E., Geydebrekht O. V., Hill E. A., Reed J. L., Konopka A. E., Beliaev A. S., Fredrickson J. K. (2011) Pyruvate and lactate metabolism by Shewanella oneidensis MR-1 under fermentation, oxygen limitation, and fumarate respiration conditions. Appl. Environ. Microbiol. 77, 8234–8240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brown T. D., Jones-Mortimer M. C., Kornberg H. L. (1977) The enzymic interconversion of acetate and acetyl-coenzyme A in Escherichia coli. J. Gen. Microbiol. 102, 327–336 [DOI] [PubMed] [Google Scholar]
- 4. Ihlenfeldt M. J., Gibson J. (1977) Acetate uptake by the unicellular cyanobacteria Synechococcus and Aphanocapsa. Arch. Microbiol. 113, 231–241 [DOI] [PubMed] [Google Scholar]
- 5. Kay W. W., Kornberg H. L. (1971) The uptake of C4-dicarboxylic acids by Escherichia coli. Eur. J. Biochem. 18, 274–281 [DOI] [PubMed] [Google Scholar]
- 6. Gutowski S. J., Rosenberg H. (1975) Succinate uptake and related proton movements in Escherichia coli K12. Biochem. J. 152, 647–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wright R. T., Hobbie J. E. (1966) Use of glucose and acetate by bacteria and algae in aquatic ecosystems. Ecology 47, 447–464 [Google Scholar]
- 8. Gibbs M., Gfeller R. P., Chen C. (1986) Fermentative metabolism of Chlamydomonas reinhardii III. Photoassimilation of acetate. Plant Physiol. 82, 160–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kornberg H. (1966) The role and control of the glyoxylate cycle in Escherichia coli. Biochem. J. 99, 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Boyle N. R., Morgan J. A. (2009) Flux balance analysis of primary metabolism in Chlamydomonas reinhardtii. BMC Syst. Biol. 3, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kornberg H. L., Krebs H. A. (1957) Synthesis of cell constituents from C2-units by a modified tricarboxylic acid cycle. Nature 179, 988–991 [DOI] [PubMed] [Google Scholar]
- 12. Kornberg H. (2000) Krebs and his trinity of cycles. Nat. Rev. Mol. Cell Biol. 1, 225–228 [DOI] [PubMed] [Google Scholar]
- 13. Dunn M. F., Ramírez-Trujillo J. A., Hernández-Lucas I. (2009) Major roles of isocitrate lyase and malate synthase in bacterial and fungal pathogenesis. Microbiology 155, 3166–3175 [DOI] [PubMed] [Google Scholar]
- 14. Zhang S., Bryant D. A. (2014) Learning new tricks from an old cycle: the TCA cycle in cyanobacteria, algae and plants. Perspect. Phycol. 1, 73–86 [Google Scholar]
- 15. Tang K.-H., Tang Y. J., Blankenship R. E. (2011) Carbon metabolic pathways in phototrophic bacteria and their broader evolutionary implications. Front. Microbiol. 2, 165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sirevåg R. (1995) Carbon metabolism in green bacteria. in Advances in Photosynthesis and Respiration (Blankenship R., Madigan M., Bauer C., eds) Vol. 2, pp. 871–883, Springer, Netherlands [Google Scholar]
- 17. Zarzycki J., Fuchs G. (2011) Coassimilation of organic substrates via the autotrophic 3-hydroxypropionate bi-cycle in Chloroflexus aurantiacus. Appl. Environ. Microbiol. 77, 6181–6188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Feng X., Tang K.-H., Blankenship R. E., Tang Y. J. (2010) Metabolic flux analysis of the mixotrophic metabolisms in the green sulfur bacterium Chlorobaculum tepidum. J. Biol. Chem. 285, 39544-39550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Madigan M. (2006) The family heliobacteriaceae. in The Prokaryotes (Dworkin M., Falkow S., Rosenberg E., Schleifer K.-H., Stackebrandt E., eds) pp. 951–964, Springer; US. [Google Scholar]
- 20. Zhang S., Bryant D. A. (2011) The tricarboxylic acid cycle in cyanobacteria. Science 334, 1551–1553 [DOI] [PubMed] [Google Scholar]
- 21. Eley J. H. (1988) Glyoxylate cycle enzyme activities in the cyanobacterium Anacystis nidulans. J. Phycol. 24, 586–588 [Google Scholar]
- 22. Pearce J., Carr N. (1967) The metabolism of acetate by the blue-green algae, Anabaena variabilis and Anacystis nidulans. J. Gen. Microbiol. 49, 301–313 [DOI] [PubMed] [Google Scholar]
- 23. Miller J. S., Allen M. M. (1972) Carbon utilization patterns in the heterotrophic blue-green alga Chlorogloea fritschii. Arch. Mikrobiol. 86, 1–12 [DOI] [PubMed] [Google Scholar]
- 24. Hoare D., Hoare S., Moore R. (1967) The photoassimilation of organic compounds by autotrophic blue-green algae. J. Gen. Microbiol. 49, 351–370 [DOI] [PubMed] [Google Scholar]
- 25. Knoop H., Gründel M., Zilliges Y., Lehmann R., Hoffmann S., Lockau W., Steuer R. (2013) Flux balance analysis of cyanobacterial metabolism: the metabolic network of Synechocystis sp. PCC 6803. PLoS Comput. Biol. 9, e1003081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bandyopadhyay A., Elvitigala T., Welsh E., Stöckel J., Liberton M., Min H., Sherman L. A., Pakrasi H. B. (2011) Novel metabolic attributes of the genus Cyanothece, comprising a group of unicellular nitrogen-fixing cyanobacteria. MBio 2, e00214–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rippka R., Deruelles J., Waterbury J. B., Herdman M., Stanier R. Y. (1979) Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J. Gen. Microbiol. 111, 1–61 [Google Scholar]
- 28. Ludwig M., Bryant D. A. (2011) Transcription profiling of the model cyanobacterium Synechococcus sp. strain PCC 7002 by Next-Gen (SOLiD) sequencing of cDNA. Front. Microbiol. 2, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Xu Y., Alvey R. M., Byrne P. O., Graham J. E., Shen G., Bryant D. A. (2011) Expression of genes in cyanobacteria: Adaptation of endogenous plasmids as platforms for high-level gene expression in Synechococcus sp. PCC 7002. Methods Mol. Biol. 684, 273–293 [DOI] [PubMed] [Google Scholar]
- 30. Frigaard N. U., Sakuragi Y., Bryant D. A. (2004) Gene inactivation in the cyanobacterium Synechococcus sp. PCC 7002 and the green sulfur bacterium Chlorobium tepidum using in vitro-made DNA constructs and natural transformation. Methods Mol. Biol. 274, 325–340 [DOI] [PubMed] [Google Scholar]
- 31. Tsang T. K., Roberson R. W., Vermaas W. F. (2013) Polyhydroxybutyrate particles in Synechocystis sp. PCC 6803: facts and fiction. Photosynth. Res. 118, 37–49 [DOI] [PubMed] [Google Scholar]
- 32. Braunegg G., Sonnleitner B., Lafferty R. M. (1978) A rapid gas chromatographic method for the determination of poly-β-hydroxybutyric acid in microbial biomass. Eur. J. Appl. Microbiol. Biotechnol. 6, 29–37 [Google Scholar]
- 33. Gan F., Zhang S., Rockwell N. C., Martin S. S., Lagarias J. C., Bryant D. A. (2014) Extensive remodeling of a cyanobacterial photosynthetic apparatus in far-red light. Science 345, 1312–1317 [DOI] [PubMed] [Google Scholar]
- 34. Gan F., Shen G., Bryant D. A. (2014) Occurrence of far-red light photoacclimation (FaRLiP) in diverse cyanobacteria. Life 5, 4–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Oh M.-K., Rohlin L., Kao K. C., Liao J. C. (2002) Global expression profiling of acetate-grown Escherichia coli. J. Biol. Chem. 277, 13175–13183 [DOI] [PubMed] [Google Scholar]
- 36. Eastmond P. J., Graham I. A. (2001) Re-examining the role of the glyoxylate cycle in oilseeds. Trends Plant Sci. 6, 72–78 [DOI] [PubMed] [Google Scholar]
- 37. Kunze M., Pracharoenwattana I., Smith S. M., Hartig A. (2006) A central role for the peroxisomal membrane in glyoxylate cycle function. Biochim. Biophys. Acta 1763, 1441–1452 [DOI] [PubMed] [Google Scholar]
- 38. Davis W. L., Jones R. G., Farmer G. R., Dickerson T., Cortinas E., Cooper O. J., Crawford L., Goodman D. B. (1990) Identification of glyoxylate cycle enzymes in chick liver: the effect of vitamin D3: cytochemistry and biochemistry. Anat. Rec. 227, 271–284 [DOI] [PubMed] [Google Scholar]
- 39. Nakazawa M., Nishimura M., Inoue K., Ueda M., Inui H., Nakano Y., Miyatake K. (2011) Characterization of a bifunctional glyoxylate cycle enzyme, malate synthase/isocitrate lyase, of Euglena gracilis. J. Eukaryot. Microbiol. 58, 128–133 [DOI] [PubMed] [Google Scholar]
- 40. Kondrashov F. A., Koonin E. V., Morgunov I. G., Finogenova T. V., Kondrashova M. N. (2006) Evolution of glyoxylate cycle enzymes in Metazoa: evidence of multiple horizontal transfer events and pseudogene formation. Biol. Direct 1, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Plancke C., Vigeolas H., Höhner R., Roberty S., Emonds-Alt B., Larosa V., Willamme R., Duby F., Onga Dhali D., Thonart P., Hiligsmann S., Franck F., Eppe G., Cardol P., Hippler M., Remacle C. (2014) Lack of isocitrate lyase in Chlamydomonas leads to changes in carbon metabolism and in the response to oxidative stress under mixotrophic growth. Plant J. 77, 404–417 [DOI] [PubMed] [Google Scholar]
- 42. Wood B. J. B., Grimson P. H. K., German J. B., Turner M. (1999) Photoheterotrophy in the production of phytoplankton organisms. J. Biotechnol. 70, 175–183 [Google Scholar]
- 43. Graham I. A. (2008) Seed storage oil mobilization. Annu. Rev. Plant Biol. 59, 115–142 [DOI] [PubMed] [Google Scholar]
- 44. Chen Z.-H., Walker R. P., Acheson R. M., Técsi L. I., Wingler A., Lea P. J., Leegood R. C. (2000) Are isocitrate lyase and phosphoenolpyruvate carboxykinase involved in gluconeogenesis during senescence of barley leaves and cucumber cotyledons? Plant Cell Physiol. 41, 960–967 [DOI] [PubMed] [Google Scholar]
- 45. Pickett M. W., Williamson M. P., Kelly D. J. (1994) An enzyme and 13C-NMR study of carbon metabolism in heliobacteria. Photosynth. Res. 41, 75–88 [DOI] [PubMed] [Google Scholar]
- 46. Tang K.-H., Yue H., Blankenship R. E. (2010) Energy metabolism of Heliobacterium modesticaldum during phototrophic and chemotrophic growth. BMC Microbiol. 10, 150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Korotkova N., Chistoserdova L., Kuksa V., Lidstrom M. E. (2002) Glyoxylate regeneration pathway in the methylotroph Methylobacterium extorquens AM1. J. Bacteriol. 184, 1750–1758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Erb T. J., Berg I. A., Brecht V., Müller M., Fuchs G., Alber B. E. (2007) Synthesis of C5-dicarboxylic acids from C2-units involving crotonyl-CoA carboxylase/reductase: the ethylmalonyl-CoA pathway. Proc. Natl. Acad. Sci. U.S.A. 104, 10631–10636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Khomyakova M., Bükmez Ö., Thomas L. K., Erb T. J., Berg I. A. (2011) A methylaspartate cycle in haloarchaea. Science 331, 334–337 [DOI] [PubMed] [Google Scholar]
- 50. Ensign S. A. (2011) Another microbial pathway for acetate assimilation. Science 331, 294–295 [DOI] [PubMed] [Google Scholar]
- 51. McNeely K., Xu Y., Ananyev G., Bennette N., Bryant D. A., Dismukes G. C. (2011) Synechococcus sp. strain PCC 7002 nifJ mutant lacking pyruvate:ferredoxin oxidoreductase. Appl. Environ. Microbiol. 77, 2435–2444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Shastri A. A., Morgan J. A. (2005) Flux balance analysis of photoautotrophic metabolism. Biotechnol. Prog. 21, 1617–1626 [DOI] [PubMed] [Google Scholar]
- 53. You L., Berla B., He L., Pakrasi H. B., Tang Y. J. (2014) 13C-MFA delineates the photomixotrophic metabolism of Synechocystis sp. PCC 6803 under light- and carbon-sufficient conditions. Biotechnol. J. 9, 684–692 [DOI] [PubMed] [Google Scholar]
- 54. Gimenez R., Nuñez M. F., Badia J., Aguilar J., Baldoma L. (2003) The gene yjcG, cotranscribed with the gene acs, encodes an acetate permease in Escherichia coli. J. Bacteriol. 185, 6448–6455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Becker H. M., Hirnet D., Fecher-Trost C., Sültemeyer D., Deitmer J. W. (2005) Transport activity of MCT1 expressed in Xenopus oocytes is increased by interaction with carbonic anhydrase. J. Biol. Chem. 280, 39882–39889 [DOI] [PubMed] [Google Scholar]
- 56. Perez-Garcia O., Escalante F. M., de-Bashan L. E., Bashan Y. (2011) Heterotrophic cultures of microalgae: metabolism and potential products. Water Res. 45, 11–36 [DOI] [PubMed] [Google Scholar]
- 57. Combres C., Laliberté G., Sevrin Reyssac J., de la Noüe J. (1994) Effect of acetate on growth and ammonium uptake in the microalga Scenedesmus obliquus. Physiol. Plant. 91, 729–734 [Google Scholar]
- 58. Bedhomme M., Zaffagnini M., Marchand C. H., Gao X.-H., Moslonka-Lefebvre M., Michelet L., Decottignies P., Lemaire S. D. (2009) Regulation by glutathionylation of isocitrate lyase from Chlamydomonas reinhardtii. J. Biol. Chem. 284, 36282–36291 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.