Background: Iron-sulfur cluster biogenesis requires the coordinated delivery of iron and sulfur.
Results: Deletion of IscA/SufA or depletion of intracellular iron produces a red-colored cysteine desulfurase IscS in Escherichia coli cells.
Conclusion: Deficiency of accessible iron results in the accumulation of red IscS in cells.
Significance: IscA/SufA may work in concert with IscS in delivering iron and sulfur for iron-sulfur cluster biogenesis.
Keywords: enzyme mechanism, iron metabolism, iron-sulfur protein, metalloenzyme, pyridoxal phosphate, sulfur
Abstract
In Escherichia coli, sulfur in iron-sulfur clusters is primarily derived from l-cysteine via the cysteine desulfurase IscS. However, the iron donor for iron-sulfur cluster assembly remains elusive. Previous studies have shown that, among the iron-sulfur cluster assembly proteins in E. coli, IscA has a unique and strong iron-binding activity and that the iron-bound IscA can efficiently provide iron for iron-sulfur cluster assembly in proteins in vitro, indicating that IscA may act as an iron chaperone for iron-sulfur cluster biogenesis. Here we report that deletion of IscA and its paralog SufA in E. coli cells results in the accumulation of a red-colored cysteine desulfurase IscS under aerobic growth conditions. Depletion of intracellular iron using a membrane-permeable iron chelator, 2,2′-dipyridyl, also leads to the accumulation of red IscS in wild-type E. coli cells, suggesting that the deletion of IscA/SufA may be emulated by depletion of intracellular iron. Purified red IscS has an absorption peak at 528 nm in addition to the peak at 395 nm of pyridoxal 5′-phosphate. When red IscS is oxidized by hydrogen peroxide, the peak at 528 nm is shifted to 510 nm, which is similar to that of alanine-quinonoid intermediate in cysteine desulfurases. Indeed, red IscS can also be produced in vitro by incubating wild-type IscS with excess l-alanine and sulfide. The results led us to propose that deletion of IscA/SufA may disrupt the iron delivery for iron-sulfur cluster biogenesis, therefore impeding sulfur delivery by IscS, and result in the accumulation of red IscS in E. coli cells.
Introduction
In the past decade, a group of highly conserved proteins has been identified as essential for iron-sulfur cluster biogenesis in bacteria (1, 2) and eukaryotes (3). Among these identified proteins, cysteine desulfurases (NifS (4), IscS (5, 6), and SufS (7)) catalyze the desulfurization of l-cysteine and provide sulfur for iron-sulfur cluster assembly in the scaffold proteins IscU (8–10) or the SufBCD complex (11, 12). The scaffold proteins then transfer the assembled clusters to target proteins. Two heat shock cognate proteins, HscB and HscA, interact with the scaffold protein IscU and regulate the iron-sulfur cluster transfer from IscU to target proteins (10, 13, 14). Specific glutaredoxins are also involved in storing and transporting iron-sulfur clusters from scaffold proteins to target proteins (15–17). Nevertheless, the iron donor for iron-sulfur cluster biogenesis remains elusive. Frataxin, a mitochondrial protein associated with the human neurodegenerative disease Friedreich ataxia (18), has been proposed previously as a putative iron chaperone for iron-sulfur cluster biogenesis (19, 20). Frataxin is highly conserved from bacteria to humans and interacts with iron-sulfur protein aconitase (21), mitochondrial electron transfer components (22), and the iron-sulfur cluster assembly proteins IscS (23–27) and IscU (28). However, frataxin has a weak iron-binding activity under physiological conditions (29–31), and deletion of frataxin has little or no effect on iron-sulfur proteins in Saccharomyces cerevisiae (32), Salmonella enterica (33), and Escherichia coli (34). Only in iron-rich medium does the deletion of frataxin have a mild effect on iron-sulfur proteins in E. coli cells (26). It has therefore been postulated that frataxin may act as a gatekeeper to regulate iron-sulfur cluster biogenesis by activating cysteine desulfurase (27) or directing the sulfur flux from cysteine desulfurase (23, 25).
Among the iron-sulfur cluster assembly proteins encoded by the gene cluster iscSUA-HscBA-fdx-iscX in E. coli (1), IscX has also been proposed as a possible iron donor for iron-sulfur cluster biogenesis (35). However, like frataxin, IscX has a weak iron-binding activity (36), and deletion of IscX has very little effect on iron-sulfur proteins in E. coli cells (26), indicating that IscX may have functions other than directly providing iron for iron-sulfur cluster assembly under physiological conditions. In contrast, IscA, a proposed alternative scaffold protein (37–39), has a unique and strong iron-binding activity (40–44) with an iron association constant of ∼3.0 × 1019 m−1 (45, 46). The iron center in IscA can be readily mobilized by l-cysteine (43, 44, 47) and transferred for iron-sulfur cluster assembly in proteins in vitro (43, 48). These results led us to postulate that IscA may recruit intracellular iron and deliver iron for iron-sulfur cluster biogenesis. It has also been reported that depletion of IscA in Azotobacter vinelandii results in a null-growth phenotype in modified Burks minimal medium under elevated oxygen conditions (49). In E. coli cells, deletion of IscA and its paralog SufA has the same null-growth phenotype in M9 minimal medium under aerobic conditions (50, 51). In S. cerevisiae, depletion of IscA homologs leads to accumulation of iron in mitochondria and dependence on lysine and glutamate for cell growth (52, 53). In cultured human cells, depletion of IscA1 significantly decreases iron-sulfur cluster assembly activity in mitochondria and in the cytosol (54). Recent studies also revealed that IscA and its homologs have an essential role in [4Fe-4S] cluster assembly in E. coli (55), S. cerevisiae (42), and human cells (56) under aerobic conditions.
If IscA and its paralog SufA act as iron chaperones (48), deletion of IscA and SufA would disrupt iron delivery for iron-sulfur cluster biogenesis, therefore impeding sulfur delivery by the cysteine desulfurase IscS, a major sulfur donor in E. coli cells (6). In this study, we find that deletion of IscA and SufA indeed results in the accumulation of a red-colored IscS in E. coli cells under aerobic growth conditions. Red IscS also accumulates in wild-type E. coli cells in which intracellular iron is depleted by using a membrane-permeable iron chelator 2,2′-dipyridyl, suggesting that deletion of IscA/SufA can be emulated by depleting intracellular iron in E. coli cells. Purified red IscS has a distinct absorption peak at 528 nm in addition to the absorption peak at 395 nm of pyridoxal 5′-phosphate (5, 6). When purified red IscS is oxidized by H2O2, the absorption peak at 528 nm is shifted to 510 nm, which is reminiscent of an alanine-quinonoid intermediate in cysteine desulfurases from other organisms (4, 57). The results suggest that deletion of IscA/SufA or depletion of intercellular iron in E. coli cells leads to accumulation of red IscS, which likely contains a highly conjugated quinonoid intermediate, and that the proposed iron chaperones IscA/SufA and cysteine desulfurase IscS may work in concert in delivering iron and sulfur for iron-sulfur cluster biogenesis.
Experimental Procedures
Protein Purification
Recombinant E. coli IscS was expressed in the E. coli iscA/sufA mutant or its parental wild-type strain MC4100 cells. E. coli iscA/sufA mutant was constructed as described previously (50). The E. coli gene encoding IscS was cloned to an expression plasmid, pBAD (Life Technologies). The E. coli cells hosting the expression plasmid were grown in LB (Luria-Bertani) medium to A600 nm of 0.6 under aerobic conditions. Arabinose (at a final concentration of 0.02%) was then added to the cell cultures to induce the expression of recombinant IscS in the cells. IscS was purified as described in Ref. 48. The purity of purified IscS was over 95%, judging from the SDS-polyacrylamide gel electrophoresis staining with Coomassie Brilliant Blue. The protein concentration of IscS was measured from the absorption peak at 280 nm using an extinction coefficient of 39.8 mm−1cm−1. The UV-visible absorption spectra were recorded using the Beckman DU640 UV-visible spectrometer equipped with a temperature controller.
Site-directed Mutagenesis of E. coli IscS
Two pairs of primers (IscS-K206A-1, 5′-CTTTCTCCGGTCACGCAATCTATGGCCCG-3′; IscS-K206A-2, 5′-CGGGCCATAGATTGCGTGACCGGAGAAAG-3′; IscS-C328S-1, 5′-CAGTTTCTTCAGGTTCCGCCTCTACGTCAGCAAGCC-3′; IscS-C328S-2, 5′-GAGGCGGAACCTGAAGAAACTGCGAGGTCTTTCAGC-3′) were synthesized for construction of IscS mutants IscS-K206A and IscS-C328S, respectively. The site-directed mutagenesis of IscS was carried out using the QuikChange kit (Agilent Technologies), and the mutations were confirmed by direct sequencing (Eurofins Co.).
Treatment of 2,2′-Dipyridyl
Different amounts of 2,2′-dipyridyl were added to LB medium before the overnight E. coli culture was inoculated. Cells were grown at 37 °C under aerobic conditions. The cell growth was measured after cells were washed once with fresh LB medium to remove 2,2′-dipyridyl.
Cysteine Desulfurase Activity Assay
Cysteine desulfurase activity of purified E. coli IscS was measured using the sulfide detection method by Siegel (58). Briefly, purified cysteine desulfurase (1 μm) was incubated with l-cysteine (1 mm) in buffer containing Tris (20 mm (pH 8.0)), NaCl (200 mm), and dithiothreitol (2 mm) at 37 °C for 10 min. Reactions were terminated by addition of N,N-dimethyl-p-phenylene-diamine sulfate (20 mm) (in 7.2 N HCl) and FeCl3 (30 mm) (in 1.2 N HCl). Color was allowed to develop for 20 min at room temperature before quantifying methylene blue at 670 nm. Freshly prepared Na2S was used as the standard for sulfide quantification in the enzyme reaction.
Results
Deletion of IscA/SufA Results in the Accumulation of a Red-colored IscS in E. coli Cells
To explore the physiological interplay between IscA, a putative iron donor (44), and IscS, a major sulfur donor for iron-sulfur cluster biogenesis (6), we expressed recombinant IscS in the E. coli wild type and iscA/sufA mutant cells grown in LB medium under aerobic conditions. Fig. 1 shows that, although IscS purified from wild-type E. coli cells is yellow (indicative of pyridoxal 5′-phosphate) (5), IscS purified from the iscA/sufA mutant cells is bright red. In parallel experiments, IscS was also expressed in E. coli mutant cells in which the scaffold protein IscU (8–10) was deleted. Unlike in iscA/sufA mutant cells, IscS expressed in iscU mutant cells has the same yellow color as that produced in wild-type E. coli cells (data not shown), suggesting that deletion of IscU does not significantly affect IscS in E. coli cells under aerobic conditions.
FIGURE 1.
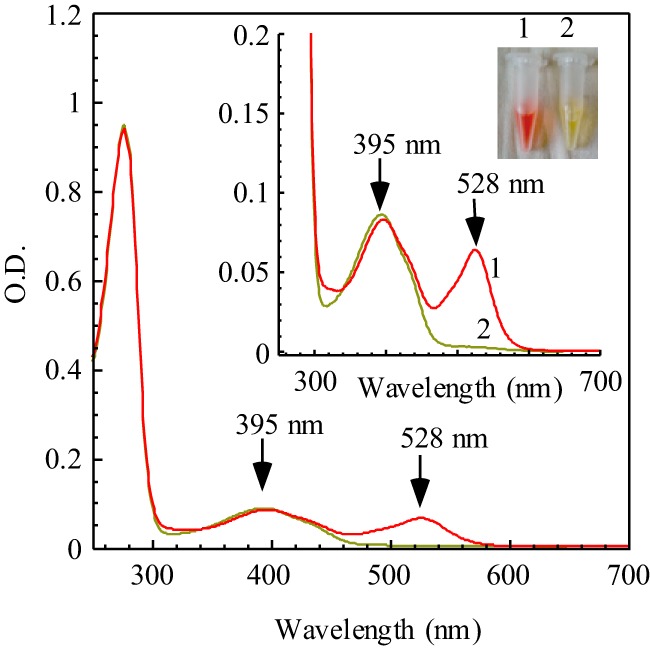
UV-visible spectra of IscS purified from the iscA/sufA mutant and parental wild-type E. coli cells. Recombinant E. coli IscS was expressed in E. coli iscA/sufA mutant cells (spectrum 1) or parental wild-type cells (spectrum 2). The protein concentration was calibrated to about 22 μm in buffer containing NaCl (500 mm) and Tris (20 mm (pH 8.0)). Inset, a photograph of IscS proteins purified from the E. coli iscA/sufA mutant cells (1) and parental wild-type cells (2).
UV-visible absorption measurements showed that IscS purified from E. coli iscA/sufA mutant cells has a distinct absorption peak at 528 nm in addition to the absorption peak at 395 nm of pyridoxal 5′-phosphate (5, 6) (Fig. 1). Metal content analyses showed that IscS proteins purified from wild-type or iscA/sufA mutant cells did not contain any detectable amounts of transition metals (data not shown), suggesting that the red color of IscS is not due to the metal binding in the protein.
Formation of Red IscS in E. coli iscA/sufA Mutant Cells Requires an Active Catalytic Site in IscS
To examine whether the formation of red IscS in the E. coli iscA/sufA mutant cells is an enzymatic process, we constructed two IscS mutants, IscS-C328S and IscS-K206A, by site-directed mutagenesis. Cys-328 in IscS directly attacks substrate l-cysteine (4), whereas Lys-206 is the ligand for pyridoxal 5′-phosphate (5). Both mutant proteins were expressed in wild-type E. coli cells grown in LB medium under aerobic conditions. As expected, purified IscS-C328S and IscS-K206A are not active to catalyze the desulfurization of l-cysteine (data not shown). UV-visible absorption measurements showed that purified IscS-C328S has an absorption peak at 395 nm of pyridoxal 5′-phosphate (Fig. 2A), which is identical to that of wild-type IscS (Fig. 1). To our surprise, purified IscS-K206A has two new absorption peaks, at 338 nm and 428 nm (Fig. 2A). Nevertheless, binding of pyridoxal 5′-phosphate to the enzyme without the ligand lysine is not unprecedented because substitution of the active-site Lys-313 with alanine in E. coli aminolevulinate synthase produces external aldimine intermediates (59). Because the absorption peaks at 338 nm and 428 nm have been assigned to the Cys-ketimine and Cys-aldimine intermediates in other cysteine desulfurases, respectively (4, 57), it is likely that Cys-ketimine and Cys-aldimine intermediates are trapped in the IscS-K206A mutant when expressed in wild-type E. coli cells.
FIGURE 2.
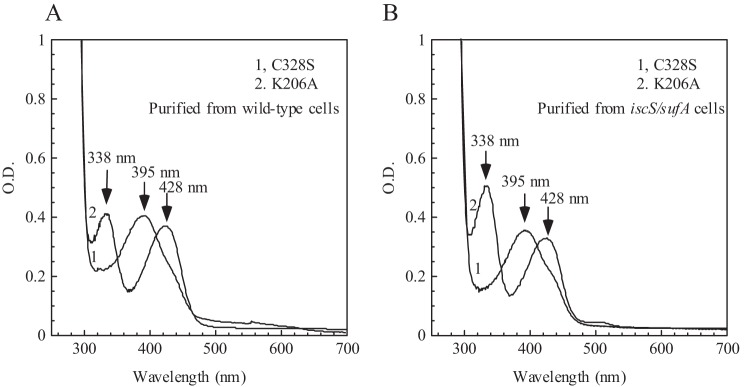
An active catalytic site is required for the formation of red IscS in E. coli iscA/sufA mutant cells. IscS-C328S and IscS-K206A were expressed in wild-type (A) or iscA/sufA mutant E. coli cells (B). Purified proteins (50 μm) were subjected to UV-visible absorption measurements. Spectrum 1, IscS-C328S; spectrum 2, IscS-K206A.
IscS-C328S and IscS-K206A were then expressed in E. coli iscA/sufA mutant cells grown in LB medium under aerobic conditions. As shown in Fig. 2B, purified IscS-C328S and IscS-K206A have essentially the same absorption peaks as those purified from wild-type E. coli cells (Fig. 2A), suggesting that the formation of red IscS in the iscA/sufA mutant cells requires an active catalytic center in IscS.
Red IscS Also Accumulates in Wild-type E. coli Cells with Depletion of Intracellular Iron
If deletion of IscA/SufA disrupts iron delivery for iron-sulfur cluster biogenesis and results in the accumulation of red IscS in E. coli cells under aerobic growth conditions (Fig. 1), then we reasoned that depletion of intracellular iron would also produce a red-colored IscS in wild-type E. coli cells under the same experimental conditions.
To test this idea, wild-type E. coli cells expressing recombinant IscS were grown in LB medium supplemented with or without 2,2′-dipyridyl, a membrane-permeable iron chelator (31), under aerobic growth conditions. Fig. 3 shows that IscS purified from wild-type E. coli cells grown in LB medium supplemented with 0.4 mm 2,2′-dipyridyl is indeed red, with an absorption peak at 528 nm (spectrum 2). The addition of 0.4 mm ferrous ammonium sulfate to the LB medium containing 0.4 mm 2,2′-dipyridyl completely suppresses the formation of red IscS in wild-type E. coli cells (spectrum 3). Therefore, depletion of intracellular iron can also result in the accumulation of red IscS in wild-type E. coli cells.
FIGURE 3.
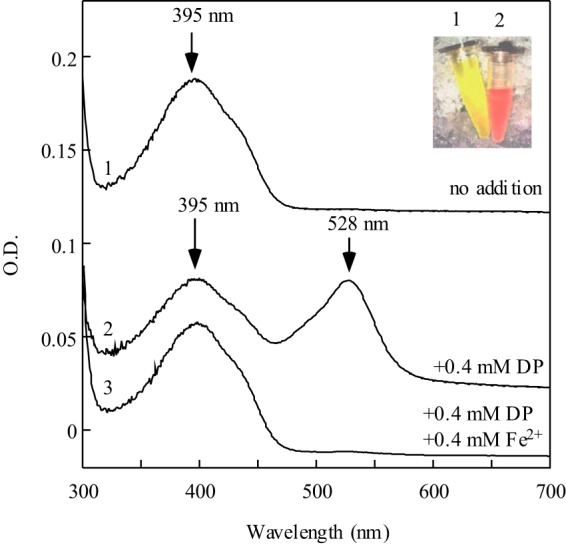
Red IscS also accumulates in wild-type E. coli cells with depletion of intracellular iron. Recombinant IscS was expressed in wild-type E. coli cells grown in LB medium supplemented with 0 (spectrum 1), 0.4 mm 2,2′-dipyridyl (spectrum 2), or 0.4 mm 2,2′-dipyridyl and 0.4 mm Fe(NH4)2(SO4)2 (spectrum 3). Purified IscS proteins (20 μm) were subjected to UV-visible absorption measurements. Inset, a photograph of IscS proteins purified from wild-type E. coli cells grown in LB medium supplemented with 0 (1) or 0.4 mm 2,2′-dipyridyl (2).
E. coli cells expressing recombinant IscS were then grown in LB medium supplemented with different concentrations of 2,2′-dipyridyl under aerobic growth conditions. As shown in Fig. 4A, when the concentration of 2,2′-dipyridyl in LB medium increases from 0 to 1.0 mm, cell growth is severely inhibited to a level close to that of the E. coli iscA/sufA mutant grown in LB medium without 2,2′-dipyridyl. Recombinant IscS was also purified from the wild-type E. coli cells grown in LB medium supplemented with different concentrations of 2,2′-dipyridyl. As the concentration of 2,2′-dipyridyl in LB medium increases from 0 to 1.0 mm, the absorption peak at 528 nm of purified IscS increases gradually (Fig. 4B). Therefore, deletion of IscA/SufA can be emulated by depletion of intracellular iron in inhibiting cell growth and in producing red IscS in E. coli cells under aerobic growth conditions.
FIGURE 4.
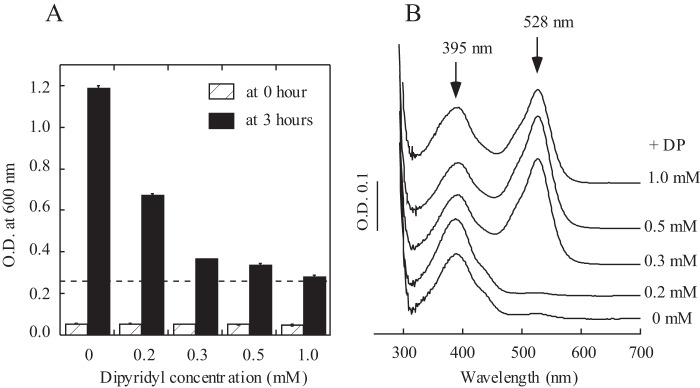
Effects of depletion of intracellular iron on cell growth of wild-type E. coli cells and on recombinant IscS expressed in wild-type E. coli cells. A, inhibition of cell growth of wild-type E. coli by 2,2′-dipyridyl. The same amount of overnight E. coli cultures was inoculated in fresh LB medium supplemented with the indicated concentrations of 2,2′-dipyridyl (0–1.0 mm). Cell growth was measured at 0 (white columns) and 3 h (black columns) under aerobic growth conditions. The dotted line represents the cell growth of the E. coli iscA/sufA mutant in LB medium without 2,2′-dipyridyl for 3 h under aerobic growth conditions. B, recombinant IscS was expressed in wild-type E. coli cells grown in LB medium supplemented with the indicated concentrations of 2,2′-dipyridyl (0–1.0 mm). Purified IscS proteins (30 μm) were subjected to UV-visible absorption measurements. The results are representatives from three independent experiments.
Redox Property of Red IscS
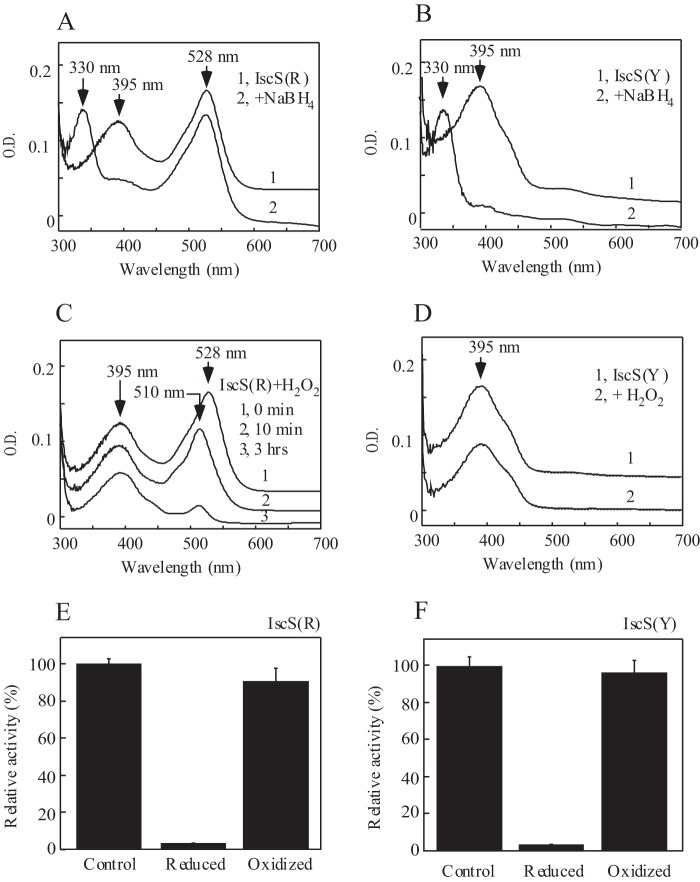
To the best of our knowledge, the red-colored IscS has not been previously purified or characterized. To explore the redox property of red IscS, we incubated the purified protein with sodium borohydride, a strong reducing reagent. As shown in Fig. 5, A and B, the absorption peak at 395 nm of pyridoxal 5′-phosphate in red IscS and wild-type IscS is shifted to 330 nm upon reduction with sodium borohydride. However, the absorption peak at 528 nm of red IscS is not affected by sodium borohydride (Fig. 5A). This result suggests that purified red IscS may contain two distinct IscS: one with an absorption peak at 395 nm of pyridoxal 5′-phosphate and the other with an absorption peak at 528 nm of an unknown intermediate. Unfortunately, attempts to separate the IscS with an absorption peak at 395 nm from that with an absorption peak at 528 nm using various chromatographic approaches were not successful (data not shown), likely because of the subtle difference between these intermediates of IscS.
FIGURE 5.
Redox property of red IscS. A, reduction of red IscS by sodium borohydride. Purified red IscS (IscS(R), 30 μm, spectrum 1) was incubated with NaBH4 (100 μm, spectrum 2) at 4 °C for 30 min. B, reduction of wild-type IscS by sodium borohydride. Purified wild-type IscS (IscS(Y), 30 μm, spectrum 1) was incubated with NaBH4 (100 μm, spectrum 2) at 4 °C for 30 min. C, oxidation of red IscS by H2O2. Purified red IscS (30 μm, spectrum 1) was incubated with H2O2 (10 mm, spectrum 2) at 4 °C for 10 min (spectrum 2) or 3 h (spectrum 3). D, oxidation of wild-type IscS by H2O2. Purified wild-type IscS (30 μm, spectrum 1) was incubated with 10 mm H2O2 (spectrum 2) at 4 °C for 3 h. E, relative cysteine desulfurase activity of red IscS after being treated with sodium borohydride (reduced) or H2O2 (oxidized). F, relative cysteine desulfurase activity of wild-type IscS after being treated with sodium borohydride or H2O2. The enzyme activity of purified red IscS varied in different preparations and closely correlated with the amplitudes of the absorption peak at 395 nm of pyridoxal 5′-phosphate. The total enzyme activity of red IscS shown was ∼75% of that of wild-type IscS. The results are the representatives from three independent different experiments.
Purified red IscS was also oxidized with H2O2. As shown in Fig. 5, C and D, although the absorption peak at 395 nm of red IscS and wild-type IscS is not changed upon addition of H2O2, the absorption peak at 528 nm of red IscS is quickly shifted to 510 nm (Fig. 5C). The results further suggest that the absorption peaks at 395 nm and 528 nm of purified red IscS represent two distinct IscS. It should be pointed out that the absorption peak at 510 nm of red IscS is not stable at 37 °C and decreases quickly (Fig. 5C). However, when dithiothreitol is added to the incubation solution right after the addition of H2O2, the absorption peak at 510 nm is partially reversed back to 528 nm (data not shown), suggesting that oxidation of red IscS by H2O2 is at least partially reversible.
The cysteine desulfurase activity was also measured for purified red IscS and wild-type IscS when the proteins were oxidized or reduced. The enzyme activity of purified red IscS and wild-type IscS varies in different preparations and closely correlates with the amplitude of the absorption peak at 395 nm of the prepared proteins. Interestingly, although oxidation with H2O2 has very little or no effect on the enzyme activity of red IscS and wild-type IscS, reduction with sodium borohydride completely inactivates red IscS and wild-type IscS (Fig. 5, E and F). Because the reduced red IscS is inactive (Fig. 5E) but still has the absorption peak at 528 nm (Fig. 5A), and the oxidized red IscS remains largely active (Fig. 5E) but without 528 nm (Fig. 5C), we propose that the IscS fraction with the absorption peak at 528 nm is inactive to catalyze desulfurization of l-cysteine.
Formation of Red IscS in Vitro
The transient absorption peak at 510 nm of purified red IscS after oxidization with H2O2 (Fig. 5C) is very close to the absorption peak at 506 nm of the proposed alanine-quinonoid intermediate in cysteine desulfurases from other organisms (4, 57). Conceivably, the absorption peak at 528 nm of purified red IscS could represent an alanine-quinonoid-like intermediate. To test this idea, we incubated wild-type IscS with l-alanine and sulfide, the two products produced from l-cysteine by IscS (4).
Fig. 6A shows that, when wild-type IscS is incubated with excess l-alanine and sulfide under aerobic conditions, red IscS is gradually formed. In parallel experiments, incubation of wild-type IscS with d-alanine and sulfide or glycine and sulfide fails to produce red IscS (Fig. 6B), demonstrating that l-alanine is essential for the formation of red IscS in vitro. Red IscS formed in vitro has an absorption peak at 510 nm (Fig. 6A), which is the same as that of red IscS purified from E. coli cells after oxidation with H2O2 (Fig. 5C). Similar to that shown in Fig. 5C, the absorption peak at 510 nm of red IscS produced in vitro is not stable at 37 °C and disappears quickly (data not shown). Taken together, the results suggest that the absorption peaks at 528 nm and 510 nm of red IscS purified from E. coli cells with deletion of IscA/SufA represent the quinonoid-like intermediates at different redox states.
FIGURE 6.
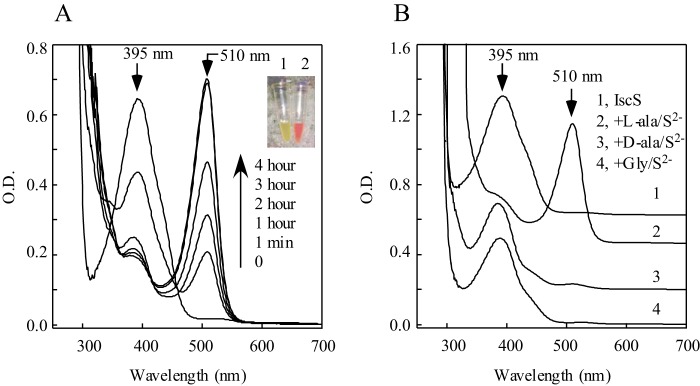
Formation of red IscS in vitro. A, purified wild-type IscS (100 μm) was incubated with l-alanine (100 mm) and Na2S (100 mm) in the presence of Tris (120 mm (pH 8.0)) at room temperature. The UV-visible absorption spectra were taken at 0, 1 min, 1 h, 2 h, 3 h, and 4 h. Inset, a photograph of IscS proteins before (1) and after (2) incubation with l-alanine and Na2S at room temperature for 3 h. B, l-alanine is essential for formation of red IscS in vitro. Purified wild-type IscS (100 μm) was incubated with buffer (spectrum 1), l-alanine (100 mm, spectrum 2), d-alanine (100 mm, spectrum 3), or glycine (100 mm, spectrum 4) in the presence of Na2S (100 mm) and Tris (120 mm (pH 8.0)) at room temperature. Spectra were taken after incubation at room temperature for 3 h.
Discussion
In this study, we report that deletion of the proposed iron chaperones IscA/SufA or depletion of intracellular iron results in the accumulation of a red-colored cysteine desulfurase IscS in E. coli cells under aerobic growth conditions. Red IscS can also be produced in vitro by incubating wild-type IscS with excess l-alanine and sulfide under aerobic conditions. We propose that deletion of IscA/SufA may block iron delivery for iron-sulfur cluster biogenesis, therefore impeding the sulfur delivery of cysteine desulfurase IscS, and result in the accumulation of red IscS intermediate in E. coli cells. The results also represent the first evidence for the physiological interplay between the proposed iron chaperones IscA/SufA and major cysteine desulfurase IscS in iron-sulfur cluster biogenesis in E. coli cells under aerobic growth conditions.
IscA is highly conserved from bacteria to humans (41). Although IscA and its homologs have been characterized previously as alternative scaffold proteins or carriers (15, 37–39), IscA has a unique and strong iron-binding activity in vitro (40–44) and in vivo (31) under aerobic conditions. The three invariant cysteine residues (45), and possibly an oxygen ligand (43), are likely involved in iron binding in IscA. Furthermore, the iron center in IscA can be readily mobilized by l-cysteine and transferred for iron-sulfur cluster assembly in vitro under aerobic conditions (43, 44, 47). Recently, we also reported that E. coli IscA has its unique activity to bind copper in vivo and in vitro and that excess copper can compete with iron for the metal binding sites in IscA and block iron-sulfur cluster biogenesis (60). These results led us to propose that IscA and its homologs may act as iron chaperones for iron-sulfur cluster biogenesis. Interestingly, although IscA and its homologs are essential for iron-sulfur cluster biogenesis in bacteria and eukaryotic cells under aerobic conditions (42, 49–56), deletion of IscA and its homolog SufA has very little or no effect on iron-sulfur proteins in E. coli cells under anaerobic growth conditions (31). One of the simplest explanation is that, under anaerobic conditions, the intracellular iron concentration may be sufficient to facilitate iron-sulfur cluster assembly in proteins without IscA and its homologs (31). Consistent with this idea, IscA and its homologs are absent in most anaerobic organisms (61). Here we found that, although deletion of IscA/SufA results in the accumulation of red IscS in E. coli cells under aerobic conditions, deletion of IscA/SufA has no apparent effects on IscS in E. coli cells under anaerobic growth conditions (data not shown). Therefore, IscA and its homologs may have a crucial role in recruiting intracellular iron and delivering iron for iron-sulfur cluster biogenesis under aerobic conditions but not under anaerobic conditions.
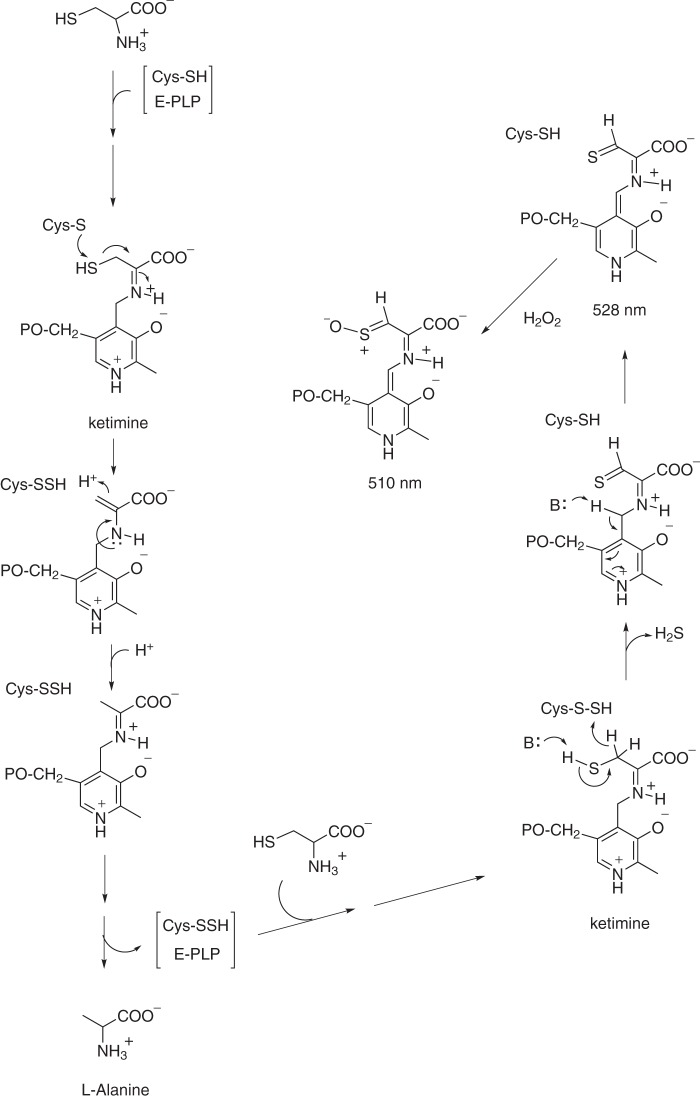
In E. coli cells, IscS is not only a major sulfur donor for iron-sulfur cluster biogenesis (6) but also provides sulfur for biogenesis of thiamine, tRNA thiolation (62), DNA phosphorothioation (63), molybdopterin (64), and other biological thiolation processes (65). Although the chemical nature of the red chromophore in IscS accumulated in the E. coli cells with deletion of IscA/SufA or depletion of intracellular iron could not be ascertained, a similar absorption peak at 521 nm of the serine-glyoxylate aminotransferase intermediate from Hyphomicrobium methylovorum has been reported previously (66). The absorption peak at 521 nm has been attributed to a highly conjugated quinonoid intermediate produced from hydroxyaminoacrylate and pyridoxal 5′-phosphate (66). In this context, we propose that the absorption peak at 528 nm of red IscS may also represent a highly conjugated quinonoid intermediate. With deficiency of accessible iron for iron-sulfur cluster biogenesis in E. coli cells, IscS may accumulate persulfide on the catalytic residue Cys-328 (67). Accumulated persulfide in Cys-328 may promote the transfer of a hydride from substrate l-cysteine to produce hydrogen sulfide and a thiocarbonyl group in l-cysteine that forms a highly conjugated quinonoid represented by an absorption peak at 528 nm (Fig. 7). When red IscS is oxidized by H2O2, the thiocarbonyl group could be modified to form a new intermediate with an absorption peak at 510 nm. Such an intermediate IscS may also be produced in vitro by incubating wild-type IscS with excess l-alanine and sulfide under aerobic conditions. Nevertheless, the exact nature of red chromophores in IscS formed in E. coli cells remains to be further investigated.
FIGURE 7.
Proposed intermediates of IscS formed in E. coli cells with deficiency of accessible iron for iron-sulfur cluster biogenesis. In E. coli cells with deletion of IscA/SufA or depletion of intracellular iron, IscS accumulates persulfide in Cys-328, which may transfer a hydride from the thiol group of l-cysteine to produce H2S and a thiocarbonyl group in l-cysteine. The thiocarbonyl group may form a highly conjugated quinonoid intermediate in red IscS that has an absorption peak at 528 nm. Oxidation of red IscS with H2O2 may modify the thiocarbonyl group and generate an unstable intermediate with an absorption peak at 510 nm.
In E. coli, the cysteine desulfurase IscS, the scaffold protein IscU, and the proposed iron chaperone IscA are encoded by the same operon (1), which is regulated by the global transcription factor IscR (68) and the small regulatory RNA RyhB (69). It is likely that IscS, IscU, and IscA work in concert for iron-sulfur cluster biogenesis (44, 48). The physical interaction between IscS and IscU has been well characterized (70). Dimeric IscU interacts with dimeric IscS to form a functional protein complex (9). However, the protein-protein interactions between IscA and IscS or IscA and IscU have not been reported. The finding that deletion of IscA and its paralog SufA results in the accumulation of red IscS in E. coli cells under aerobic conditions (Fig. 1) provides the first evidence of the physiological interplay between the sulfur donor IscS and the proposed iron chaperones IscA/SufA during iron-sulfur cluster biogenesis under aerobic growth conditions.
Acknowledgments
We thank the members of our group for discussions.
Footnotes
This work was supported, in whole or in part, by National Institutes of Health Award GM109399. This work was also supported by American Heart Association Grant 13GRNT16890014, Chinese National Natural Science Foundation Grants 31228006 and 31200587, and Natural Science Foundation of Zhejiang Province Grant LY12C05003.
References
- 1. Zheng L., Cash V. L., Flint D. H., Dean D. R. (1998) Assembly of iron-sulfur clusters: identification of an iscSUA-hscBA-fdx gene cluster from Azotobacter vinelandii. J. Biol. Chem. 273, 13264–13272 [DOI] [PubMed] [Google Scholar]
- 2. Bandyopadhyay S., Chandramouli K., Johnson M. K. (2008) Iron-sulfur cluster biosynthesis. Biochem. Soc. Trans. 36, 1112–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lill R. (2009) Function and biogenesis of iron-sulphur proteins. Nature 460, 831–838 [DOI] [PubMed] [Google Scholar]
- 4. Zheng L., White R. H., Cash V. L., Dean D. R. (1994) Mechanism for the desulfurization of l-cysteine catalyzed by the nifS gene product. Biochemistry 33, 4714–4720 [DOI] [PubMed] [Google Scholar]
- 5. Cupp-Vickery J. R., Urbina H., Vickery L. E. (2003) Crystal structure of IscS, a cysteine desulfurase from Escherichia coli. J. Mol. Biol. 330, 1049–1059 [DOI] [PubMed] [Google Scholar]
- 6. Schwartz C. J., Djaman O., Imlay J. A., Kiley P. J. (2000) The cysteine desulfurase, IscS, has a major role in in vivo Fe-S cluster formation in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 97, 9009–9014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kurihara T., Mihara H., Kato S., Yoshimura T., Esaki N. (2003) Assembly of iron-sulfur clusters mediated by cysteine desulfurases, IscS, CsdB and CSD, from Escherichia coli. Biochim. Biophys. Acta 1647, 303–309 [DOI] [PubMed] [Google Scholar]
- 8. Yan R., Kelly G., Pastore A. (2014) The scaffold protein IscU retains a structured conformation in the Fe-S cluster assembly complex. ChemBioChem. 15, 1682–1686 [DOI] [PubMed] [Google Scholar]
- 9. Marinoni E. N., de Oliveira J. S., Nicolet Y., Raulfs E. C., Amara P., Dean D. R., Fontecilla-Camps J. C. (2012) (IscS-IscU)2 complex structures provide insights into Fe2S2 biogenesis and transfer. Angew. Chem. Int. Ed. Engl. 51, 5439–5442 [DOI] [PubMed] [Google Scholar]
- 10. Bonomi F., Iametti S., Morleo A., Ta D., Vickery L. E. (2011) Facilitated transfer of IscU-[2Fe2S] clusters by chaperone-mediated ligand exchange. Biochemistry 50, 9641–9650 [DOI] [PubMed] [Google Scholar]
- 11. Chahal H. K., Dai Y., Saini A., Ayala-Castro C., Outten F. W. (2009) The SufBCD Fe-S scaffold complex interacts with SufA for Fe-S cluster transfer. Biochemistry 48, 10644–10653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wollers S., Layer G., Garcia-Serres R., Signor L., Clemancey M., Latour J. M., Fontecave M., Ollagnier de Choudens S. (2010) Iron-sulfur (Fe-S) cluster assembly: the SufBCD complex is a new type of Fe-S scaffold with a flavin redox cofactor. J. Biol. Chem. 285, 23331–23341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kim J. H., Füzéry A. K., Tonelli M., Ta D. T., Westler W. M., Vickery L. E., Markley J. L. (2009) Structure and dynamics of the iron-sulfur cluster assembly scaffold protein IscU and its interaction with the cochaperone HscB. Biochemistry 48, 6062–6071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kim J. H., Alderson T. R., Frederick R. O., Markley J. L. (2014) Nucleotide-dependent interactions within a specialized Hsp70/Hsp40 complex involved in Fe-S cluster biogenesis. J. Am. Chem. Soc. 136, 11586–11589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mapolelo D. T., Zhang B., Randeniya S., Albetel A. N., Li H., Couturier J., Outten C. E., Rouhier N., Johnson M. K. (2013) Monothiol glutaredoxins and A-type proteins: partners in Fe-S cluster trafficking. Dalton Trans. 42, 3107–3115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shakamuri P., Zhang B., Johnson M. K. (2012) Monothiol glutaredoxins function in storing and transporting [Fe2S2] clusters assembled on IscU scaffold proteins. J. Am. Chem. Soc. 134, 15213–15216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li H., Mapolelo D. T., Randeniya S., Johnson M. K., Outten C. E. (2012) Human glutaredoxin 3 forms [2Fe-2S]-bridged complexes with human BolA2. Biochemistry 51, 1687–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Campuzano V., Montermini L., Moltò M. D., Pianese L., Cossée M., Cavalcanti F., Monros E., Rodius F., Duclos F., Monticelli A., Zara F., Cañizares J., Koutnikova H., Bidichandani S. I., Gellera C., Brice A., Trouillas P., De Michele G., Filla A., De Frutos R., Palau F., Patel P. I., Di Donato S., Mandel J. L., Cocozza S., Koenig M., Pandolfo M. (1996) Friedreich's ataxia: autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science 271, 1423–1427 [DOI] [PubMed] [Google Scholar]
- 19. Yoon T., Cowan J. A. (2003) Iron-sulfur cluster biosynthesis. Characterization of frataxin as an iron donor for assembly of [2Fe-2S] clusters in ISU-type proteins. J. Am. Chem. Soc. 125, 6078–6084 [DOI] [PubMed] [Google Scholar]
- 20. Layer G., Ollagnier-de Choudens S., Sanakis Y., Fontecave M. (2006) Iron-sulfur cluster biosynthesis: characterization of Escherichia coli CyaY as an iron donor for the assembly of [2Fe-2S] clusters in the scaffold IscU. J. Biol. Chem. 281, 16256–16263 [DOI] [PubMed] [Google Scholar]
- 21. Bulteau A. L., O'Neill H. A., Kennedy M. C., Ikeda-Saito M., Isaya G., Szweda L. I. (2004) Frataxin acts as an iron chaperone protein to modulate mitochondrial aconitase activity. Science 305, 242–245 [DOI] [PubMed] [Google Scholar]
- 22. González-Cabo P., Vázquez-Manrique R. P., García-Gimeno M. A., Sanz P., Palau F. (2005) Frataxin interacts functionally with mitochondrial electron transport chain proteins. Hum. Mol. Genet. 14, 2091–2098 [DOI] [PubMed] [Google Scholar]
- 23. Adinolfi S., Iannuzzi C., Prischi F., Pastore C., Iametti S., Martin S. R., Bonomi F., Pastore A. (2009) Bacterial frataxin CyaY is the gatekeeper of iron-sulfur cluster formation catalyzed by IscS. Nat. Struct. Mol. Biol. 16, 390–396 [DOI] [PubMed] [Google Scholar]
- 24. Kim J. H., Frederick R. O., Reinen N. M., Troupis A. T., Markley J. L. (2013) [2Fe-2S]-Ferredoxin binds directly to cysteine desulfurase and supplies an electron for iron-sulfur cluster assembly but is displaced by the scaffold protein or bacterial frataxin. J. Am. Chem. Soc. 135, 8117–8120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yan R., Konarev P. V., Iannuzzi C., Adinolfi S., Roche B., Kelly G., Simon L., Martin S. R., Py B., Barras F., Svergun D. I., Pastore A. (2013) Ferredoxin competes with bacterial frataxin in binding to the desulfurase IscS. J. Biol. Chem. 288, 24777–24787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Roche B., Huguenot A., Barras F., Py B. (2015) The iron-binding CyaY and IscX proteins assist the ISC-catalyzed Fe-S biogenesis in Escherichia coli. Mol. Microbiol. 95, 605–623 [DOI] [PubMed] [Google Scholar]
- 27. Pandey A., Gordon D. M., Pain J., Stemmler T. L., Dancis A., Pain D. (2013) Frataxin directly stimulates mitochondrial cysteine desulfurase by exposing substrate-binding sites, and a mutant Fe-S cluster scaffold protein with frataxin-bypassing ability acts similarly. J. Biol. Chem. 288, 36773–36786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gerber J., Mühlenhoff U., Lill R. (2003) An interaction between frataxin and Isu1/Nfs1 that is crucial for Fe/S cluster synthesis on Isu1. EMBO Rep. 4, 906–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bou-Abdallah F., Adinolfi S., Pastore A., Laue T. M., Dennis Chasteen N. (2004) Iron binding and oxidation kinetics in frataxin CyaY of Escherichia coli. J. Mol. Biol. 341, 605–615 [DOI] [PubMed] [Google Scholar]
- 30. Ding H., Yang J., Coleman L. C., Yeung S. (2007) Distinct iron binding property of two putative iron donors for the iron-sulfur cluster assembly: IscA and the bacterial frataxin ortholog CyaY under physiological and oxidative stress conditions. J. Biol. Chem. 282, 7997–8004 [DOI] [PubMed] [Google Scholar]
- 31. Wang W., Huang H., Tan G., Si F., Liu M., Landry A. P., Lu J., Ding H. (2010) In vivo evidence for the iron-binding activity of an iron-sulfur cluster assembly protein IscA in Escherichia coli. Biochem. J. 432, 429–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Duby G., Foury F., Ramazzotti A., Herrmann J., Lutz T. (2002) A non-essential function for yeast frataxin in iron-sulfur cluster assembly. Hum. Mol. Genet. 11, 2635–2643 [DOI] [PubMed] [Google Scholar]
- 33. Vivas E., Skovran E., Downs D. M. (2006) Salmonella enterica strains lacking the frataxin homolog CyaY show defects in Fe-S cluster metabolism in vivo. J. Bacteriol. 188, 1175–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li D. S., Ohshima K., Jiralerspong S., Bojanowski M. W., Pandolfo M. (1999) Knock-out of the cyaY gene in Escherichia coli does not affect cellular iron content and sensitivity to oxidants. FEBS Lett. 456, 13–16 [DOI] [PubMed] [Google Scholar]
- 35. Kim J. H., Bothe J. R., Frederick R. O., Holder J. C., Markley J. L. (2014) Role of IscX in iron-sulfur cluster biogenesis in Escherichia coli. J. Am. Chem. Soc. 136, 7933–7942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pastore C., Adinolfi S., Huynen M. A., Rybin V., Martin S., Mayer M., Bukau B., Pastore A. (2006) YfhJ, a molecular adaptor in iron-sulfur cluster formation or a frataxin-like protein? Structure 14, 857–867 [DOI] [PubMed] [Google Scholar]
- 37. Gupta V., Sendra M., Naik S. G., Chahal H. K., Huynh B. H., Outten F. W., Fontecave M., Ollagnier de Choudens S. (2009) Native Escherichia coli SufA, coexpressed with SufBCDSE, purifies as a [2Fe-2S] protein and acts as an Fe-S transporter to Fe-S target enzymes. J. Am. Chem. Soc. 131, 6149–6153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mapolelo D. T., Zhang B., Naik S. G., Huynh B. H., Johnson M. K. (2012) Spectroscopic and functional characterization of iron-sulfur cluster-bound forms of Azotobacter vinelandii (Nif)IscA. Biochemistry 51, 8071–8084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Banci L., Brancaccio D., Ciofi-Baffoni S., Del Conte R., Gadepalli R., Mikolajczyk M., Neri S., Piccioli M., Winkelmann J. (2014) [2Fe-2S] cluster transfer in iron-sulfur protein biogenesis. Proc. Natl. Acad. Sci. U.S.A. 111, 6203–6208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ding H., Clark R. J. (2004) Characterization of iron binding in IscA, an ancient iron-sulphur cluster assembly protein. Biochem. J. 379, 433–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lu J., Bitoun J. P., Tan G., Wang W., Min W., Ding H. (2010) Iron binding activity of human iron-sulfur cluster assembly protein hIscA-1. Biochem. J. 428 125–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mühlenhoff U., Richter N., Pines O., Pierik A. J., Lill R. (2011) Specialized function of yeast Isa1 and Isa2 in the maturation of mitochondrial [4fe-4s] proteins. J. Biol. Chem. 286, 41205–41216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mapolelo D. T., Zhang B., Naik S. G., Huynh B. H., Johnson M. K. (2012) Spectroscopic and functional characterization of iron-bound forms of Azotobacter vinelandii (Nif)IscA. Biochemistry 51, 8056–8070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Landry A. P., Cheng Z., Ding H. (2013) Iron binding activity is essential for the function of IscA in iron-sulphur cluster biogenesis. Dalton Trans. 42, 3100–3106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ding H., Harrison K., Lu J. (2005) Thioredoxin reductase system mediates iron binding in IscA and iron delivery for the iron-sulfur cluster assembly in IscU. J. Biol. Chem. 280, 30432–30437 [DOI] [PubMed] [Google Scholar]
- 46. Bitoun J. P., Wu G., Ding H. (2008) Escherichia coli FtnA acts as an iron buffer for re-assembly of iron-sulfur clusters in response to hydrogen peroxide stress. Biometals 21, 693–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ding B., Smith E. S., Ding H. (2005) Mobilization of the iron centre in IscA for the iron-sulphur cluster assembly in IscU. Biochem. J. 389, 797–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yang J., Bitoun J. P., Ding H. (2006) Interplay of IscA and IscU in biogenesis of iron-sulfur clusters. J. Biol. Chem. 281, 27956–27963 [DOI] [PubMed] [Google Scholar]
- 49. Johnson D. C., Unciuleac M. C., Dean D. R. (2006) Controlled expression and functional analysis of iron-sulfur cluster biosynthetic components within Azotobacter vinelandii. J. Bacteriol. 188, 7551–7561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lu J., Yang J., Tan G., Ding H. (2008) Complementary roles of SufA and IscA in the biogenesis of iron-sulfur clusters in Escherichia coli. Biochem. J. 409, 535–543 [DOI] [PubMed] [Google Scholar]
- 51. Mettert E. L., Outten F. W., Wanta B., Kiley P. J. (2008) The impact of O2 on the Fe-S cluster biogenesis requirements of Escherichia coli FNR. J. Mol. Biol. 384, 798–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Jensen L. T., Culotta V. C. (2000) Role of Saccharomyces cerevisiae ISA1 and ISA2 in iron homeostasis. Mol. Cell. Biol. 20, 3918–3927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kaut A., Lange H., Diekert K., Kispal G., Lill R. (2000) Isa1p is a component of the mitochondrial machinery for maturation of cellular iron-sulfur proteins and requires conserved cysteine residues for function. J. Biol. Chem. 275, 15955–15961 [DOI] [PubMed] [Google Scholar]
- 54. Song D., Tu Z., Lee F. S. (2009) Human IscA1 interacts with IOP1/NARFL and functions in both cytosolic and mitochondrial iron-sulfur protein biogenesis. J. Biol. Chem. 284, 35297–35307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Tan G., Lu J., Bitoun J. P., Huang H., Ding H. (2009) IscA/SufA paralogs are required for the [4Fe-4S] cluster assembly in enzymes of multiple physiological pathways in Escherichia coli under aerobic growth conditions. Biochem. J. 420, 463–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sheftel A. D., Wilbrecht C., Stehling O., Niggemeyer B., Elsässer H. P., Mühlenhoff U., Lill R. (2012) The human mitochondrial ISCA1, ISCA2, and IBA57 proteins are required for [4Fe-4S] protein maturation. Mol. Biol. Cell 23, 1157–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Behshad E., Bollinger J. M., Jr. (2009) Kinetic analysis of cysteine desulfurase CD0387 from Synechocystis sp. PCC 6803: formation of the persulfide intermediate. Biochemistry 48, 12014–12023 [DOI] [PubMed] [Google Scholar]
- 58. Siegel L. M. (1965) A direct microdetermination of sulfide. Anal. Biochem. 11, 126–132 [DOI] [PubMed] [Google Scholar]
- 59. Ferreira G. C., Vajapey U., Hafez O., Hunter G. A., Barber M. J. (1995) Aminolevulinate synthase: lysine 313 is not essential for binding the pyridoxal phosphate cofactor but is essential for catalysis. Protein Sci. 4, 1001–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Tan G., Cheng Z., Pang Y., Landry A. P., Li J., Lu J., Ding H. (2014) Copper binding in IscA inhibits iron-sulphur cluster assembly in Escherichia coli. Mol. Microbiol. 93, 629–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Vinella D., Brochier-Armanet C., Loiseau L., Talla E., Barras F. (2009) Iron-sulfur (Fe/S) protein biogenesis: phylogenomic and genetic studies of A-type carriers. PLoS Genet. 5, e1000497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Shi R., Proteau A., Villarroya M., Moukadiri I., Zhang L., Trempe J.-F., Matte A., Armengod M. E., Cygler M. (2010) Structural Basis for Fe-S cluster assembly and tRNA thiolation mediated by IscS protein-protein interactions. PLoS Biol. 8, e1000354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. An X., Xiong W., Yang Y., Li F., Zhou X., Wang Z., Deng Z., Liang J. (2012) A novel target of IscS in Escherichia coli: participating in DNA phosphorothioation. PLoS ONE 7, e51265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Zhang W., Urban A., Mihara H., Leimkühler S., Kurihara T., Esaki N. (2010) IscS functions as a primary sulfur-donating enzyme by interacting specifically with MoeB and MoaD in the biosynthesis of molybdopterin in Escherichia coli. J. Biol. Chem. 285, 2302–2308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Lauhon C. T., Kambampati R. (2000) The iscS gene in Escherichia coli is required for the biosynthesis of 4-thiouridine, thiamin, and NAD. J. Biol. Chem. 275, 20096–20103 [DOI] [PubMed] [Google Scholar]
- 66. Karsten W. E., Cook P. F. (2009) Detection of a gem-diamine and a stable quinonoid intermediate in the reaction catalyzed by serine-glyoxylate aminotransferase from Hyphomicrobium methylovorum. Biochim. Biophys. Acta 1790, 575–580 [DOI] [PubMed] [Google Scholar]
- 67. Smith A. D., Agar J. N., Johnson K. A., Frazzon J., Amster I. J., Dean D. R., Johnson M. K. (2001) Sulfur transfer from IscS to IscU: the first step in iron-sulfur cluster biosynthesis. J. Am. Chem. Soc. 123, 11103–11104 [DOI] [PubMed] [Google Scholar]
- 68. Fleischhacker A. S., Stubna A., Hsueh K. L., Guo Y., Teter S. J., Rose J. C., Brunold T. C., Markley J. L., Münck E., Kiley P. J. (2012) Characterization of the [2Fe-2S] cluster of Escherichia coli transcription factor IscR. Biochemistry 51, 4453–4462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Desnoyers G., Morissette A., Prévost K., Massé E. (2009) Small RNA-induced differential degradation of the polycistronic mRNA iscRSUA. EMBO J. 28, 1551–1561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Nuth M., Cowan J. (2009) Iron-sulfur cluster biosynthesis: characterization of IscU-IscS complex formation and a structural model for sulfide delivery to the [2Fe-2S] assembly site. J. Biol. Inorg. Chem. 14, 829–839 [DOI] [PubMed] [Google Scholar]