Background: A-kinase anchoring proteins position signaling enzymes to control neuronal phosphorylation events.
Results: Biochemical and cellular approaches confirm that the AKAP79/150 signaling complex interfaces with the cytoplasmic tail of Roundabout (Robo) receptors.
Conclusion: AKAP79/150-associated protein kinase C facilitates the phosphorylation of Ser-1330 on the Robo3.1 isoform.
Significance: Kinase anchoring is a mechanism to control the phosphorylation of Robo3.1 within macromolecular assemblies.
Keywords: A-kinase-anchoring protein (AKAP), phosphorylation, protein kinase, protein-protein interaction, signal transduction
Abstract
Anchoring proteins direct protein kinases and phosphoprotein phosphatases toward selected substrates to control the efficacy, context, and duration of neuronal phosphorylation events. The A-kinase anchoring protein AKAP79/150 interacts with protein kinase A (PKA), protein kinase C (PKC), and protein phosphatase 2B (calcineurin) to modulate second messenger signaling events. In a mass spectrometry-based screen for additional AKAP79/150 binding partners, we have identified the Roundabout axonal guidance receptor Robo2 and its ligands Slit2 and Slit3. Biochemical and cellular approaches confirm that a linear sequence located in the cytoplasmic tail of Robo2 (residues 991–1070) interfaces directly with sites on the anchoring protein. Parallel studies show that AKAP79/150 interacts with the Robo3 receptor in a similar manner. Immunofluorescent staining detects overlapping expression patterns for murine AKAP150, Robo2, and Robo3 in a variety of brain regions, including hippocampal region CA1 and the islands of Calleja. In vitro kinase assays, peptide spot array mapping, and proximity ligation assay staining approaches establish that human AKAP79-anchored PKC selectively phosphorylates the Robo3.1 receptor subtype on serine 1330. These findings imply that anchored PKC locally modulates the phosphorylation status of Robo3.1 in brain regions governing learning and memory and reward.
Introduction
Axonal guidance cues influence neuronal development by modulating local cell signaling events at growth cones of migrating neurons and at the postsynaptic densities of mature neurons (1–3). A common feature is the dissemination of extracellular signals through transmembrane receptor-associated complexes that include protein kinases and phosphatases (4–6). These anchored enzymes often reside on the inner face of the plasma membrane, where they respond to the generation of chemical second messengers, such as cyclic nucleotides, calcium, and phospholipids (7, 8). Scaffolding and anchoring proteins enhance the fidelity of this process by sequestering second messenger-responsive kinases and phosphatases within range of particular substrates (9). In some cases, the receptors are the substrates themselves.
For example, A-kinase anchoring proteins (AKAPs),2 a burgeoning family of intracellular proteins that sequester protein kinase A, have been shown to modulate the phosphorylation status and activity of several neuronal transmembrane receptors (10, 11). Subsequent work has shown that AKAPs coordinate higher order macromolecular complexes of PKA and other signaling molecules at defined subcellular locations (12, 13). Consequently, AKAPs augment the specificity of local cell signaling events by scaffolding protein kinases, phosphatases, phosphodiesterases, and small GTPases in proximity to upstream activators and within range of their downstream targets (14–17). Products of the AKAP5 gene encode a multivalent anchoring protein known as AKAP79/150 (note that AKAP79 is the human form and AKAP150 is the murine ortholog). AKAP79/150 organizes signaling enzymes that participate in the regulation of synaptic ionotropic and metabotropic glutamate receptors as well as other neuronal G-protein-coupled receptors (18–21). A principal molecular role of AKAP79/150 is to tether PKA, PKC, and PP2B within range of these receptors to control changes in synaptic strength. These local signaling events underlie aspects of hippocampal learning and memory and a myriad of other cellular functions (22, 23).
In this report, we show that AKAP79/150 binds the Roundabout axon guidance receptors Robo2 and Robo3. AKAP150 co-distributes with both of these Robo receptors in the hippocampus and olfactory tubercle of adult mice. Biochemical and cell-based experiments confirm that the AKAP-Robo subcomplex provides a platform for the assembly of larger signaling complexes that include kinases and phosphatases. Accordingly, we demonstrate that AKAP79/150-associated PKC regulates the phosphorylation status of Robo3 in vitro and inside cells.
Experimental Procedures
Antibodies
Antibodies used for immunoblotting included mouse anti-FLAG (Sigma-Aldrich), mouse anti-V5-HRP (Invitrogen), rabbit anti-Robo2 (Abcam), rabbit anti-AKAP150 (V088), mouse anti-RIIα (BD Biosciences), mouse anti-PP2B B subunit (Abcam), mouse anti-PKCα (BD Biosciences), mouse anti-His-HRP (GenScript), and rabbit anti-phospho-Ser PKC substrate (Cell Signaling).
Plasmid Constructs
Full-length and truncated Robo1, Robo2, Robo3.1, and Robo3.2 were generated by PCR amplification and subcloned into pcDNA3.1/V5-His (Invitrogen). FLAG-AKAP79, GST-AKAP79 fragment (14–17), HA-muscarinic M1 receptor, pSilencer, and AKAP79 shRNA constructs have been described previously (24). Robo1, Robo2, and Robo3.1 C termini were cloned into pGEX6P-1 (GE Healthcare) for bacterial expression.
Mass Spectrometry
Bands of interest from silver-stained gels were excised and analyzed using MALDI-TOF by the Oregon Health and Science University Proteomics Shared Resource.
Immunoprecipitations
HEK293 cells were transiently transfected using Mirus Transit-LT1. After 48 h, cells were washed with cold PBS and lysed in HSE buffer (20 mm HEPES (pH 7.4), 150 mm NaCl, 5 mm EDTA, 1% Triton X-100, and protease inhibitors). For PKC phosphorylation experiments, cells were treated with DMSO or 2 μm phorbol 12,13-dibutyrate (PDBu) for 10 min prior to harvesting, and lysis buffer also contained NaF (Sigma-Aldrich) and okadaic acid (Millipore). Lysates were centrifuged at 15,000 × g for 20 min. Supernatants were incubated on a nutator overnight at 4 ºC with either FLAG-agarose (Sigma-Aldrich) or anti-V5 antibody (Invitrogen) and Protein A/G-agarose (Millipore). Beads were washed two times with HSE buffer supplemented with 10% glycerol and 500 mm NaCl and another two times with regular HSE buffer. LDS buffer (Invitrogen) was added, and samples were run on NuPage gels (Invitrogen).
HEK293 Staining
HEK293 cells were transfected with V5-Robo2 and FLAG-AKAP79 constructs. After 24 h, coverslips were washed three times in PBS and fixed for 20 min in 4% paraformaldehyde (in PBS) at room temperature. Cells were permeabilized in PBS containing 0.1% Triton X-100 and blocked for 1 h in PBS supplemented with 10% donkey serum and 0.1% fish gelatin (Sigma). Cells were incubated with antibodies against V5 (Invitrogen; 1:1000) and FLAG (Sigma; 1:1000) overnight at 4 ºC. Coverslips were washed three times with PBS and then incubated with Alexa Fluor-conjugated secondary antibodies for 1 h prior to mounting. Imaging was performed using ×40 and ×63 objectives on an LSM 510 META confocal microscope (Zeiss).
Immunofluorescent Staining of Day in Vitro (DIV) 4 Mouse Hippocampal Neurons
For preparation of cultured hippocampal neurons, hippocampi were dissected from postnatal day 1 mice and plated at a density of 100,000 cells/well (25). After culture for 4 DIV, neurons were fixed in 4% paraformaldehyde for 10 min, washed four times with PBS, permeabilized, and then blocked overnight in 10% donkey serum plus PBS. Cells were stained with goat anti-AKAP150 (1:200; Santa Cruz Biotechnology, Inc.) and rabbit anti-Robo2 (1:500; Abcam) antibodies. Cells were washed three times with PBS and incubated for 1 h at room temperature with Alexa Fluor-conjugated secondary antibodies before mounting. Neurons were imaged using an Axiovert 200M microscope (Zeiss) with a ×63 objective (1.4 numerical aperture; plan-Apo) and a CoolSNAP2 (Photometrics) charge-coupled device camera. Acquisition and off-line processing were conducted using Slidebook version 5.5 (Intelligent Imaging Innovations, Denver, CO). Focal plane z-stacks (spaced 0.2 μm apart) were acquired and deconvolved to discard out of focus light. Deconvolved three-dimensional z-stacks were then collapsed to generate two-dimensional maximum intensity projections.
Transfection of Hippocampal Neurons
DIV 14 mouse hippocampal neurons were transfected with V5-Robo using calcium phosphate. Cells were incubated for 24 h prior to fixation and staining with anti-V5 (1:1000; Invitrogen) and anti-AKAP150 (1:1000; V088) antibodies. Coverslips were imaged on a LSM 510 META confocal microscope (Zeiss) using ×40 and ×63 objectives.
GST Pull-downs
GST-Robo fusion proteins were purified from Escherichia coli and left on glutathione beads. Mouse brains were homogenized in cold HSE buffer using a Polytron homogenizer and cleared by centrifuging at 15,000 × g for 30 min. Brain lysate or purified protein was added and rocked overnight at 4 ºC. Beads were washed three times with HSE buffer supplemented to contain 1 m NaCl, two times with regular HSE, and finally with PBS prior to the addition of sample buffer. For PP2B pull-downs, lysates were prepared as described above and rocked with glutathione beads for 1 h at 4 ºC. Beads were washed three times with buffer containing 150 mm NaCl, 50 mm Tris (pH 7.5), 0.5% Nonidet P-40, and protease inhibitors before adding sample buffer. For PKC pull-downs, glutathione beads were rocked overnight with brain lysate and washed with HSE buffer as described above. 100 ng of PKCβII was added to beads and incubated in 100 μl of buffer containing 150 mm NaCl, 50 mm Tris (pH 7.5), 0.5% Nonidet P-40, and protease inhibitors for 1 h. Samples were washed three times with the same buffer prior to the addition of LDS sample buffer.
RII Overlay
RII overlays were performed as described previously (26, 27). In this far-Western technique, proteins from GST pull-downs were resolved by SDS-PAGE and transferred to nitrocellulose, and blots were subsequently overlaid with digoxigenin-labeled RII subunit. Proteins that bound the labeled RII were identified following incubation with anti-digoxigenin antibody (Abcam).
Immunohistochemistry of Spinal Sections
Embryonic day 11.5 embryos were fixed in 4% paraformaldehyde for 2 h, washed four times with PBS, and cryoprotected in 30% sucrose in PBS overnight prior to embedding in OCT. Frozen sections were cut at 20 μm on a cryostat (Leica) and mounted onto Superfrost-plus slides (Fisher). The sections were blocked and permeabilized for 30 min in PBS + 0.1% Triton X-100 + 1% normal donkey serum (Jackson ImmunoResearch) and stained overnight at 4 °C with the following primary antibodies: anti-L1Cam (1:200; Chemicon), anti-AKAP150 (1:500; Santa Cruz Biotechnology), and anti-Robo2 (1:500; Abcam). The slides were then washed three times in PBS and incubated with the appropriate fluorescent secondary antibody (Jackson ImmunoResearch) for 2 h at 4 °C. Finally, slides were washed three times in PBS and mounted with Fluromount-G (Southern Biotech). Images were collected using a DMI6000B inverted microscope equipped for both wide field and spinning disc confocal fluorescent microscopy (Leica).
Immunohistochemistry of Sagittal Brain Sections
Brains from 16-week-old wild type and AKAP150−/− mice were fixed in formalin for 48 h prior to paraffin embedding and sagittal sectioning at a thickness of 4 μm by the University of Washington Pathology Research Services Laboratory. Deparaffinized sections underwent antigen retrieval in a pressure cooker before blocking and staining with anti-AKAP150 (1:1000 (V088) and 1:500 (Santa Cruz Biotechnology)), anti-Robo2 (1:500; Abcam), or anti-Robo3 (1:500; R&D Systems) antibodies. The sections were then washed three times in PBS prior to incubation with Alexa Fluor-conjugated secondary antibodies for 1 h at room temperature. Sections were imaged using a DMI6000B inverted microscope equipped for both wide field and spinning disc confocal fluorescent microscopy (Leica).
Kinase Assays
For PKA kinase assays, GST-Robo fusion proteins were phosphorylated in kinase assay buffer containing 25 mm Tris, pH 7.5, 0.1 mm EGTA, 10 mm MgCl2, 50 μm ATP, and 0.2 μg of PKA. PKC kinase assays were performed using 0.2 μg of PKC, and the above buffer was supplemented with PKC activation mix (Millipore), 1 μg/ml diacylglycerol, and 10 μg/ml phosphatidylserine. Kinase reactions were stopped by washing three times with cold HSE buffer prior to the addition of LDS sample buffer.
Peptide Array
Peptides corresponding to regions of Robo3 containing potential PKC phosphosites were synthesized using an Auto-Spot Robot ASP 222 as described previously (28). Prior to phosphorylation, membranes were briefly wetted in ethanol and then incubated overnight in preincubation buffer (20 mm HEPES (pH 7.2), 100 mm NaCl, 2 mm MgCl2, 1 mm EDTA, 1 mm DTT, and 0.2 mg/ml BSA). Membranes were then blocked for 1 h at 30 ºC in the same buffer supplemented with 1 mg/ml BSA and 30 μm ATP. Phosphorylation was carried out by incubating membrane in kinase assay buffer (25 mm Tris, pH 7.5, 0.1 mm EGTA, 10 mm MgCl2, 50 μm ATP, PKC activation mix (Millipore), 1 μg/ml diacylglycerol, 10 μg/ml phosphatidylserine, and 0.2 μg of PKC) for 30 min at 30 ºC. Following phosphorylation, membranes were washed four times for 15 min in 1 m NaCl, three times for 5 min in H2O, three times for 15 min in 5% phosphoric acid, and finally three times for 5 min in H2O. Membranes were blocked for 1 h in 5% nonfat milk and 1% BSA and incubated overnight with anti-phospho-Ser PKC substrate antibody (Cell Signaling).
Proximity Ligation Assay
HEK293 cells were transfected with V5-Robo3 and cultured for 24 h prior to fixation and permeabilization in buffer containing 20 mm PIPES (pH 6.8), 10 mm EGTA, 1 mm MgCl, 0.2% Triton X-100, and 4% paraformaldehyde. Coverslips were blocked for 1 h in PBS containing 10% donkey serum and 0.1% fish gelatin (Sigma) and then incubated overnight with anti-Robo3 (R&D Systems; 1:500), and anti-phospho-Ser PKC substrates (Cell Signaling; 1:1000) antibodies. The Duolink in situ proximity ligation reaction (Sigma-Aldrich) was carried out according to the manufacturer's instructions. To identify transfected cells, coverslips were then incubated overnight with anti-V5 antibody, washed, and incubated for 1 h at room temperature with Alexa Fluor-conjugated secondary antibody (Invitrogen). Coverslips were mounted using Prolong antifade mounting medium containing DAPI (Molecular Probes) and imaged using a ×63 objective lens on a DMI6000B inverted confocal fluorescent microscope (Leica). The amount of proximity ligation assay (PLA) signal for each condition was quantified using Metamorph software (Molecular Devices). Individual cells expressing V5-Robo3 were outlined, and the integrated intensities of both the V5-Robo3 and PLA signals were recorded. The average ratio of PLA signal to V5-Robo3 was calculated for each experiment for both control and PDBu treatments. The -fold change in normalized PLA/Robo3 ratio was determined and analyzed using a one-sample t test with 0.05 as the level of significance.
shRNA Knockdown of AKAP79
HEK293 cells were transfected with V5-Robo3, HA-m1, and either pSilencer or AKAP79 pSilencer constructs. Cells were incubated for 72 h post-transfection and then treated with either vehicle (H2O) or 10 μm oxotremorine-M for 2 min at 37 ºC. Robo3 immunoprecipitations and Western blotting were performed as described above. Levels of phospho-Robo3 were determined by densitometric analysis and normalized to total Robo3 expression. Comparisons between pSilencer- and AKAP79 pSilencer-expressing cells were made using an unpaired Student's t test with 0.05 as the level of significance.
Results
Mass Spectrometry Identifies Robo2 as a Putative AKAP79/150 Binding Partner
Previous work has shown that AKAP79/150 is a multifunctional anchoring protein that interacts with a range of neuronal binding partners (14, 26, 29, 30). Therefore, we conducted a proteomic screen to look for additional proteins interacting with the murine ortholog, AKAP150. Protein complexes were immunoprecipitated from mouse brain extracts using a polyclonal antibody against AKAP150 and separated by SDS-gel electrophoresis (31) (Fig. 1A, lane 2). Control immunoprecipitations were performed with IgG (Fig. 1A, lane 1). Silver-stained bands that were enriched in the AKAP150 immunoprecipitation were excised and subjected to protein identification using MALDI-TOF mass spectrometry. Known members of the AKAP150 complex were detected, including the NR2A subunit of the NMDA receptor, thereby validating this approach (18–21). In addition, we identified several previously unknown AKAP150 binding partners (Fig. 1B). These included key elements of neuronal guidance pathways, such as the Roundabout receptor Robo2 and its ligands Slit2 and Slit3 (32, 33). These results imply that AKAP150 signaling complexes have the potential to interface with the axon guidance machinery and dendritic ion channels.
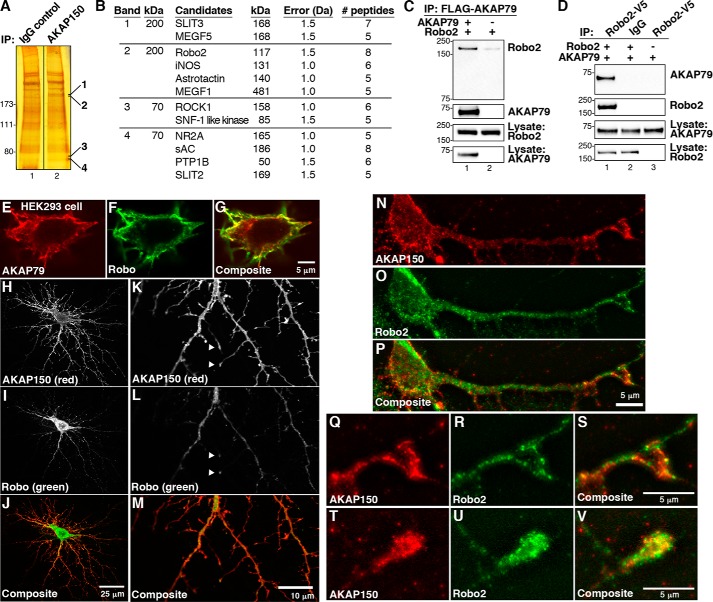
FIGURE 1.
AKAP79/150 interacts with Robo2. A, mass spectrometry screening to identify AKAP150 binding proteins. Immune complex complexes were isolated from mouse brain lysates. Silver-stained SDS-polyacrylamide gels of control (lane 1) and AKAP150 (lane 2) immune complexes are shown. Molecular weight markers are indicated. Bands present in AKAP150 immunoprecipitations (IP) were excised (the bands shown), and protein determination was by MALDI-TOF mass spectrometry. B, table of proteins identified in AKAP150 complexes. Columns indicate the name, molecular weight, mass error, number of peptides detected, and percentage coverage of each protein. C and D, validation of AKAP150 association with Robo2. HEK293 cells were transfected with FLAG-AKAP79 and V5-Robo2. FLAG immune complexes were immunoblotted for Robo2 (top) and AKAP79 (top middle). Loading controls for Robo2 (bottom middle) and AKAP79 (bottom) are included. D, reciprocal immunoprecipitation of Robo2 immune complexes blotted for AKAP79 (top) and Robo2 (top middle). E–G, confocal imaging of AKAP79 (E and G, red) and Robo (F and G, green) in HEK293 cells. H–P, DIV 15 mouse hippocampal neurons were transfected with Robo and immunostained for Robo and endogenous AKAP150. The staining patterns of AKAP150 (H and K (grayscale) and J and M (red)) and Robo (I and L (grayscale) and J and M (green)) were assessed by confocal microscopy. Details of the dendritic arbor are shown at higher magnification, and co-distribution of AKAP150 and Robo at tips of neuronal processes is indicated with arrows (K–M). N–P, neonatal mouse hippocampal neurons cultured for 4 DIV were stained for endogenous AKAP150 (N and P, red) and endogenous Robo2 (O and P, green). The staining patterns of both proteins were assessed by digital deconvolution microscopy. Q–V, higher magnification images of dendritic processes showing co-clustering (yellow) of AKAP150 (Q, S, T, and V; red) and Robo2 (R, S, U, and V; green) in putative growth cones.
AKAP79 Interacts with the Robo2 Receptor
In order to validate the results from our proteomics screen, it was important to determine whether AKAP79/150 interacts with Robo2 inside cells. Therefore, HEK293 cells were transfected with plasmids encoding FLAG-tagged AKAP79, the human ortholog of the anchoring protein, and V5-tagged Robo2. Cells were harvested after 48 h, and cell lysates were subjected to immunoprecipitation with anti-FLAG antibody. Western blot analysis of AKAP79 immune complexes detected the Robo2 receptor (Fig. 1C, top panel, lane 1). Control experiments performed from cells transfected with only Robo2 plasmid were negative (Fig. 1C, top panel, lane 2). Immunoblot analysis evaluated the expression levels of both proteins in HEK293 cell lysates (Fig. 1C, bottom two panels). Further validation of this protein-protein interaction was provided by reciprocal immunoprecipitation experiments. Immunoblots revealed the presence of AKAP79 in Robo2 immune complexes but not control immunoprecipitations (Fig. 1D, top panel). Immunoblot analyses evaluated the expression levels of both proteins in HEK293 cell lysates (Fig. 1D, bottom two panels). Collectively, these results allow us to conclude that heterologously expressed AKAP79 and Robo2 interact in HEK293 cells.
Immunofluorescence staining further suggests that Robo2 and AKAP79 reside in the same subcellular compartment (Fig. 1, E–G). Confocal imaging revealed that recombinant AKAP79 (Fig. 1E, red) and Robo2 (Fig. 1F, green) have overlapping regions of expression and accumulate near the plasma membrane in HEK293 cells. This is best observed in the composite image of the merged signals (Fig. 1G). These results imply that AKAP79 and Robo2 co-distribute inside cells.
Robo2 is expressed throughout the developing murine hippocampus, a region of the brain that is enriched with AKAP79/150 (22, 34). Thus, a more pertinent validation of the interaction between these two proteins was to be found in primary cultures of mouse hippocampal neurons grown for 15 DIV. Endogenous AKAP150 (Fig. 1, H and J, red) was found to overlap with recombinant Robo (Fig. 1, I and J, green). Higher magnification confocal images reveal that both proteins accumulate at the tips of neuronal processes (Fig. 1, K–M, arrows). Further experiments revealed that endogenous AKAP150 (Fig. 1, N and P, red) co-distributed with endogenous Robo2 (Fig. 1, O and P, green) in discrete clusters. These regions of signal overlap were located in both the cell body and dendrites of mouse hippocampal neurons that had been cultured for 4 days. At higher magnification, it was evident that both proteins co-clustered at the tips of neuronal processes, including putative dendritic growth cones (Fig. 1, Q–V).
Mapping the AKAP79-Robo2 Binding Interface
On the basis of mass spectrometry identification and biochemical analyses, we postulate that AKAP79/150 and Robo2 can exist as a macromolecular complex. The next phase of these studies was to determine what regions of AKAP79 and Robo2 participate in this protein-protein interaction. To map the Robo2 binding site on AKAP79, we split the anchoring protein into N-terminal (residues 1–153), central (residues 154–296), and C-terminal (residues 297–427) fragments. Each AKAP79 fragment was expressed as a GST fusion protein (Fig. 2A, middle panel). This family of deletion fragments was used to pull down Robo2 from HEK293 cell lysates. Immunoblot analysis revealed that Robo2 selectively bound to the N-terminal fragment of AKAP79, suggesting that its principal binding site lies between residues 1 and 153 of the anchoring protein (Fig. 2A, top panel, lane 2). It should be noted that Robo2 weakly interacted with the C-terminal regions of the anchoring protein (Fig. 2A, top panel, lane 4). Control experiments confirmed that equivalent amounts of Robo2 were used in all binding experiments (Fig. 2A, bottom panel). These experiments suggest that multiple regions of AKAP79 may interface with Robo2.
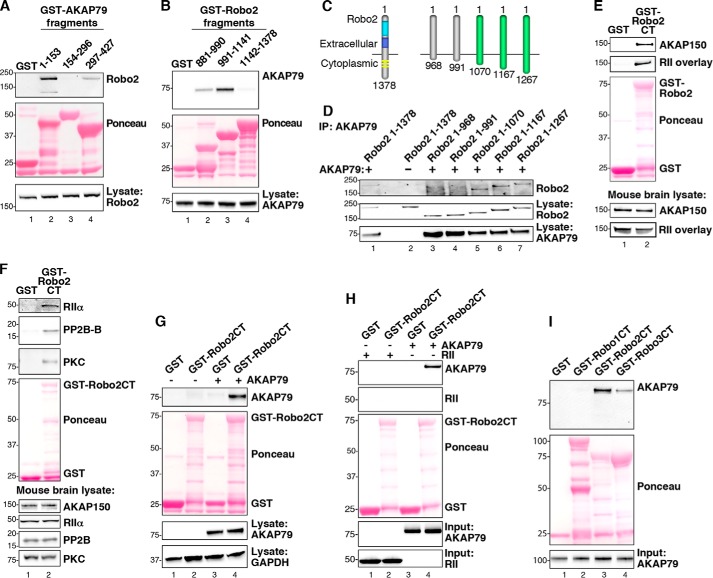
FIGURE 2.
Biochemical characterization of the Robo2-AKAP79/150 interaction. A, purified GST-AKAP79 fragments containing GST alone (lane 1) and amino acids 1–153 (lane 2), 154–296 (lane 3), and 297–427 (lane 4) were used to isolate Robo2 from HEK293 cell lysates. Molecular weight markers are indicated. B, GST-tagged fragments, including GST alone (lane 1) and amino acids 881–990 of Robo2 (lane 2), 991–1141 of Robo2 (lane 3), and 1142–1378 of Robo2 (lane 4), were used to pull down overexpressed AKAP79 from HEK293 cell lysates. C, diagram depicting progressive truncation of the Robo2 receptor used for fine mapping of the AKAP79 binding site. The first and last residues of each fragment are denoted. D, AKAP79 and truncated Robo2 receptors were overexpressed in HEK293 cells. AKAP79 immune complexes were immunoprecipitated (IP), and Robo2 binding was assessed by immunoblot. E, purified Robo2 C terminus (Robo2-CT) fused to GST was incubated with mouse brain lysate. Immunoblots were probed for AKAP150 (top) and overlaid with digoxigenin-labeled RII (top middle). F, GST-Robo2CT pull-downs from mouse brain lysate were immunoblotted for RIIα (top), PP2B (second from top), and PKC (third from top). G, GST-Robo2CT pull-downs from HEK293 cell lysate containing overexpressed AKAP79 were probed for AKAP79 (top). H, GST-Robo2CT was incubated with either purified His-AKAP79 or His-RII. GST-Robo2CT complexes were immunoblotted for AKAP79 (top) and RII (top middle). I, the C termini of Robo1, Robo2, and Robo3 were each fused to GST and purified from bacteria. Purified His-AKAP79 was incubated with the GST-RoboCT proteins, and immunoblots of the GST-Robo complexes were probed for AKAP79 (top).
In reciprocal experiments, a similar approach was used to map the region of Robo2 that binds to the anchoring protein. The cytoplasmic tail of Robo2 was split into three fragments (Fig. 2B, middle panel). These GST fusion proteins were then used to pull down FLAG-AKAP79 from HEK293 cell lysate. The anchoring protein bound strongly to a central portion of Robo2 that encompasses residues 991–1141 (Fig. 2B, top panel, lane 3), although weaker binding of AKAP79 was also detected to a fragment of Robo2 that is immediately proximal to the membrane-spanning segment (residues 881–990; Fig. 2B, top panel, lane 2). This led us to conclude that the primary AKAP79 binding site lies between residues 991 and 1141 of Robo2. Control experiments confirmed that equivalent amounts of AKAP79 were used in all binding experiments (Fig. 2B, bottom panel).
To further narrow down the AKAP79 binding sites, we generated an additional family of truncations in the cytoplasmic tail of Robo2 (Fig. 2C). AKAP79 binding was analyzed as described above. These studies revealed that a Robo2 receptor expressing the first 1070 residues was still competent to bind AKAP79. This represents a region of ∼190 residues that reside in the cytoplasm of cells (Fig. 2C). Further truncation of the cytoplasmic tail of Robo2 prevented interaction with the anchoring protein (Fig. 2D, top panel, lane 5). Thus, we can conclude that residues 991–1070 of Robo2 are necessary for interaction with AKAP79. Control experiments confirmed that equivalent amounts of Robo2 and AKAP79 were used in all binding experiments (Fig. 2D, middle and bottom panels).
AKAP79/150 Binds Directly to the Cytoplasmic Tails of Robo2 and Robo3
Studies described above indicate that human AKAP79 and its murine ortholog AKAP150 have the capacity to interact with Robo2. Next, we investigated whether Robo2 is capable of interacting with endogenous AKAP150 and its anchored enzymes. A GST-tagged Robo2 881–1378 fragment was used to perform pull-downs from mouse brain lysate. Overlay with labeled RII revealed that an AKAP with the molecular weight of AKAP150 was bound to this Robo2 fragment (Fig. 2E, upper panel, lane 2). Western blotting confirmed that this band corresponded to endogenous AKAP150 (Fig. 2E, top, lane 2). The anchoring protein was not detected in control pull-downs using GST alone (Fig. 2E, top two panels, lane 1). Analysis of mouse brain lysates verified that equivalent amounts of AKAP150 were present in both samples (Fig. 2E, bottom two panels). Related experiments established that the AKAP79/150 binding partners RIIα, the phosphatase PP2B (B subunit), and conventional PKCs co-purify with the Robo2 881–1378 fragment (Fig. 2F, top three panels, lane 2). Collectively, these findings strengthen our view that Robo2 associates with AKAP150 complexes containing a range of anchored signaling enzymes. Additional experiments revealed that GST-Robo2 also binds human AKAP79 from HEK293 cell lysates (Fig. 2G, top panel, lane 4).
AKAP79/150 binds directly to certain membrane proteins, such as the L-type calcium channel and the KCNQ2 subunits of the M-channel (22, 30, 35–38). In contrast, NMDA and AMPA receptors interact with AKAP79/150 through a bridging interaction with membrane-associated guanylate kinase (MAGUK) proteins (18, 19, 39). To determine whether AKAP79 can interface directly with Robo2, we repeated our GST-Robo2 pull-downs using His-tagged AKAP79 purified from E. coli. These experiments confirmed that AKAP79 binds directly to Robo2, without the need for an adaptor protein (Fig. 2H, top panel, lane 4). Purified recombinant RII was used as a negative control for these binding studies (Fig. 2H, bottom panel, lane 2).
There are four members of the Robo family of chemotactic guidance receptors (40). Therefore, it seemed logical to explore the possibility that AKAP79/150 may associate with other Robo receptor isoforms. However, only Robo1, -2, and -3 are expressed in neurons (40). Therefore, AKAP79 pull-down experiments were conducted with GST fusion proteins encompassing the cytoplasmic tails of Robo1, Robo2, and Robo3. These binding studies revealed that Robo3 also associates with AKAP79 (Fig. 2I, top panel, lane 4). GST-Robo2 binding to AKAP79 served as an internal control (Fig. 2I, top panel, lane 3). Immunoblot analysis confirmed that equivalent amounts of each binding partner were present in these experiments (Fig. 2I, middle and bottom panels). Sequence homology between Robo2 and Robo3 within the region that spans the AKAP binding site may explain why the AKAP is capable of binding both of these receptors. In contrast, the Robo1 isoform contains an insertion within this region, potentially explaining why it does not associate with this anchoring protein (Fig. 2I, top panel, lane 2).
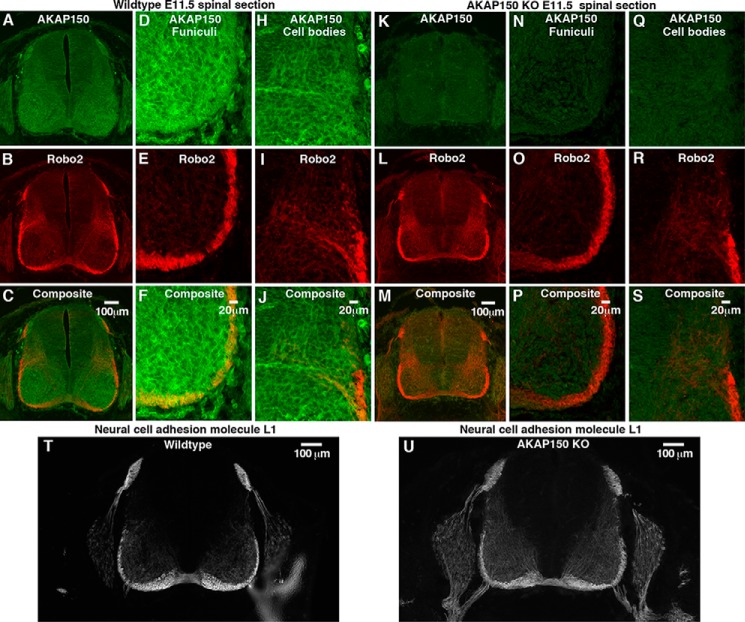
Much of the knowledge of Roundabout receptors has been gleaned from the investigation of axonal guidance in the developing spinal cord. Signals processed by Robo1 and Robo2 expel axons from the floor plate of the spinal cord and prevent inappropriate recrossing of the midline (41). In contrast, signaling through Robo3 promotes midline crossing (42). Therefore, we reasoned that association with AKAP150 might influence aspects of axonal guidance in mouse spinal commissural axons. As a prelude to these studies, we examined whether AKAP150 is expressed in the developing spinal cord of embryonic day 11.5 mice. Immunostaining revealed that AKAP150 is broadly expressed throughout the developing spinal cord (Fig. 3A, green). In keeping with published reports, Robo2 expression was restricted to the ventral and lateral funiculi (43) (Fig. 3, B and C, red). These two regions of the spinal cord were reexamined at higher magnification by confocal microscopy (Fig. 3, D–J). Co-distribution of both signals was most apparent in the ventral and lateral funiculi (Fig. 3, D–F, yellow), but some overlap of staining was also observed in the cell bodies of commissural neurons (Fig. 3, H–J, yellow). Control experiments using the same antibodies confirmed that the AKAP150 signal was absent in equivalent sections from embryonic day 11.5 AKAP150−/− mice (22) (Fig. 3, K, N, and Q). Robo2 staining appeared normal in these sections (Fig. 3, L–P). Normal patterns of midline crossing were observed when wild type and AKAP150−/− sections were stained for neural cell adhesion molecule L1, a postcrossing axonal marker (44) (Fig. 3, T and U). Thus, it would appear that although AKAP150 and Robo2 are co-distributed in the developing spinal cord, their association does not influence midline crossing of spinal commissural axons in mouse embryos.
FIGURE 3.
AKAP150 is expressed in the developing spinal cord, where it co-distributes with Robo2 in specialized regions. Transverse sections of embryonic day 11.5 spinal cords from wild type and AKAP150−/− mice were stained for AKAP150 and Robo2 and imaged using wide field fluorescence microscopy (wild type (A–C) and AKAP150−/− (K–M)). AKAP150 is shown in green, and Robo2 is shown in red. Higher magnification spinning disc confocal images provide more detailed localization of these proteins in the funiculi (wild type (D–F) and AKAP150−/− (N–P)) and commissural neuron cell bodies (wild type (H–J) and AKAP150−/− (Q–S)). T and U, sections of spinal cord from wild type (T) and AKAP150−/− mice (U) stained for neural cell adhesion molecule L1, a marker for midline crossing.
Robo2 and Robo3 Co-distribute with AKAP150 in the Adult Mouse Brain
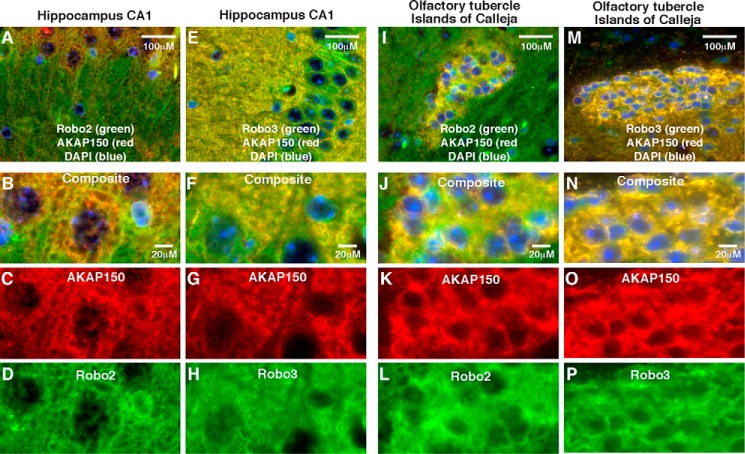
Robo receptors are expressed in the adult brain (45). Likewise, neuronal AKAP150 expression peaks later in development (20). Therefore, we wondered whether AKAP150 co-distributed with Robo receptors in the brains of mature mice. To test this possibility, brains from 4-month-old wild type and AKAP150−/− mice were isolated and paraffin-embedded. Sagittal sections were prepared, and immunofluorescent staining with antibodies to AKAP150, Robo2, and Robo3 was completed. Fluorescent microscopy of intact sagittal sections at low magnification revealed distinct but overlapping staining patterns for AKAP150 and Robo2 and Robo3 in a variety of brain regions (data not shown). Wide field fluorescent images (×40 objective) show overlapping expression patterns for AKAP150 (red) with both Roundabout receptors (green) in hippocampal sections (Fig. 4, A and E). Upon higher magnification (×63) analysis on a confocal microscope, the co-distribution of the anchoring protein with Robo2 (Fig. 4, B–D) and Robo3 (Fig. 4, F–H) was evident in the CA1 region of the hippocampus. Likewise, analysis of lower magnification images detected both Roundabout receptors co-distributed with AKAP150 in the islands of Calleja in the olfactory tubercle (Fig. 4, I and M, yellow). Overlapping staining patterns between AKAP150 and both Robo2 and Robo3 were also evident in this same tissue at higher magnification (Fig. 4, J–L and N–P). Staining patterns of Robo2 and Robo3 were unchanged in brain sections from AKAP150−/− mice (data not shown).
FIGURE 4.
AKAP150 co-distributes with Robo2 and Robo3 in the adult brain. Sagittal paraffin-embedded sections from 16-week-old mouse brains were immunostained for AKAP150 (A–C, E–G, I–K, and M–O; red) and either Robo2 (A, B, D, I, J, and L; green)) or Robo3 (E, F, H, M, N, and O; green). A–H, staining in the CA1 region of the hippocampus. I–P, staining of the islands of Calleja in the olfactory tubercle. Images from the top panel were collected at low magnification, whereas the bottom panels represent high magnification confocal images from the same region.
Robo3 Is Phosphorylated by Anchored PKC
AKAP150 expression in the hippocampus peaks at postnatal day 14 in mice (20), a developmental stage when Robo receptors, their Slit ligands, and other axon guidance molecules are also present in the hippocampus (43). For instance, the Netrin receptor DCC (deleted in colorectal cancer) is enriched in the postsynaptic density, where it is required to facilitate long term potentiation, an electrophysiological paradigm for learning and memory (46). Therefore, it seemed plausible that Robo receptors might contribute to acute neuronal signaling events that control synaptic strength. An attractive feature of this hypothesis is that AKAP79/150 signaling complexes also populate the postsynaptic density, where they provide local control of synaptic strength (31, 47). In keeping with this notion, deletion of AKAP150 in mice impairs long term depression due to the loss of PKA, PKC, and PP2B anchoring (22, 24, 25).
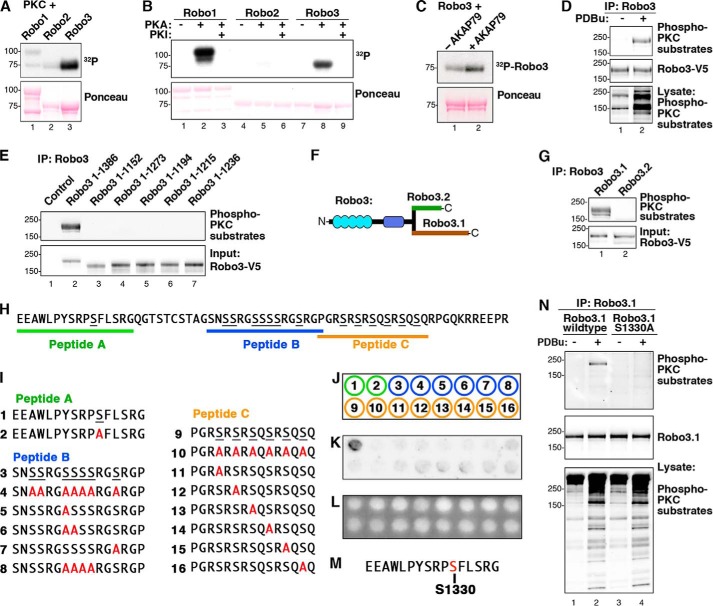
In light of these findings, we performed in vitro kinase assays to determine whether any Robo receptors are targets of AKAP150-anchored kinases. Purified GST-tagged cytoplasmic tails of Robo1, Robo2, and Robo3 were immobilized on beads and phosphorylated in vitro with a mixture of conventional PKCs and [32P]ATP. Autoradiography revealed that only Robo3 was phosphorylated by PKC (Fig. 5A, top panel, lane 3). Analogous experiments showed that Robo1 was phosphorylated by protein kinase A, whereas Robo3 was labeled to a lesser extent (Fig. 5B, top panel, lanes 2 and 8). However, we were unable to recapitulate PKA phosphorylation of either Robo receptor inside cells. Because Slit/Robo signaling can mobilize intracellular calcium (48), we reasoned that the selective phosphorylation of Robo3 by PKC might contribute to the unique function of this receptor. For these reasons, we focused on investigating PKC phosphorylation of Robo3.
FIGURE 5.
PKC phosphorylates Ser-1330 in the cytoplasmic tail of Robo3.1. A, GST-Robo1CT, -Robo2CT, and -Robo3CT were phosphorylated by PKC in vitro. 32P incorporation was assessed by autoradiography. B, GST-Robo1CT, -Robo2CT, and -Robo3CT were phosphorylated using PKA in vitro, and 32P incorporation was assessed by autoradiography. C, GST-Robo3CT was phosphorylated by PKC in vitro plus or minus purified AKAP79. D, Robo3 was overexpressed in HEK293 cells. Cells were stimulated with vehicle (DMSO) or 2 μm PDBu for 10 min. Robo3-V5 immunoprecipitations (IP) were probed using an antibody that recognizes phosphorylated consensus (phospho-Ser) PKC phosphorylation sites (top). E, Robo3 receptors containing progressive C-terminal truncations were expressed in HEK293 cells and stimulated with PDBu. V5 immunoprecipitations were immunoblotted with the phospho-PKC substrate antibody. F, diagram depicting the differing distal C termini of Robo3.1 and Robo3.2. G, Robo3.1 and Robo3.2 were expressed in HEK293 cells. Cells were treated with PDBu, and V5 immunoprecipitations of cell lysates were immunoblotted with the PKC substrate antibody (top). H, unique Robo3.1 C-terminal amino acid sequence. Three Robo3.1 peptides containing putative PKC phosphorylation sites are indicated. I, peptides containing putative PKC sites from Robo3.1 and serine to alanine-substituted peptides. J, peptides 1–16 in I were synthesized using peptide spot array according to template. K, peptide array membrane was in vitro phosphorylated with PKC and [32P]ATP. The autoradiograph shows phosphorylated peptide. L, UV illumination of membrane shown in K validates equal spotting efficiency for all peptides. M, sequence surrounding the Ser-1330 phosphosite from peptide A. N, HEK293 cells expressing full-length wild type or S1330A Robo3.1 were treated with vehicle alone (DMSO) or PDBu. Robo3.1 immunoprecipitates were immunoblotted using the phospho-PKC substrate antibody to show phosphorylation of Robo3.1.
Next, we performed experiments in the presence of AKAP79 to investigate the role of the anchoring protein in this phosphorylation event. Formation of a Robo3-AKAP79-PKC ternary complex enhanced phosphorylation of the substrate (Fig. 5C, top panel, lane 2) as compared with experiments performed in the absence of the anchoring protein (Fig. 5C, top panel, lane 1). This suggests that AKAP79-anchored PKC preferentially phosphorylates Robo3. A more stringent validation of this concept was performed in HEK293 cells as they express high levels of endogenous AKAP79. Cells were transfected with plasmids encoding full-length V5-tagged Robo3, and PKC activity was stimulated upon application of PDBu. Robo3 immune complexes were probed with an antibody that detects phosphorylated PKC substrates. Immunoblot analysis revealed robust phorbol ester-dependent phosphorylation of Robo3 (Fig. 5D, top panel, lane 2) as compared with unstimulated controls (Fig. 5D, top panel, lane 1). Immunoblot analysis of whole cell lysates using the phospho-PKC substrate antibody confirmed kinase activation, and additional controls established that equivalent levels of Robo3 were present in each sample (Fig. 5D, middle and bottom panels).
We subsequently probed a family of Robo3 truncation fragments in an attempt to locate the sites of PKC phosphorylation. These mapping studies indicate that the extreme C terminus of Robo3 (residues 1236–1386) contained the site(s) of PKC phosphorylation (Fig. 5E, top panel, lane 2). This region of the Robo3 gene is subject to differential splicing and results in the expression of two subtypes, Robo3.1 and Robo3.2 (Fig. 5F). Plasmids encoding both Robo3 subtypes were expressed in HEK293 cells (Fig. 5G). Cell-based phosphorylation experiments demonstrate that only the Robo3.1 form is phosphorylated by PKC inside cells (Fig. 5G, top panel, lane 1).
Unfortunately, quantitative analysis by mass spectrometry was unsuccessful in obtaining more precise information about the identity of these putative phosphorylation sites. Therefore, we utilized peptide spot arrays as an alternate means to approach this problem (Fig. 5, H–L). There are 20 serine or threonine residues in the unique cytoplasmic region of Robo3.1 (Fig. 5H). Given the number of possible phosphosite combinations, we focused on three segments of Robo3.1 sequence that contain consensus PKC substrate motifs (peptides A, B, and C are denoted in Fig. 5, H and I). A total of 16 immobilized peptides were evaluated (Fig. 5, I–K). These included wild type sequences and peptides where alanine was substituted for putative target serines (Fig. 5I). UV analysis confirmed equivalent spotting efficiency for all peptides (Fig. 5L).
In vitro phosphorylation of wild type peptides with purified PKC revealed that only peptide A was labeled (Fig. 5I). This peptide contains Ser-1330, which conforms to a consensus PKC substrate site (Fig. 5M). Importantly, an alanine-substituted peptide analog of this sequence was not phosphorylated by this kinase (Fig. 5, I–K). Cell-based validation of this result was provided by analysis of a Robo3.1 S1330A mutant. As expected, wild type Robo3.1 was efficiently phosphorylated upon stimulation of PKC (Fig. 5N, top panel, lane 2). In contrast, the Robo3.1 S1330A mutant was not phosphorylated by phorbol ester-mediated stimulation of endogenous PKC (Fig. 5N, top panel, lane 4). Loading controls confirmed equal expression of both Robo3.1 forms, and immunoblot analysis using phosphosubstrate antibodies detected phorbol ester stimulation of PKCs (Fig. 5N, middle and bottom panels). Taken together, the data in Fig. 5 identify Ser-1330 on Robo3.1 as a target for PKC in vitro and inside cells.
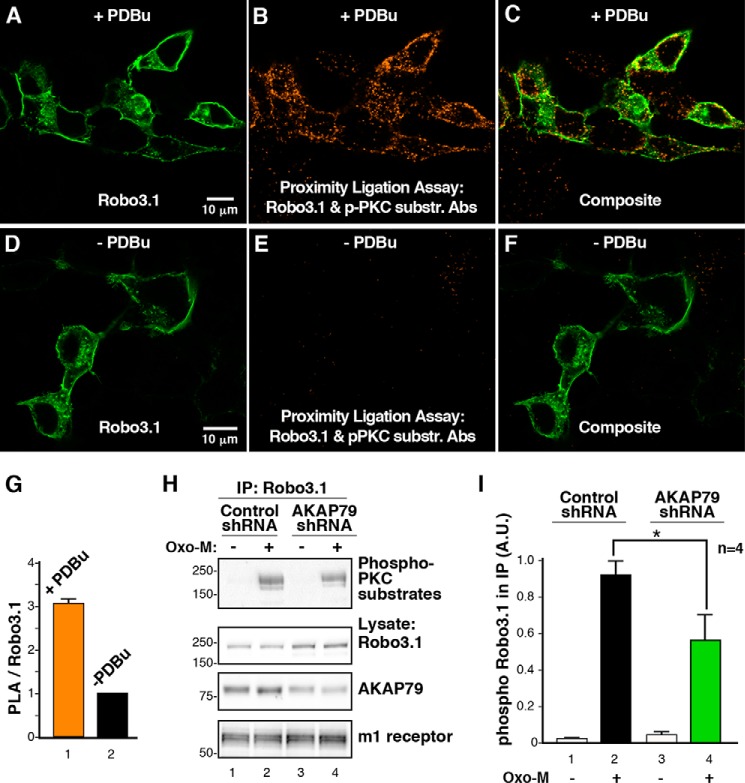
Cellular analysis of Robo3.1 phosphorylation was further examined by PLA, a sensitive method that detects protein-protein interactions or local posttranslational modification of proteins in situ (49). The conceptual basis of this approach depends on the dual proximal binding of distinct antibody probes that are conjugated to complementary oligonucleotides. When the probes are within range of each other, ligation occurs to generate an amplifiable DNA circle. The resulting incorporation of fluorescent nucleotides following multiple rounds of amplification serves as a visible marker for protein-protein interactions or covalent modification of proteins that occur within a radius of 40–60 nm. Cells were transfected with Robo3.1 and stimulated with PDBu to activate PKCs. Proximity ligation allowed us to use the phospho-PKC substrate antibody to selectively monitor PKC activity in the vicinity of Robo3.1. Accordingly, we were able to show that Robo3.1 was efficiently phosphorylated by PKC (Fig. 6, A–C and G) as compared with unstimulated controls (Fig. 6, D–F and G). Quantitation of this result is shown as the ratio of PLA signal to the single stain for Robo3.1 (Fig. 6G).
FIGURE 6.
AKAP79 regulates the phosphorylation of Robo3.1 by PKC. A–F, HEK293 cells were transfected with Robo3 and treated with PDBu (A–C) or vehicle (DMSO; D–F) for 10 min. Cells were incubated with a Robo3 antibody and the phospho-PKC substrate antibody, and a PLA reaction was carried out between these two antibodies (B, C, E, and F; orange). Cells were subsequently stained using a V5 antibody to recognize Robo3.1 (A, C, D, and F; green). G, analysis of PLA signal normalized to Robo3 expression (means ± S.E. (error bars), n = 3, >100 cells, p ≤ 0.05, one-sample Student's t test). H, Robo3 and the m1 muscarinic receptor were coexpressed in HEK293 cells with control or AKAP79 shRNA. Cells were treated with vehicle (H2O) or 10 μm Oxo-M for 2 min. V5 immunoprecipitations (IP) of cell lysates were immunoblotted with the PKC substrate antibody (top). Robo3.1 expression (top middle), AKAP79 knockdown (bottom middle), and m1 receptor expression (bottom) were confirmed by immunoblotting. I, quantification of phospho-Robo3 signal present in immunoprecipitations normalized to Robo3 expression (means ± S.E., n = 4, p ≤ 0.05, unpaired Student's t test). A.U., arbitrary units.
Finally, gene silencing was utilized to discern a role for the anchoring protein in the phosphorylation of Robo3.1 (Fig. 6, H and I). A well characterized shRNA to AKAP79 was used to deplete the anchoring protein in HEK293 cells (24) (Fig. 6H, bottom middle panel, lanes 3 and 4). In order to accentuate the physiological relevance of this anchored phosphorylation event, we used the m1 muscarinic receptor agonist oxotremorine-M (Oxo-M) as an activator of anchored PKC (35). Cells expressing both Robo3 and the m1 receptor and transfected with either control or AKAP79 shRNA were incubated for 72 h. Following treatment with vehicle or Oxo-M, Robo3 immune complexes were isolated. Phospho-Robo3.1 levels were evaluated by immunoblotting (Fig. 6H, top panel, lanes 2 and 4). Analysis of data from four independent experiments showed that depletion of AKAP79 significantly reduced PKC phosphorylation of Robo3.1 as assessed by densitometry (Fig. 6I, columns 2 and 4). Overall, the data in Fig. 6 demonstrate that AKAP79 facilitates PKC phosphorylation of Robo3.1 in response to a physiological agonist.
Discussion
Protein kinase A-anchoring proteins modulate membrane proximal signaling events, principally by tethering protein kinases and phosphatases in the vicinity of G-protein-coupled receptors, ion channels, and adenylyl cyclases (7, 25, 51–54). Taken together, the data in Fig. 1 imply that an additional role for AKAPs may be to provide a molecular interface between signaling enzymes and the axon guidance machinery. Axon guidance receptors, including the Robo family, are transmembrane proteins that are enriched in the growth cones of migrating axons (55). However, it has also been shown that Robo family members are expressed in the forebrain, hippocampus, and other regions of the developing brain (34, 45).
Certain pools of guidance receptors can associate with lipid rafts, where they respond to chemotactic signals that instruct directional movement of axons (56). Interestingly, these guidance cues can either attract or repel axons (57). For example, binding of the Slit ligand is a repulsive cue that causes the growth cone to collapse and reverses the direction of the migrating axon (34). Robo receptors are also expressed in dendrites, where Slit binding has been shown to regulate dendritic branching (58). Likewise, the ephrin and plexin classes of guidance receptors have been implicated in growth cone repulsion and the propagation of dendritic arborization (59). It is also worthy of note that second messenger signaling events impact neuronal migration. Classic studies have shown that altering the levels of cyclic nucleotides in the growth cone can interfere with axon guidance signals (60). For example, increasing the ratio of cAMP/cGMP can switch the action of guidance molecules from neuronal repulsion to attraction (61). Added to this, it has been demonstrated that elevation of intracellular cAMP and the subsequent activation of PKA can suppress Robo signaling (62).
Kinase anchoring contributes to various modes of directional neurite outgrowth control (10). For example, the AKAP WAVE1 is expressed at the leading edge of growth cones, where it controls actin dynamics to facilitate axonal protrusions (63–66). In addition, the Drosophila anchoring protein Nervy coordinates signaling at the plexin family of semaphorin receptors (67). Nervy is purported to cluster PKA at the plexin receptor, thereby facilitating the termination of semaphorin signaling through the phosphorylation of these receptors (67). Taken together, these studies provide a precedent for the notion that local signaling mechanisms govern dynamic aspects of neuronal guidance and development. The data presented in this report provide the first evidence that AKAP-associated enzymes may regulate Robo receptors. We propose that the interaction of AKAP79/150 with Robo receptors may position PKC for a role in the fine tuning of axonal and dendritic guidance.
The accumulated data presented in Fig. 2 demonstrate that the cytoplasmic tail of Robo receptors primarily interfaces with the N-terminal region of AKAP79/150. Previous studies have shown that this segment of the anchoring protein contains three polybasic regions that participate in membrane targeting through interaction with negatively charged phospholipids (29). More recent studies have implicated the palmitoylation of conserved cysteines in the proximal and distal polybasic regions as a means to guide AKAP79/150 to lipid rafts (21, 68). These latter findings raise the intriguing possibility that AKAP-Robo2 assemblies might be sequestered in specialized submembrane compartments. This may be particularly relevant in regions of the hippocampus where AKAP79/150 appears to co-distribute with Robo2 and Robo3, particularly at the growth cones of developing neurons. Although the immunofluorescence images presented in Figs. 1 and 4 are consistent with this notion, superresolution approaches will be necessary to locate precisely where AKAP-Robo signaling units are sequestered in neuronal membranes. In addition, it remains possible that ancillary protein-protein or protein-lipid interactions may further secure AKAP-Robo macromolecular assemblies at synaptic locations.
We also demonstrate that AKAP79-bound signaling enzymes, such as PKA, PKC, and PP2B, can remain associated with the anchoring protein when in complex with Robo receptors. The data presented in Fig. 2F suggest that kinase and phosphatase anchoring may coordinate the reversible phosphorylation of Robo receptors in a manner analogous to the GluA1 subunit of the AMPA receptor (19). Thus, several distinct targeting mechanisms may act synergistically with the AKAP-Robo interface to stably sequester AKAP-associated signaling enzymes with these transmembrane receptors.
Protein kinase C requires basic residues in the vicinity of the phosphorylated amino acid, but its specificity is less well defined than that of other kinases, such as PKA or Akt (69). Taken together, the results from Fig. 5 suggest that AKAP79 directs PKC proximal to Robo3.1 to enhance phosphorylation of its cytoplasmic tail. Analysis of peptide spot arrays when combined with site-directed mutagenesis has identified Ser-1330 as a primary site of PKC phosphorylation. This region is rich in basic and bulky hydrophobic residues, hallmarks of a consensus site for PKCs (69).
Data from Fig. 6 confirm and extend these observations by demonstrating that AKAP79 facilitates the phosphorylation of Robo3.1 by PKC. Perhaps the strongest support for this latter statement is provided by the gene silencing experiments presented in Fig. 6, H and I. Depletion of the anchoring protein suppresses phosphorylation of Robo3.1 in response to muscarinic agonists. Although little is known about Robo3 receptor subtypes, there is reason to believe that Robo3.1 and Robo3.2 have divergent functions. In the developing spinal cord, Robo3.1 antagonizes repulsive axonal migration events that proceed through Robo1 and Robo2 receptors (70). This ability to repress canonical Slit/Robo signals is not shared by the Robo3.2 isoform. Although the mechanism that accounts for the opposing functions of Robo3.1 and Robo3.2 is not clear, our data raise the possibility that PKC phosphorylation at Ser-1330 of Robo3.1 may play a role in this process.
In conclusion, our data suggest that PKC anchoring through AKAP79 may confer an added element of spatiotemporal control to Slit/Robo signaling. AKAP79 can be thought of as an adapter molecule that brings enzyme and substrate together. Not only does this mechanism provide exquisite local modulation of Robo3.1 phosphorylation, but the immediate proximity of each component ensures that signals have the capacity to be instantaneously transduced. Furthermore, AKAP79/150 has the ability to bind all isoforms of PKC, which are activated by different combinations of calcium and phospholipid (8). This anchoring protein can also self-associate and complex with ion channels (17, 71). Finally, Robo receptor subtypes homo- and heterodimerize (50). These factors suggest that incorporation of the PKC-AKAP-Robo3.1 subcomplexes into higher order macromolecular assemblies may expand the potential for signal integration and enzyme cross-talk between second messenger cascades and the neuronal guidance machinery.
Acknowledgments
We thank Donelson Smith and other members of the Scott laboratory for helpful discussion and suggestions. We also thank Jennifer Sanderson for providing cultured mouse hippocampal neurons.
This work was supported, in whole or in part, by National Institutes of Health Grants DK105542 and DK54441 (to J. D. S.), MH094536 (to L. S. Z.), and NS40701 (to M. L. D.).
- AKAP
- A-kinase anchoring protein
- PP2B
- protein phosphatase 2B
- RII
- PKA regulatory subunit type II
- DIV
- day(s) in vitro
- MAGUK
- membrane-associated guanylate kinase
- PDBu
- phorbol 12,13-dibutyrate
- PLA
- proximity ligation assay
- Oxo-M
- oxotremorine-M.
References
- 1. Song H., Poo M. (2001) The cell biology of neuronal navigation. Nat. Cell Biol. 3, E81–E88 [DOI] [PubMed] [Google Scholar]
- 2. Jan Y. N., Jan L. Y. (2010) Branching out: mechanisms of dendritic arborization. Nat. Rev. Neurosci. 11, 316–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Scott J. D., Pawson T. (2009) Cell signaling in space and time: where proteins come together and when they're apart. Science 326, 1220–1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dessauer C. W. (2009) Adenylyl cyclase–A-kinase anchoring protein complexes: the next dimension in cAMP signaling. Mol. Pharmacol. 76, 935–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lefkowitz R. J. (2004) Historical review: a brief history and personal retrospective of seven-transmembrane receptors. Trends Pharmacol. Sci. 25, 413–422 [DOI] [PubMed] [Google Scholar]
- 6. Scott J. D., Dessauer C. W., Taskén K. (2013) Creating order from chaos: cellular regulation by kinase anchoring. Annu. Rev. Pharmacol. Toxicol. 53, 187–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bauman A. L., Soughayer J., Nguyen B. T., Willoughby D., Carnegie G. K., Wong W., Hoshi N., Langeberg L. K., Cooper D. M., Dessauer C. W., Scott J. D. (2006) Dynamic regulation of cAMP synthesis through anchored PKA-adenylyl cyclase V/VI complexes. Mol. Cell 23, 925–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Faux M. C., Rollins E. N., Edwards A. S., Langeberg L. K., Newton A. C., Scott J. D. (1999) Mechanism of A-kinase-anchoring protein 79 (AKAP79) and protein kinase C interaction. Biochem. J. 343, 443–452 [PMC free article] [PubMed] [Google Scholar]
- 9. Zalatan J. G., Coyle S. M., Rajan S., Sidhu S. S., Lim W. A. (2012) Conformational control of the Ste5 scaffold protein insulates against MAP kinase misactivation. Science 337, 1218–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Welch E. J., Jones B. W., Scott J. D. (2010) Networking with AKAPs: context-dependent regulation of anchored enzymes. Mol. Interv. 10, 86–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Smith F. D., Reichow S. L., Esseltine J. L., Shi D., Langeberg L. K., Scott J. D., Gonen T. (2013) Intrinsic disorder within an AKAP-protein kinase A complex guides local substrate phosphorylation. eLife 2, e01319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Taskén K., Aandahl E. M. (2004) Localized effects of cAMP mediated by distinct routes of protein kinase A. Physiol. Rev. 84, 137–167 [DOI] [PubMed] [Google Scholar]
- 13. Dodge-Kafka K. L., Langeberg L., Scott J. D. (2006) Compartmentation of cyclic nucleotide signaling in the heart: the role of A-kinase anchoring proteins. Circ. Res. 98, 993–1001 [DOI] [PubMed] [Google Scholar]
- 14. Klauck T. M., Faux M. C., Labudda K., Langeberg L. K., Jaken S., Scott J. D. (1996) Coordination of three signaling enzymes by AKAP79, a mammalian scaffold protein. Science 271, 1589–1592 [DOI] [PubMed] [Google Scholar]
- 15. Taskén K. A., Collas P., Kemmner W. A., Witczak O., Conti M., Taskén K. (2001) Phosphodiesterase 4D and protein kinase a type II constitute a signaling unit in the centrosomal area. J. Biol. Chem. 276, 21999–22002 [DOI] [PubMed] [Google Scholar]
- 16. Logue J. S., Whiting J. L., Tunquist B., Langeberg L. K., Scott J. D. (2011) Anchored protein kinase A recruitment of active Rac GTPase. J. Biol. Chem. 286, 22113–22121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gold M. G., Stengel F., Nygren P. J., Weisbrod C. R., Bruce J. E., Robinson C. V., Barford D., Scott J. D. (2011) Architecture and dynamics of an A-kinase anchoring protein 79 (AKAP79) signaling complex. Proc. Natl. Acad. Sci. U.S.A. 108, 6426–6431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Colledge M., Dean R. A., Scott G. K., Langeberg L. K., Huganir R. L., Scott J. D. (2000) Targeting of PKA to glutamate receptors through a MAGUK-AKAP complex. Neuron 27, 107–119 [DOI] [PubMed] [Google Scholar]
- 19. Tavalin S. J., Colledge M., Hell J. W., Langeberg L. K., Huganir R. L., Scott J. D. (2002) Regulation of GluR1 by the A-kinase anchoring protein 79 (AKAP79) signaling complex shares properties with long-term depression. J. Neurosci. 22, 3044–3051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Robertson H. R., Gibson E. S., Benke T. A., Dell'Acqua M. L. (2009) Regulation of postsynaptic structure and function by an A-kinase anchoring protein-membrane-associated guanylate kinase scaffolding complex. J. Neurosci. 29, 7929–7943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Keith D. J., Sanderson J. L., Gibson E. S., Woolfrey K. M., Robertson H. R., Olszewski K., Kang R., El-Husseini A., Dell'acqua M. L. (2012) Palmitoylation of A-kinase anchoring protein 79/150 regulates dendritic endosomal targeting and synaptic plasticity mechanisms. J. Neurosci. 32, 7119–7136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tunquist B. J., Hoshi N., Guire E. S., Zhang F., Mullendorff K., Langeberg L. K., Raber J., Scott J. D. (2008) Loss of AKAP150 perturbs distinct neuronal processes in mice. Proc. Natl. Acad. Sci. U.S.A. 105, 12557–12562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Weisenhaus M., Allen M. L., Yang L., Lu Y., Nichols C. B., Su T., Hell J. W., McKnight G. S. (2010) Mutations in AKAP5 disrupt dendritic signaling complexes and lead to electrophysiological and behavioral phenotypes in mice. PLoS One 5, e10325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hoshi N., Langeberg L. K., Scott J. D. (2005) Distinct enzyme combinations in AKAP signalling complexes permit functional diversity. Nat. Cell Biol. 7, 1066–1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sanderson J. L., Gorski J. A., Gibson E. S., Lam P., Freund R. K., Chick W. S., Dell'Acqua M. L. (2012) AKAP150-anchored calcineurin regulates synaptic plasticity by limiting synaptic incorporation of Ca2+-permeable AMPA receptors. J. Neurosci. 32, 15036–15052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lester L. B., Coghlan V. M., Nauert B., Scott J. D. (1996) Cloning and characterization of a novel A-kinase anchoring protein: AKAP220, association with testicular peroxisomes. J. Biol. Chem. 271, 9460–9465 [DOI] [PubMed] [Google Scholar]
- 27. Carr D. W., Hausken Z. E., Fraser I. D., Stofko-Hahn R. E., Scott J. D. (1992) Association of the type II cAMP-dependent protein kinase with a human thyroid RII-anchoring protein: cloning and characterization of the RII-binding domain. J. Biol. Chem. 267, 13376–13382 [PubMed] [Google Scholar]
- 28. Carlson C. R., Lygren B., Berge T., Hoshi N., Wong W., Taskén K., Scott J. D. (2006) Delineation of type I protein kinase A-selective signaling events using an RI anchoring disruptor. J. Biol. Chem. 281, 21535–21545 [DOI] [PubMed] [Google Scholar]
- 29. Dell'Acqua M. L., Faux M. C., Thorburn J., Thorburn A., Scott J. D. (1998) Membrane-targeting sequences on AKAP79 bind phosphatidylinositol-4,5- bisphosphate. EMBO J. 17, 2246–2260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hoshi N., Langeberg L. K., Gould C. M., Newton A. C., Scott J. D. (2010) Interaction with AKAP79 modifies the cellular pharmacology of PKC. Mol. Cell 37, 541–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Snyder E. M., Colledge M., Crozier R. A., Chen W. S., Scott J. D., Bear M. F. (2005) Role for A kinase-anchoring proteins (AKAPS) in glutamate receptor trafficking and long term synaptic depression. J. Biol. Chem. 280, 16962–16968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kidd T., Brose K., Mitchell K. J., Fetter R. D., Tessier-Lavigne M., Goodman C. S., Tear G. (1998) Roundabout controls axon crossing of the CNS midline and defines a novel subfamily of evolutionarily conserved guidance receptors. Cell 92, 205–215 [DOI] [PubMed] [Google Scholar]
- 33. Kidd T., Bland K. S., Goodman C. S. (1999) Slit is the midline repellent for the Robo receptor in Drosophila. Cell 96, 785–794 [DOI] [PubMed] [Google Scholar]
- 34. Nguyen Ba-Charvet K. T., Brose K., Marillat V., Kidd T., Goodman C. S., Tessier-Lavigne M., Sotelo C., Chédotal A. (1999) Slit2-mediated chemorepulsion and collapse of developing forebrain axons. Neuron 22, 463–473 [DOI] [PubMed] [Google Scholar]
- 35. Hoshi N., Zhang J. S., Omaki M., Takeuchi T., Yokoyama S., Wanaverbecq N., Langeberg L. K., Yoneda Y., Scott J. D., Brown D. A., Higashida H. (2003) AKAP150 signaling complex promotes suppression of the M-current by muscarinic agonists. Nat. Neurosci. 6, 564–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Oliveria S. F., Dell'Acqua M. L., Sather W. A. (2007) AKAP79/150 anchoring of calcineurin controls neuronal L-type Ca2+ channel activity and nuclear signaling. Neuron 55, 261–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cheng E. P., Yuan C., Navedo M. F., Dixon R. E., Nieves-Cintrón M., Scott J. D., Santana L. F. (2011) Restoration of normal L-type Ca2+ channel function during Timothy syndrome by ablation of an anchoring protein. Circ. Res. 109, 255–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nystoriak M. A., Nieves-Cintrón M., Nygren P. J., Hinke S. A., Nichols C. B., Chen C. Y., Puglisi J. L., Izu L. T., Bers D. M., Dell'acqua M. L., Scott J. D., Santana L. F., Navedo M. F. (2014) AKAP150 contributes to enhanced vascular tone by facilitating large-conductance Ca2+-activated K+ channel remodeling in hyperglycemia and diabetes mellitus. Circ. Res. 114, 607–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tavalin S. J. (2008) AKAP79 selectively enhances protein kinase C regulation of GluR1 at a Ca2+-calmodulin-dependent protein kinase II/protein kinase C site. J. Biol. Chem. 283, 11445–11452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ypsilanti A. R., Zagar Y., Chédotal A. (2010) Moving away from the midline: new developments for Slit and Robo. Development 137, 1939–1952 [DOI] [PubMed] [Google Scholar]
- 41. Jaworski A., Long H., Tessier-Lavigne M. (2010) Collaborative and specialized functions of Robo1 and Robo2 in spinal commissural axon guidance. J. Neurosci. 30, 9445–9453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sabatier C., Plump A. S., Le Ma, Brose K., Tamada A., Murakami F., Lee E. Y., Tessier-Lavigne M. (2004) The divergent Robo family protein rig-1/Robo3 is a negative regulator of slit responsiveness required for midline crossing by commissural axons. Cell 117, 157–169 [DOI] [PubMed] [Google Scholar]
- 43. Long H., Sabatier C., Ma L., Plump A., Yuan W., Ornitz D. M., Tamada A., Murakami F., Goodman C. S., Tessier-Lavigne M. (2004) Conserved roles for Slit and Robo proteins in midline commissural axon guidance. Neuron 42, 213–223 [DOI] [PubMed] [Google Scholar]
- 44. Kenwrick S., Watkins A., De Angelis E. (2000) Neural cell recognition molecule L1: relating biological complexity to human disease mutations. Hum. Mol. Genet. 9, 879–886 [DOI] [PubMed] [Google Scholar]
- 45. Marillat V., Cases O., Nguyen-Ba-Charvet K. T., Tessier-Lavigne M., Sotelo C., Chédotal A. (2002) Spatiotemporal expression patterns of slit and robo genes in the rat brain. J. Comp. Neurol. 442, 130–155 [DOI] [PubMed] [Google Scholar]
- 46. Horn K. E., Glasgow S. D., Gobert D., Bull S. J., Luk T., Girgis J., Tremblay M. E., McEachern D., Bouchard J. F., Haber M., Hamel E., Krimpenfort P., Murai K. K., Berns A., Doucet G., Chapman C. A., Ruthazer E. S., Kennedy T. E. (2013) DCC expression by neurons regulates synaptic plasticity in the adult brain. Cell Rep. 3, 173–185 [DOI] [PubMed] [Google Scholar]
- 47. Dell'Acqua M. L., Smith K. E., Gorski J. A., Horne E. A., Gibson E. S., Gomez L. L. (2006) Regulation of neuronal PKA signaling through AKAP targeting dynamics. Eur. J. Cell Biol. 85, 627–633 [DOI] [PubMed] [Google Scholar]
- 48. Guan C. B., Xu H. T., Jin M., Yuan X. B., Poo M. M. (2007) Long-range Ca2+ signaling from growth cone to soma mediates reversal of neuronal migration induced by slit-2. Cell 129, 385–395 [DOI] [PubMed] [Google Scholar]
- 49. Ke R., Nong R. Y., Fredriksson S., Landegren U., Nilsson M. (2013) Improving precision of proximity ligation assay by amplified single molecule detection. PLoS One 8, e69813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Evans T. A., Bashaw G. J. (2010) Functional diversity of Robo receptor immunoglobulin domains promotes distinct axon guidance decisions. Curr. Biol. 20, 567–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Fraser I. D., Cong M., Kim J., Rollins E. N., Daaka Y., Lefkowitz R. J., Scott J. D. (2000) Assembly of an A kinase-anchoring protein-β(2)-adrenergic receptor complex facilitates receptor phosphorylation and signaling. Curr. Biol. 10, 409–412 [DOI] [PubMed] [Google Scholar]
- 52. Gao T., Yatani A., Dell'Acqua M. L., Sako H., Green S. A., Dascal N., Scott J. D., Hosey M. M. (1997) cAMP-dependent regulation of cardiac L-type Ca2+ channels requires membrane targeting of PKA and phosphorylation of channel subunits. Neuron 19, 185–196 [DOI] [PubMed] [Google Scholar]
- 53. Efendiev R., Samelson B. K., Nguyen B. T., Phatarpekar P. V., Baameur F., Scott J. D., Dessauer C. W. (2010) AKAP79 interacts with multiple adenylyl cyclase (AC) isoforms and scaffolds AC5 and -6 to α-amino-3-hydroxyl-5-methyl-4-isoxazole-propionate (AMPA) receptors. J. Biol. Chem. 285, 14450–14458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hinke S. A., Navedo M. F., Ulman A., Whiting J. L., Nygren P. J., Tian G., Jimenez-Caliani A. J., Langeberg L. K., Cirulli V., Tengholm A., Dell'Acqua M. L., Santana L. F., Scott J. D. (2012) Anchored phosphatases modulate glucose homeostasis. EMBO J. 31, 3991–4004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Seeger M., Tear G., Ferres-Marco D., Goodman C. S. (1993) Mutations affecting growth cone guidance in Drosophila: genes necessary for guidance toward or away from the midline. Neuron 10, 409–426 [DOI] [PubMed] [Google Scholar]
- 56. Guirland C., Suzuki S., Kojima M., Lu B., Zheng J. Q. (2004) Lipid rafts mediate chemotropic guidance of nerve growth cones. Neuron 42, 51–62 [DOI] [PubMed] [Google Scholar]
- 57. Yu T. W., Bargmann C. I. (2001) Dynamic regulation of axon guidance. Nat. Neurosci. 4, 1169–1176 [DOI] [PubMed] [Google Scholar]
- 58. Whitford K. L., Marillat V., Stein E., Goodman C. S., Tessier-Lavigne M., Chédotal A., Ghosh A. (2002) Regulation of cortical dendrite development by Slit-Robo interactions. Neuron 33, 47–61 [DOI] [PubMed] [Google Scholar]
- 59. Fenstermaker V., Chen Y., Ghosh A., Yuste R. (2004) Regulation of dendritic length and branching by semaphorin 3A. J. Neurobiol. 58, 403–412 [DOI] [PubMed] [Google Scholar]
- 60. Song H. J., Ming G. L., Poo M. M. (1997) cAMP-induced switching in turning direction of nerve growth cones. Nature 388, 275–279 [DOI] [PubMed] [Google Scholar]
- 61. Nishiyama M., Hoshino A., Tsai L., Henley J. R., Goshima Y., Tessier-Lavigne M., Poo M. M., Hong K. (2003) Cyclic AMP/GMP-dependent modulation of Ca2+ channels sets the polarity of nerve growth-cone turning. Nature 423, 990–995 [DOI] [PubMed] [Google Scholar]
- 62. Chalasani S. H., Sabelko K. A., Sunshine M. J., Littman D. R., Raper J. A. (2003) A chemokine, SDF-1, reduces the effectiveness of multiple axonal repellents and is required for normal axon pathfinding. J. Neurosci. 23, 1360–1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Soderling S. H., Guire E. S., Kaech S., White J., Zhang F., Schutz K., Langeberg L. K., Banker G., Raber J., Scott J. D. (2007) A WAVE-1 and WRP signaling complex regulates spine density, synaptic plasticity, and memory. J. Neurosci. 27, 355–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Carlson B. R., Lloyd K. E., Kruszewski A., Kim I. H., Rodriguiz R. M., Heindel C., Faytell M., Dudek S. M., Wetsel W. C., Soderling S. H. (2011) WRP/srGAP3 facilitates the initiation of spine development by an inverse F-BAR domain, and its loss impairs long-term memory. J. Neurosci. 31, 2447–2460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Westphal R. S., Soderling S. H., Alto N. M., Langeberg L. K., Scott J. D. (2000) Scar/WAVE-1, a Wiskott-Aldrich syndrome protein, assembles an actin- associated multi-kinase scaffold. EMBO J. 19, 4589–4600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Soderling S. H., Langeberg L. K., Soderling J. A., Davee S. M., Simerly R., Raber J., Scott J. D. (2003) Loss of WAVE-1 causes sensorimotor retardation and reduced learning and memory in mice. Proc. Natl. Acad. Sci. U.S.A. 100, 1723–1728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Terman J. R., Kolodkin A. L. (2004) Nervy links protein kinase a to plexin-mediated semaphorin repulsion. Science 303, 1204–1207 [DOI] [PubMed] [Google Scholar]
- 68. Willoughby D., Masada N., Wachten S., Pagano M., Halls M. L., Everett K. L., Ciruela A., Cooper D. M. (2010) AKAP79/150 interacts with AC8 and regulates Ca2+-dependent cAMP synthesis in pancreatic and neuronal systems. J. Biol. Chem. 285, 20328–20342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Newton A. C. (2003) Regulation of the ABC kinases by phosphorylation: protein kinase C as a paradigm. Biochem. J. 370, 361–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Chen Z., Gore B. B., Long H., Ma L., Tessier-Lavigne M. (2008) Alternative splicing of the Robo3 axon guidance receptor governs the midline switch from attraction to repulsion. Neuron 58, 325–332 [DOI] [PubMed] [Google Scholar]
- 71. Fraser I. D., Scott J. D. (1999) Modulation of ion channels: a “current” view of AKAPs. Neuron 23, 423–426 [DOI] [PubMed] [Google Scholar]