Abstract
The genes (xylA) encoding xylose isomerase (XI) from two Lactococcus lactis subsp. lactis strains, 210 (Xyl−) and IO-1 (Xyl+), were cloned, and the activities of their expressed proteins in recombinant strains of Escherichia coli were investigated. The nucleotide and amino acid sequence homologies between the xylA genes were 98.4 and 98.6%, respectively, and only six amino acid residues differed between the two XIs. The purified IO-1 XI was soluble with Km and kcat being 2.25 mM and 184/s, respectively, while the 210 XI was insoluble and inactive. Site-directed mutagenesis on 210 xylA showed that a triple mutant possessing R202M/Y218D/V275A mutations regained XI activity and was soluble. The Km and kcat of this mutant were 4.15 mM and 141/s, respectively. One of the IO-1 XI mutants, S388T, was insoluble and showed negligible activity similar to that of 210 XI. The introduction of a K407E mutation to the IO-1 S388T XI mutant restored its activity and solubility. The dissolution of XI activity in L. lactis subsp. lactis involves a series of mutations that collectively eliminate enzyme activity by reducing the solubility of the enzyme.
d-Xylose, upon transport in prokaryotes, is isomerized to d-xylulose and then converted to xylulose-5-phosphate, which is further metabolized via the phosphoenolpyruvate pathway or the pentose phosphate pathway (16, 26). The specifics of the transport mechanism and metabolism of d-xylose to xylulose-5-phosphate depend on the genus and species (8, 11, 23). Isomerization and phosphorylation are catalyzed by d-xylose isomerase (XI; encoded by xylA) and d-xylulose kinase (encoded by xylB), respectively. In most cases, these genes are clustered into an operon (xyl operon) (14, 22, 24, 33, 39).
The XIs can be classified into two groups, based on their size, amino acid sequence similarity, and divalent cation preference. Group I includes XIs from genera such as Streptomyces (15, 35, 38, 39), Actinoplanes (21, 29, 36), Thermus (9), and Arthrobacter (27, 37). The average length of their polypeptide chains is 380 to 390 amino acids, and they have about 60% amino acid sequence identity, with the active-site residues being highly conserved. The enzymes from Klebsiella (14), Escherichia (22, 34), Lactobacillus (3, 4, 24, 40), Lactococcus (13), Clostridium (25), Bacillus (28), Staphylococcus (33), and Thermoanaerobacter (23) are classified in group II. They are 440 to 460 amino acids long and show more than 50% amino acid sequence identity among the group members. Although they share only 20 to 30% amino acid sequence identity with group I XIs, the active-site residues in group I and group II enzymes are highly conserved (Table 1). The structures of monomeric, homodimeric, and homotetrameric XIs from a number of microbial sources including Streptomyces (5-7), Actinoplanes (1, 21, 29, 36), Arthrobacter (37), Bacillus (28), and Thermus (9) were solved by X-ray crystallography. A structural feature of monomeric XIs is that they consist of an (α/β)8 barrel having an active site and a C-terminal loop region. There are two ways of forming homodimers (15, 17, 27). One is the barrel-to-barrel facing way, in which the C-terminal loop of one subunit extends to the barrel domain of the other subunit, forming an active dimer. The active site of XI is completed when a conserved amino acid residue, Phe, is provided from the other subunit (37). A dimer can also be formed when two subunits are parallel to and leaning against each other, forming an inactive dimer. In the inactive dimer conformation, the active sites of two subunits face in the opposite direction, with the result that they cannot share the Phe residue, while their C-terminal loops are forming the active dimers in the manner described above. The quaternary structure of a tetrameric enzyme is a dimer of an active dimer and an inactive dimer, and it can be dissociated into the dimers by mild treatment with a denaturant such as urea.
TABLE 1.
Active-site and subunit-interacting residues in groups I and II and their corresponding residues in IO-1 and 210 XIs
| Conserved residue in groups I and II | IO-1 XI | 210 XI |
|---|---|---|
| Active site | ||
| W16 | W48 | W48 |
| F26 | F59 | F59 |
| H54 | H101 | H101 |
| D57 | D104 | D104 |
| F94 | F145 | F145 |
| W137 | W188 | W188 |
| E181 | E232 | E232 |
| K183 | K234 | K234 |
| E186 | E237 | E237 |
| N215 | N266 | N266 |
| E217 | E268 | E268 |
| H220 | H271 | H271 |
| D245 | D296 | D296 |
| D255 | ||
| D257 | ||
| Subunit interacting | ||
| D24 | D57 | D57 |
| R140 | R191 | R191 |
| L200 | L251 | L251 |
| A201 | A252 | A252 |
| A224 | A275 | V275 |
In the lactic acid bacteria, the ability to metabolize xylose is a function of their original habitats. Most of the xylose-metabolizing lactic acid bacterial genera such as Leuconostoc and Lactobacillus were isolated from fruits, vegetables, and fermented vegetables, in which d-xylose is the primary constituent of xylan and, therefore, is the primary carbon source for the microorganisms. On the other hand, only a few strains of Lactococcus lactis subsp. lactis, which has been used for dairy fermentation, have been reported as xylose-utilizing bacteria (18). Some L. lactis subsp. lactis and Lactococcus lactis subsp. cremoris strains show only a trace of XI (13). In previous studies, genotypic and phenotypic investigations into d-xylose metabolism were conducted with several L. lactis strains. L. lactis subsp. lactis strains CM56, MS39, 210, FB61, and 61 possessed a xylA gene but could not utilize d-xylose as a sole carbon source. Also L. lactis subsp. cremoris strains 160, BO32, and MS44 had a xylA gene of a larger than usual size due to an insertion, and they were all Xyl−. Two strains, L. lactis subsp. lactis 210 and L. lactis subsp. lactis IO-1, were selected for further study.
L. lactis subsp. lactis 210, which is a commercial starter culture, does not utilize d-xylose but does produce an inactive XI, while L. lactis subsp. lactis IO-1, isolated from a kitchen sink, can grow on d-xylose as a sole carbon source. It produces an active XI whose expression is regulated by d-xylose (19). In this study, the amino acid sequence differences between the XIs from strains IO-1 (XylA+) and 210 (XylA−) were examined, and mutants of these enzymes were constructed to characterize the effects of the different residues on enzymatic activity and solubility.
MATERIALS AND METHODS
Bacterial strains and plasmids.
L. lactis subsp. lactis IO-1 was obtained from P. Stanbury (University of Hertfordshire, Hatfield, United Kingdom), while L. lactis subsp. lactis 210 was obtained from Marshall Products (Madison, Wis.). Escherichia coli BL21(DE3)pLysS [F−] ompT hsdSB(rB−mB−) gal dcm (DE3) pLysS was used for the overproduction of wild-type and mutant XIs with the use of pET-19b (Novagen, Madison, Wis.).
Media and culture conditions.
E. coli was grown in Luria broth (Sigma, St. Louis, Mo.) plus chloramphenicol or ampicillin at 37°C with shaking. For induction, E. coli transformants were grown to an optical density at 600 nm of 0.5 to 0.6, and then isopropyl-β-d-thiogalactopyranoside (IPTG) was added to the culture to a final concentration of 100 μM. The culture was incubated for another 3 h, and then cells were harvested. Lactococci were grown at 30°C without agitation. M17 medium (Difco, Detroit, Mich.) supplemented with 0.5% glucose was used to culture lactococcal strains. A mixture of 0.4% d-xylose and 0.1% d-glucose was used for induction of XI overexpression in lactococcal strains.
Reagents and chemicals.
All restriction enzymes were purchased from New England Biolabs (Beverly, Mass.). T4 DNA ligase was from Gibco BRL Life Technologies (Grand Island, N.Y.), and AmpliTaq was from Perkin-Elmer (Foster City, Calif.). Chemicals were obtained from Sigma Chemical Co.
PCR.
The primers used in this study are listed in Table 2. The primers (NFX and BRX) used to amplify the wild-type and mutated xylA genes were designed on the basis of the nucleotide sequence of xylA from Lactobacillus brevis (3). They included BamHI (BRX) or NcoI (NFX) restriction sites that are compatible with the multiple cloning site of pET-19b. The primers were synthesized at the Cornell University BioResource Center (Ithaca, N.Y.). The 100-μl PCR mixture contained 1× PCR buffer (10 mM Tris-HCl [pH 8.8], 50 mM KCl, 1% Triton X-100), 4 μl of IO-1 or 210 crude cell lysate, 50 pmol of each primer, 5 nmol of deoxynucleoside triphosphate, 75 nmol of MgCl2, and 4 U of AmpliTaq DNA polymerase. The PCR mixture was cycled in a Perkin-Elmer GeneAmp PCR System 2400 thermocycler with a program of one cycle of 4 min at 94°C; 30 cycles of 1 min at 94°C, 1 min at 55°C, and 1.5 min at 72°C; and a final cycle of 10 min at 72°C.
TABLE 2.
PCR and mutational primers used in this study
| Primer | Sequence (5′→3′) | Usagea | Source or reference |
|---|---|---|---|
| NFX | CATGCCATGGCTTACTTTAACGAC | Cloning | In this study |
| BRX | CGGGATCCGGTTCCTAAGTCAATT | Cloning | In this study |
| T7 promoter | CGAAATTAATACGACTCACTATAGG | Seq | Commercial |
| T7 terminator | ATGCTAGTTATTGCTCAGCGGTGG | Seq | Commercial |
| Barney2 | TGCTTATGGTGCTGCACAAG | Seq | 2 |
| F1014 | ATTACTTGCTTGGGTGGGATACAG | Seq | 2 |
| R480 | TGTTTTTCAAGAAGGTGGTCAG | Seq | 2 |
| R1014 | CTGTATCCCACCCAAGCAAGTAAT | Seq | 2 |
| IO-1 S247A | CCAATATGATTTTGACGCAGCAACAGCTCT | SDM | In this study |
| IO-1 S388T | ACGAACGTTATACTTCATACAAAAATACAG | SDM | In this study |
| IO-1 K407E | GGAACAGCAACTTTTGAAAGTCTTGCCGC | SDM | In this study |
| 210 R202M | TTGAACACAGATATGGGTCTTGAAATG | SDM | In this study |
| 210 Y218D | CCATTTGGCAATTGATTATGCAAAATCAAT | SDM | In this study |
| 210 V275A | CATGCTTGGCTGGCTGGTCACACTTTTGAACAC | SDM | In this study |
| 210 T388S | GACGAACGTTATAGTTCATACAAAAATACAG | SDM | In this study |
| 210 E407K | GGAACAGCAACTTTTAAAAGTCTTGCCGC | SDM | In this study |
Cloning, used for cloning the wild-type and mutant xylA genes; Seq, used for sequencing the xylA clones; SDM, used for site-directed mutagenesis of xylA.
Cloning of wild-type and mutated xylA genes.
The 100-μl PCR products were purified using a Qiagen purification kit (Qiagen Inc., Chatsworth, Calif.) and resuspended in 50 μl of sterile water. About 20 μg of the purified PCR products was digested by the restriction enzymes BamHI and NcoI, in a final volume of 50 μl along with 800 ng of pET-19b at 37°C. The reaction mixture was purified, lyophilized, and resuspended in 11 μl of sterile water. Ligation mixture contained the digested 100 ng of PCR product and 10 ng of pET-19b, 1 U of T4 DNA ligase (Gibco, Gaithersburg, Md.), and 3 μl of 5× ligase buffer (250 mM Tris-HCl [pH 7.6], 50 mM MgCl2, 5 mM dithiothreitol, 25% [wt/vol] polyethylene glycol 8000) and was incubated at 16°C for 20 h. The ligation mixture was transformed into competent E. coli BL21(DE3)pLysS, which was prepared by the calcium chloride method (30).
All the clones were sequenced at the Cornell University BioResource Center using an ABI 377 automated DNA sequencer. Lasergene software (DNAStar, Inc., Madison, Wis.) was used to analyze the sequences.
Site-directed mutagenesis.
A two-step PCR method using megaprimers was used to conduct site-directed mutagenesis (31). Briefly, oligonucleotides containing the mutated sequence(s) were synthesized and used in the first-round PCR with the primer BRX. The first-round PCR product, a partial xylA fragment containing the mutated sequence(s), was purified and used as a megaprimer in the second-round PCR with the forward primer NFX, generating a complete xylA gene containing the desired mutation. The conditions for the second PCR were modified by extending the annealing time from 1 to 4 min and by lowering the primer concentration from 50 to 10 pmol.
Enzyme purification.
The purification of the recombinant XIs overproduced in the E. coli strain was conducted according to the protocol of Yamanaka and Takahara (40) with modifications. The induced cells were harvested, washed, resuspended in a 50 mM triethanolamine (TEA) buffer (pH 7), and lysed by sonication (Heat Systems-Ultrasonics Inc., Farmingdale, N.Y.). The lysed cells were centrifuged at 14,300 × g for 20 min. MnCl2 was added to the supernatant to a final concentration of 70 mM, and the supernatant was held at 55°C for 10 min. The precipitate was removed by centrifugation at 12,000 × g for 20 min. The enzyme was precipitated with 60% saturation of ammonium sulfate. The precipitate was resuspended in 50 mM TEA buffer (pH 7) and dialyzed overnight against the same buffer containing 1 mM MnCl2. The dialyzed solution was applied to a Q Sepharose (Pharmacia, Uppsala, Sweden) fast flow ionic column. The fractions eluted between 0.2 M and 0.3 M NaCl were pooled and then applied to a Sephadex G-200 (1.5- by 90-cm) column (Pharmacia). The active fractions eluted with 50 mM TEA were pooled and applied to the ionic column and gel filtration again for higher purification. The active fractions after the second round of the ionic column and gel filtration were pooled, dialyzed, and concentrated using an Ultrafree-4 centrifugal filter and tube (Millipore Corporation, Bedford, Mass.).
Protein analysis.
A 1-ml aliquot of E. coli recombinant cells was centrifuged at 14,000 × g for 5 min and resuspended in 100 μl of a 50 mM TEA buffer (pH 7) containing 10 μl of lysozyme (50 mg/ml). The suspension was incubated at 37°C for 1 h and centrifuged, and then the supernatant was saved for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The total protein concentration of the supernatant was measured using a Bio-Rad kit (Bio-Rad Laboratories, Richmond, Calif.). About 5 μg of total protein was used for the SDS-PAGE and separated by electrophoresis (100 V for 90 min) on a 12% polyacrylamide gel.
For Western blotting, lactococcal strains IO-1 and 210 were harvested, washed, lysed, and used for SDS-PAGE. After separation, the proteins were transferred to a nitrocellulose membrane in a transfer buffer containing 20% (vol/vol) methanol, 3.03 g of Tris/liter, and 14.4 g of glycine/liter at 0.04 A overnight. The immunoblotting was performed in four steps with a washing step between each one. The membrane was washed in TBST (Tris-buffered saline [TBS] buffer plus 1% Tween 20) three times with agitation for 5 min each time and washed once in TBS (pH 7.5) (29.22 g of NaCl and 2.41 g of Tris per liter) for 5 min. Then, the membrane was blocked with 0.4% bovine serum albumin in TBS buffer at room temperature for an hour and washed. The blocked membrane was treated with 1:500-diluted primary antibody (rabbit anti-E. coli XI polyclonal antibody) in TBS for an hour at room temperature, washed, and incubated with 1:1,000-diluted goat anti-rabbit immunoglobulin G (alkaline phosphatase conjugate) in TBS buffer containing 0.4% bovine serum albumin. The membrane was washed and incubated in 10 ml of Sigma Fast 5-bromo-4-chloro-3-indolylphosphate-nitroblue tetrazolium solution (0.15 mg of 5-bromo-4-chloro-3-indolylphosphate/ml, 0.3 mg of nitroblue tetrazolium/ml, 100 mM Tris buffer [pH 9.5], 5 mM MgCl2) for 5 min, and the reaction was stopped by rinsing the membrane with water.
Enzyme assays.
XI activity was qualitatively measured using the cysteine-carbazole assay (12). Whole-cell lysate was used, and the amount of xylulose produced was calculated with a standard curve which was made using different concentrations of xylulose. The Km and kcat values were determined by a coupled enzymatic assay with d-sorbitol dehydrogenase (Boehringer Mannheim, Indianapolis, Ind.) (20). The reaction mixture contained 0.2 to 0.3 U of XI (1 U was defined as the amount of enzyme that converts 1 μM d-xylose to d-xylulose/min), 1 U of d-sorbitol dehydrogenase (1 U was defined as the amount of enzyme that oxidizes 1 μM NADH/min), 2 to 30 mM d-xylose, and 0.15 mM NADH (Boeringer Mannheim). The oxidation rate of NADH was measured at 340 nm with a DU Series 600 spectrophotometer (Beckman Instruments, Fullerton, Calif.). Kinetic parameters, Km (millimolar) and kcat (per second), were calculated from a 1/v versus 1/S plot.
RESULTS
Cloning and sequencing of IO-1 and 210 xylA genes.
The xylA genes of IO-1 and 210 were amplified by PCR, cloned into pET-19b, and sequenced. The recombinant pET-19b plasmids harboring each xylA gene were designated as pET-IO-xylA and pET-210-xylA, respectively. Both xylA genes showed an open reading frame of 1,320 bp encoding a protein of 439 amino acids with a calculated molecular mass of 49.7 kDa. They shared 98.4 and 98.6% identity in their nucleotide sequence and amino acid sequence, respectively. Alignment of the two XIs showed that a total of six amino acid residues were different at positions 202, 218, 247, 275, 388, and 407 (Table 3). The amino acid residues R202, Y218, and V275 of 210 XI are not conserved or predominant in group II XIs, while T388 and E407 are in nonconserved regions.
TABLE 3.
Amino acid sequences and relative activities of the wild-type and mutant XIs of IO-1 and 210
| Strain | Amino acid at position:
|
Relative activitya (%) | |||||
|---|---|---|---|---|---|---|---|
| 202 | 218 | 247 | 275 | 388 | 407 | ||
| IO-1 | |||||||
| Wild type | M | D | S | A | S | K | 100 |
| S247A | M | D | A | A | S | K | 97 |
| S388T | M | D | S | A | T | K | 8 |
| K407E | M | D | S | A | S | E | 92 |
| S247A/S388T | M | D | A | A | T | K | 27 |
| S247A/K407E | M | D | A | A | S | E | 79 |
| S388T/K407E | M | D | S | A | T | E | 50 |
| 210 | |||||||
| Wild type | R | Y | A | V | T | E | 7 |
| R202M | M | Y | A | V | T | E | 9 |
| Y218D | R | D | A | V | T | E | 14 |
| V275A | R | Y | A | A | T | E | 24 |
| T388S | R | Y | A | V | S | E | 11 |
| E407K | R | Y | A | V | T | K | 2 |
| R202M/Y218D | M | D | A | V | T | E | 9 |
| R202M/V275A | M | Y | A | A | T | E | 26 |
| Y218D/V275A | R | D | A | A | T | E | 24 |
| R202M/Y218D/V275A | M | D | A | A | T | E | 62 |
Relative activity was measured by the cysteine-carbazole method using whole-cell lysates.
Wild-type XIs of 210 and IO-1.
Cell crude extracts of lactococcal strains IO-1 and 210 induced by d-xylose were prepared, and the cell debris and supernatant were separated by centrifugation and then analyzed by SDS-PAGE. Western blotting showed that the XI enzymes were expressed in both strains (Fig. 1). Western blotting, however, showed two differences between IO-1 and 210. First, the total amount of 210 XI in the extract was less than that of IO-1. Second, immunoreactive material was found in the cell debris but not in the supernatant from L. lactis subsp. lactis 210, suggesting that the enzyme is insoluble (Fig. 1, lanes 1 and 2). Their activities were measured using the cysteine carbazole method. 210 XI displayed no activity compared to that of the blank, while IO-1 XI was active.
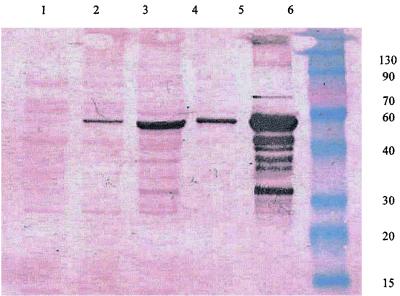
FIG. 1.
Western blotting of lactococcal strains IO-1 and 210. Lane 1, cell debris-free 210 cell lysate; lane 2, 210 cell debris; lane 3, cell debris-free IO-1 cell lysate; lane 4, IO-1 cell debris; lane 5, whole-cell lysate of recombinant E. coli harboring pET-210-xylA; lane 6, molecular mass (kilodalton) markers. The molecular mass of the immunoreactive XI is approximately 49 kDa.
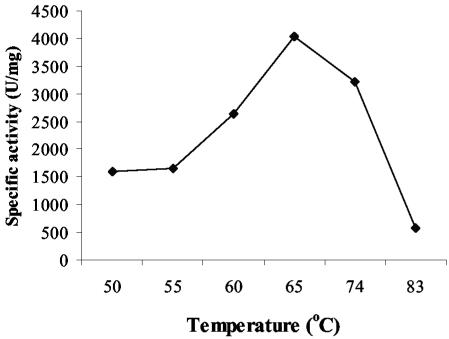
The IO-1 XI was purified, and the kinetics of the purified enzyme were measured. The native 210 XI could not be purified, because the protein always precipitated with the cell debris. The optimal temperature for IO-1 XI activity was around 65°C, and activity decreased dramatically above 74°C (Fig. 2). The Km and kcat of purified IO-1 XI were 2.25 mM and 184/s, respectively (Table 4).
FIG. 2.
Optimum temperature of IO-1 wild-type XI. The specific activity was measured by the cysteine carbazole method with the whole-cell lysate. One unit was defined as micromolar d-xylulose production in 1 min.
TABLE 4.
Kinetic values of IO-1 wild-type XI, IO-1 S388T/K407E XI, and 210 R202M/Y218D/V275A XI on d-xylose
| XI | Km (mM) | kcat (1/s) | Vmax (μM/min) |
|---|---|---|---|
| IO-1 WTa | 2.25 | 184 | 15 |
| IO-1 S388T/K407E | 4.4 | 124 | 6 |
| 210 R202M/Y218D/V275A | 3.46 | 142 | 8.1 |
WT, wild type.
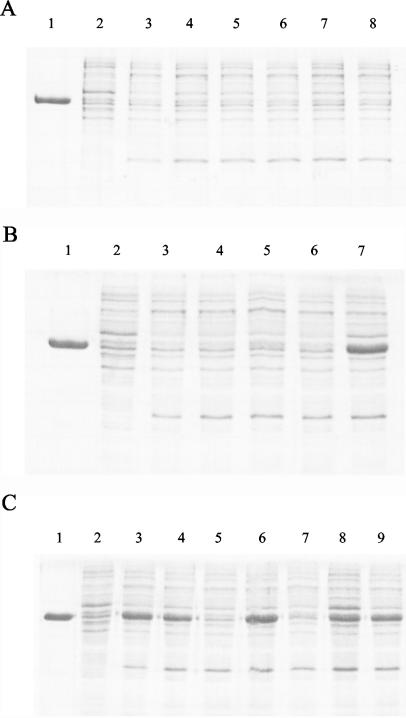
The wild-type XIs that were cloned into pET-19b, designated pET-IO-xylA and pET-210-xylA, were expressed in E. coli. 210 XI expressed in E. coli precipitated with cell debris and was inactive, similar to the enzyme found in the native host strain, while IO-1 XI was soluble and active (Table 3; Fig. 3C, lane 3). 210 XI was never found in a soluble form despite modification of induction conditions such as reducing the agitation speed, shortening the induction time, lowering the temperature, and decreasing the IPTG concentration.
FIG. 3.
SDS-PAGE of wild type and mutants of d-xylose isomerases expressed in E. coli. Only cell debris-free cell lysates were used for SDS-PAGE. Lanes 1 to 3 of each panel are purified IO-1 XI, the cell debris-free cell lysate of E. coli harboring intact plasmid pET-19b, and wild-type XI, respectively. (A) 210 recombinant XIs. Lane 4, R202M XI; lane 5, Y218D XI; lane 6, V275A XI; lane 7, T388S XI; lane 8, E407K XI. (B) 210 recombinant XIs. Lane 4, R202M/Y218D XI; lane 5, R202M/V275A XI; lane 6, Y218D/V275A XI; lane 7, R202M/Y218D/V275A XI. (C) IO-1 recombinant XIs. Lane 4, S247A XI; lane 5, S388T XI; lane 6, K407E XI; lane 7, S247A/S388T XI; lane 8, S388T/K407E XI; lane 9, S247A/K407E XI.
XI mutants of 210 and IO-1.
Among the six amino acids that differ between the 210 and IO-1 XIs, five residues (R202, Y218, V275, T388, and E407) of 210 XI were individually changed to the corresponding residue from IO-1 XI by site-directed mutagenesis, and the effects on activity and solubility of 210 XI were examined. Ala247 of 210 XI was not changed because Ala is the conserved residue in group II XIs. None of the mutants gained any detectable activity except the 210 XI V275A mutant. This mutant showed about 24% relative activity compared to the wild-type IO-1 XI activity (Table 3). Three different 210 XI double mutants, R202M/Y218D, R202M/V275A, and Y218D/V275A, were constructed. The R202M/V275A and Y218D/V275A mutants showed levels of activities similar to that of the V275A single mutant, 26 and 24%, respectively. The R202M/Y218D mutant did not gain any activity (Table 3). A triple 210 XI mutant, R202M/Y218D/V275A, was constructed. It showed about 60% relative activity and was soluble (Table 3; Fig. 3B, lane 7). The Km of the mutant was 3.46 mM, and its kcat was 142/s (Table 4).
Several IO-1 XI single mutants including S247A, S388T, and K407E were constructed. The S388T mutation in IO-1 XI resulted in a complete loss of its activity (Table 3), and the mutant protein was insoluble (Fig. 3C, lane 5). To confirm the absence of any unintended secondary mutations, the S388T mutation was mutated back to T388S, which resulted in restoration of full activity to IO-1 XI. Residues 247 and 407 are the only differences between the S388T IO-1 mutant and the 210 XI R202M/Y218D/V275A mutant (Table 3). These differences therefore define the activity and solubility of the 210 XI R202M/Y218D/V275A mutant and IO-1 XI S388T mutant. While the former is an active, soluble protein, the latter is inactive and insoluble. To examine the effect of the two residues, a second mutation (either S247A or K407E) was added to the IO-1 XI S388T mutant. Two double mutants of IO-1 XI, S247A/S388T and S388T/K407E, showed 27 and 50% relative activity, respectively (Table 3). Also, the IO-1 XI S388T/K407E mutant clearly produced a soluble protein (Fig. 3C, lane 8). The Km and kcat of the IO-1 XI S388T/K407E mutant were 4.4 mM and 124/s, respectively (Table 4). On the other hand, the single and double mutations S247A and K407E had no significant effect on the activity and solubility of IO-1 XI (Table 3; Fig. 3C, lane 9).
DISCUSSION
According to Kahl (19) and Erlandson (13), L. lactis strains IO-1 and 210 have xylA, which is induced by d-xylose. The 210 XI was inactive, however, and could be responsible for the Xyl− phenotype of the strain. In this study, it was confirmed that d-xylose induced the expression of the inactive 210 XI and also that the expression level of 210 XI in its natural host was lower than that of IO-1 in its respective lactococcal host strain (Fig. 1). The lower-level expression of 210 XI could be the result of the reduced d-xylose transport in L. lactis 210, which was not induced by d-xylose but remained at a basal level. In contrast d-xylose transport in IO-1 was fully induced by d-xylose (19).
The 210 XI always precipitated with cell debris when it was expressed in the native lactococcal strain or in E. coli (Fig. 3A, lane 3). All of the inactive 210 mutant XIs and the inactive IO-1 XI S388T mutant expressed in E. coli also precipitated with cell debris (Fig. 3). Expression in E. coli under alternative induction conditions (lower IPTG or lower temperature) did not yield soluble protein (data not shown). It is therefore difficult to separate the lack of activity of this protein from its insolubility.
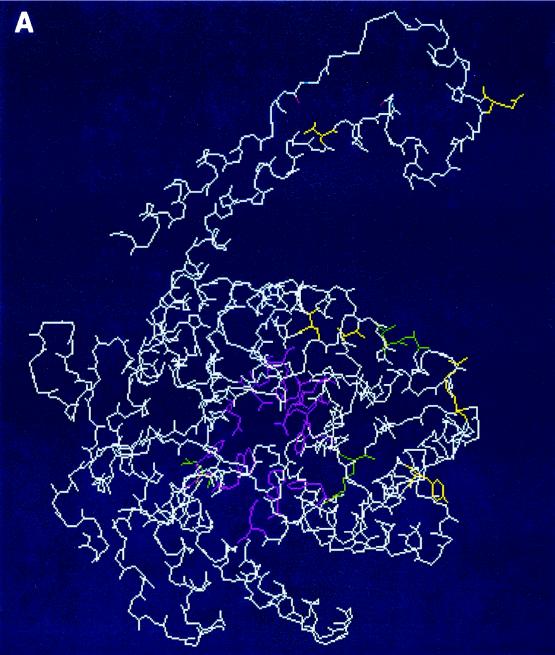
The crystallographic structures of a few group II XIs have been submitted to the Protein Data Base (PDB) (PDB accession numbers 1A0C, 1A0D, and 1A0E). The active-site residues and residues mediating subunit interactions in the group I XIs are conserved in the group II XIs (2, 14, 25, 32). The active-site residues of the Streptomyces rubiginosus XI (38) are conserved through the 20 group I and II XIs including the 210 and IO-1 XIs (Table 1). In addition, the residues (D24, R140, L200, A201, and A224) having a role in the subunit interactions are also conserved or similar, except that the 210 XI has V275 instead of A224 (Table 1). The three-dimensional structure of a monomeric 210 XI was predicted by SwissProt homology modeling (http://www.expasy.org/spdbv/text/modeling) with the XIs of Thermoanaerobacter and Thermotoga as templates (Fig. 4). Superimposition of the α-backbones of the predicted 210 XI and those of Thermoanaerobacter and Thermotoga XIs showed no significant deviation between them (data not shown). Also the six amino acids that are different between 210 and IO-1 do not appear to be directly involved in substrate or metal binding. However, these residues could affect the subunit binding. The alignment suggests that the structure of 210 and IO-1 XI is similar to those of other group I and II XIs and that the influence of the six amino acid differences on activity is not a result of a catalytic defect but more likely a significant structural perturbation.
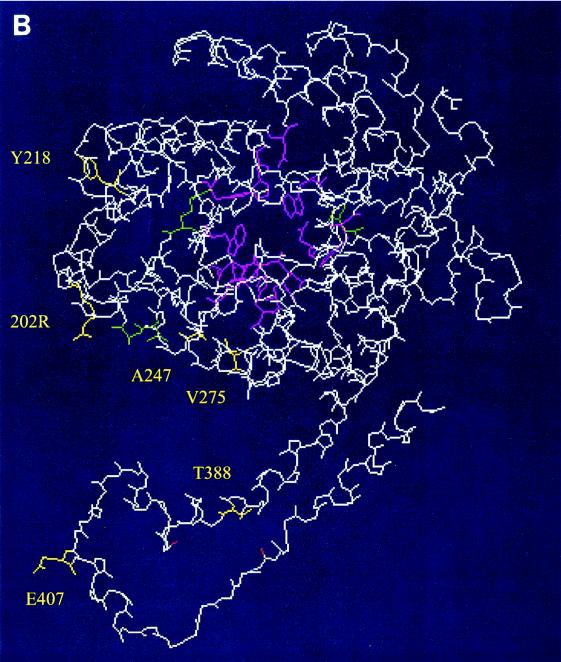
FIG. 4.
Predicted model of monomeric 210 XI. The model was produced by SwissProt homology modeling (http://www.expasy.org/spdbv/text/modeling.htm). The side chains of conserved active-site residues and subunit-interacting residues (Table 2) are shown in pink and green, respectively. The side chains of six amino acid residues different between IO-1 XI and 210 XI are shown in yellow with labeling. Panels A and B are the same molecule shown from different angles.
Residue A275 of IO-1 XI is highly conserved in group I and II XIs, and the corresponding residue in group I XIs (for example, A224 of the Streptomyces) is located in a turn structure and is involved in subunit interaction that leads to an active tetramer (5). The 210 XI has, however, Val instead of Ala at the corresponding residue, and Val is least likely to promote a turn in the protein structure (10). The V275 might destabilize the turn structure or the subunit interactions or both, inhibiting the formation of an active tetramer (Table 3).
Neither the single nor the double mutation of residues 202 and 218 of 210 XI gave rise to an active, soluble enzyme. When the two mutations were combined with the V275A mutation, however, there was a significant increase in the activity and solubility of 210 XI (Table 3; Fig. 3B, lane 7). The prevalent amino acid at position 202 in the group II XIs is Met, while an Arg is found in 210 XI. The predominant amino acid at position 218 is Asp or Asn in the group I and II XIs, while the 210 XI has Tyr. The R group of Tyr is bulky and hydrophobic compared to Asp or Asn and cannot participate in electrostatic interactions. Although these two residues do not directly contribute to the activity and solubility of the group I and II XIs, all three residues (202, 218, and 275) appear to be important for enzymatic activity and solubility and changes to them rendered the 210 XI soluble and active.
Amino acid residues 247 and 407 distinguish the 210 XI R202M/Y218D/V275A mutant from the IO-1 XI S388T mutant. When A247 and E407 are present in the 210 XI R202M/Y218D/V275A mutant, the enzyme is soluble and active, while S247 and K407 resulted in an inactive and insoluble protein in the IO-1 XI S388T mutant (Fig. 3B, lane 7, and 3C, lane 5). The S247A/K407E mutation suppressed the defective S388T mutation in IO-1, restoring the activity and solubility. To determine which of these two residues is responsible for the suppression, the S247A and K407E mutations were added to the IO-1 S388T mutant, separately. The IO-1 XI S388T/K407E mutant showed significantly increased activity and solubility, while the IO-1 XI S247A/S388T mutant showed only slightly increased activity (Table 3; Fig. 3C, lanes 7 and 8). Therefore, the K407E mutation suppressed the S388T mutation on the IO-1 XI. Amino acid residues 388 and 407 are located in the C-terminal loop that plays an important role in the homologous oligomerization of the subunits, leading to an active tetramer (29). This suggests that the S388T mutation on the IO-1 XI probably causes some critical structural defect which results in an insoluble and inactive protein. K407E did not affect the activity or the solubility of wild-type IO-1 XI (Table 3; Fig. 3C, lane 6). Probably the K407E mutation compensates for the predicted volumetric distortion in the structure as a result of the S388T mutation.
In conclusion, the 210 XI seems to have lost its activity and solubility due in part to the cumulative effect of the three mutations R202/Y218/V275. The S388 residue of the IO-1 XI appears to play a critical role in maintaining enzymatic activity and solubility. There may exist an interaction between the 388 and 407 residues, and the K407E mutation suppresses the S388T mutation.
Acknowledgments
This work was supported by the Northeast Dairy Center, whose support is derived from Dairy Management Inc.
REFERENCES
- 1.Allen, K. N., A. Lavie, A. Glasfeld, T. N. Tanada, D. P. Gerrity, S. C. Carlson, G. K. Farber, G. A. Petsko, and D. Ringe. 1994. Role of the divalent metal ion in sugar binding, ring opening, and isomerization by d-xylose isomerase: replacement of a catalytic metal by an amino acid. Biochemistry 33:1488-1494. [DOI] [PubMed] [Google Scholar]
- 2.Batt, C. A., A. C. Jamieson, and M. A. Vandeyar. 1990. Identification of essential histidine residues in the active site of Escherichia coli xylose (glucose) isomerase. Proc. Natl. Acad. Sci. USA 87:618-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bor, Y.-C. 1993. Cloning and characterization of xylose operon from Lactobacillus brevis. Ph.D. dissertation. Cornell University, Ithaca, N.Y.
- 4.Bor, Y.-C., C. Moraes, S. Lee, W. L. Crosby, A. J. Sinskey, and C. A. Batt. 1992. Cloning and sequencing the Lactobacillus brevis gene encoding xylose isomerase. Gene 114:127-131. [DOI] [PubMed] [Google Scholar]
- 5.Carrell, H. L., B. H. Rubin, T. J. Hurley, and J. P. Glusker. 1984. X-ray crystal structure of d-xylose isomerase at 4 Å resolution. J. Biol. Chem. 259:3230-3236. [PubMed] [Google Scholar]
- 6.Carrell, H. L., H. Hoier, and J. P. Glusker. 1994. Modes of binding substrates and their analogues to the enzyme d-xylose isomerase. Acta Crystallogr. D 50:113-123. [DOI] [PubMed] [Google Scholar]
- 7.Carrell, H. L., J. P. Glusker, V. Burger, F. Manfre, D. Tritsch, and J.-F. Biellmann. 1989. X-ray analysis of d-xylose isomerase at 1.9 Å: native enzyme in complex with substrate and with a mechanism-designed inactivator. Proc. Natl. Acad. Sci. USA 86:4440-4444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chaillou, S., Y.-C. Bor, C. A. Batt, P. W. Postma, and P. H. Pouwels. 1998. Molecular cloning and functional expression in Lactobacillus plantarum 80 of xylT encoding the d-xylose-H+ symporter of Lactobacillus brevis. Appl. Environ. Microbiol. 64:4720-4728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang, C., B. C. Park, D.-S. Lee, and S. W. Suh. 1999. Crystal structures of thermostable xylose isomerases from Thermus caldophilus and Thermus thermophilus: possible structural determinants of thermostability. J. Mol. Biol. 288:623-634. [DOI] [PubMed] [Google Scholar]
- 10.Creighton, T. E. 1993. Proteins, 2nd ed. W. H. Freeman and Company, New York, N.Y.
- 11.Davis, E. O., and P. J. F. Henderson. 1987. The cloning and DNA sequence of the gene xylE for xylose-proton symport in Escherichia coli K12. J. Biol. Chem. 262:13928-13932. [PubMed] [Google Scholar]
- 12.Dische, Z., and E. Borenfreund. 1951. A new spectrophotometric method for the detection and determination of keto sugars and trioses. J. Biol. Chem. 192:583-587. [PubMed] [Google Scholar]
- 13.Erlandson, K. A. 1999. Strain-specific detection and characterization of the xyl and xyn loci in Lactococcus lactis. Ph.D. dissertation. Cornell University, Ithaca, N.Y.
- 14.Feldmann, S. D., H. Sahm, and G. A. Sprenger. 1992. Cloning and expression of the genes for xylose isomerase and xylulokinase from Klebsiella pneumoniae 1033 in Escherichia coli K12. Mol. Gen. Genet. 234:201-210. [DOI] [PubMed] [Google Scholar]
- 15.Gailwad, S., H. Rao, and V. Deshpande. 1993. Structure-function relationship of glucose/xylose isomerase from Streptomyces: evidence for the occurrence of inactive dimer. Enzyme Microb. Technol. 15:155-157. [Google Scholar]
- 16.Heath, E. C., J. Hurwitz, B. L. Horecker, and A. Ginsburg. 1958. Pentose fermentation by Lactobacillus plantarum. I. The cleavage of xylulose-5-phosphate by phosphoketolase. J. Biol. Chem. 231:1008-1029. [PubMed] [Google Scholar]
- 17.Hess, J. M., V. Tchernajenko, C. Vielle, J. G. Zeikus, and R. M. Kelly. 1998. Thermotoga neapolitana homotetrameric xylose isomerase is expressed as a catalytically active and thermostable dimer in Escherichia coli. Appl. Environ. Microbiol. 64:2357-2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ishizaki, A., and T. Ohta. 1989. Batch culture kinetics of l-lactate fermentation employing Streptococcus sp. IO-1. J. Ferment. Bioeng. 67:46-51. [Google Scholar]
- 19.Kahl, W. E. 1997. Variation in d-xylose metabolism among Lactococcus lactis strains. M.S. thesis. Cornell University, Ithaca, N.Y.
- 20.Kersters-Hilderson, H., M. Callens, W. Vangrysperre, and C. K. De Bruyne. 1986. Kinetic characterization of d-xylose isomerases by enzymatic assays using d-sorbitol dehydrogenase. Enzyme Microb. Technol. 9:145-148. [Google Scholar]
- 21.Lambeir, A.-M., M. Lauwereys, P. Stanssens, N. T. Mrabet, J. Snauwaert, H. van Tilbeurgh, G. Mattyssens, I. Lasters, M. De Maeyer, S. J. Wodak, J. Jenkins, M. Chiadmi, and J. Janin. 1992. Protein engineering of xylose (glucose) isomerase from Actinoplanes missouriensis. 2. Site-directed mutagenesis of the xylose binding site. Biochemistry 31:5459-5466. [DOI] [PubMed] [Google Scholar]
- 22.Lawlis, V. B., M. S. Dennis, E. Y. Chen, D. H. Smith, and D. J. Henner. 1984. Cloning and sequencing of the xylose isomerase and xylulokinase genes of Escherichia coli. Appl. Environ. Microbiol. 47:15-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu, S.-Y., J. Wiegel, and F. C. Gherardini. 1996. Purification and cloning of a thermostable xylose (glucose) isomerase with an acidic pH optimum from Thermoanaerobacterium strain JW/SL-YS 289. J. Bacteriol. 178:5938-5945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lokman, B. C., P. Van Santen, J. C. Verdoes, J. Kruse, R. J. Leer, M. Posno, and P. H. Pouwels. 1991. Organization and characterization of the three genes involved in d-xylose catabolism in Lactobacillus pentosus. Mol. Gen. Genet. 230:161-169. [DOI] [PubMed] [Google Scholar]
- 25.Meaden, P. G., J. Aduse-Opoku, J. Reizer, A. Reizer, Y. A. Lanceman, M. F. Martin, and W. J. Mitchell. 1994. The xylose isomerase-encoding gene (xylA) of Clostridium thermosaccharolyticum: cloning, sequencing and phylogeny of XylA enzymes. Gene 141:97-101. [DOI] [PubMed] [Google Scholar]
- 26.Mortlock, R. P. 1984. The development of catabolic pathways for the uncommon aldopentose. Plenum Publishing Corporation, New York, N.Y.
- 27.Rangarajan, M., B. Asboth, and B. S. Hartley. 1992. Stability of Arthrobacter d-xylose isomerase to denaturants and heat. Biochem. J. 285:889-898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rasmussen, H., T. La Cour, J. Nyborg, and M. Schulein. 1994. Crystallization and preliminary investigation of xylose isomerase from Bacillus coagulans. Acta Crystallogr. D 50:231-233. [DOI] [PubMed] [Google Scholar]
- 29.Rey, F., J. Jenkins, J. Janin, I. Lasters, P. Alard, M. Classens, G. Mattyssens, and S. Wodak. 1988. Structural analysis of the 2.8 Å model of xylose isomerase from Actinoplanes missouriensis. Proteins Struct. Funct. Genet. 4:165-172. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 31.Sarkar, G., and S. Sommer. 1990. The “megaprimer” method of site-directed mutagenesis. BioTechniques 8:404-407. [PubMed] [Google Scholar]
- 32.Schmiedel, D., M. Kintrup, E. Kuster, and W. Hillen. 1997. Regulation of expression, genetic organization and substrate specificity of xylose uptake in Bacillus megaterium. Mol. Microbiol. 23:1053-1062. [DOI] [PubMed] [Google Scholar]
- 33.Sizemore, C., E. Buchnere, T. Rygus, C. Witke, F. Goetz, and W. Hillen. 1991. Organization, promoter analysis and transcriptional regulation of the Staphylococcus xylosus xylose utilization operon. Mol. Gen. Genet. 227:377-384. [DOI] [PubMed] [Google Scholar]
- 34.Song, S., and C. Park. 1997. Organization and regulation of the d-xylose operons in Escherichia coli K-12: XylR acts as a transcriptional activator. J. Bacteriol. 179:7025-7032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suekane, M., M. Tamura, and C. Tomimura. 1978. Physicochemical and enzymatic properties of purified glucose isomerase from Streptomyces olivochromogenes and Bacillus stearothermophilus. Agric. Biol. Chem. 42:909-917. [Google Scholar]
- 36.Tilbeurgh, H., J. Jenkins, M. Chiadmi, J. Janin, S. J. Wodak, N. T. Mrabet, and A.-M. Lambeir. 1992. Protein engineering of xylose (glucose) isomerase from Actinoplanes missouriensis. 3. Changing metal specificity and the pH profile by site-directed mutagenesis. Biochemistry 31:5467-5471. [DOI] [PubMed] [Google Scholar]
- 37.Varsani, L., T. Cui, M. Rangarajan, B. S. Hartley, J. Goldberg, C. Collyver, and D. M. Blow. 1993. Arthrobacter d-xylose isomerase: protein-engineered subunit interface. Biochem. J. 291:575-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Whitaker, R. D. 1995. Kinetic and structural studies of the Streptomyces rubiginosus d-xylose isomerase. Ph.D. dissertation. Cornell University, Ithaca, N.Y.
- 39.Wong, H. C., Y. Ting, H. C. Lin, F. Reichert, K. Myambo, K. W. K. Watt, P. L. Toy, and R. J. Drummond. 1991. Genetic organization and regulation of the xylose degradation genes in Streptomyces rubiginosus. J. Bacteriol. 173:6849-6858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamanaka, K., and N. Takahara. 1977. Purification and properties of d-xylose isomerase from Lactobacillus xylosus. Agric. Biol. Chem. 41:1909-1915. [Google Scholar]