Abstract
Fusarium graminearum and F. culmorum are the causing agents of a destructive disease known as Fusarium head blight (FHB). FHB is a re-emerging disease in small grain cereals which impairs both the grain yield and the quality. Most serious consequence is the contamination of grain with Fusarium mycotoxins that are severe threat to humans and animals. Biological control has been suggested as one of the integrated management strategies to control FHB. Paenibacillus polymyxa is considered as a promising biocontrol agent due to its unique antibiotic spectrum. P. polymyxa A26 is an efficient antagonistic agent against Fusarium spp. In order to optimize strain A26 production, formulation and application strategies traits important for its compatibility need to be revealed. Here we developed a toolbox, comprising of dual culture plate assays and wheat kernel assays, including simultaneous monitoring of FHB causing pathogens, A26, and mycotoxin production. Using this system we show that, besides generally known lipopeptide antibiotic production by P. polymyxa, biofilm formation ability may play a crucial role in the case of stain A26 F. culmorum antagonism. Application of the system for effective strain selection and maintenance is discussed.
Keywords: FHB biocontrol, Paenibacillus polymyxa, Sfp-type PPTase, lipopeptide antibiotics, bacterial biofilm
Introduction
Fusarium head blight (FHB) is a destructive disease on cereals that is caused by a group of Fusarium species including Fusarium graminearum and F. culmorum (Nazari et al., 2014). FHB is a major threat to agricultural production due to yield losses, but also constitutes a major safety concern when humans and animals consume Fusarium-contaminated products due to the accumulation of several mycotoxins (Yazar and Omurtag, 2008; Xiao et al., 2013). Both F. culmorum and F. graminearum are soil borne and cause not only FHB, but also Fusarium foot and root rot on cereals around the globe especially during wet seasons (Scherm et al., 2013). Several mycotoxins are associated with Fusarium sp. infection, including deoxynivalenol (DON) and zearalenone (ZEA; Cowger and Arellano, 2013). Higher levels of both toxins in wheat grains are usually connected to infection with F. culmorum or F. graminearum (Fredlund et al., 2013; Lindblad et al., 2013).
Crop rotation, improved cultivar resistance, or improved fungicide efficacy and timing has benefited producer in previous years on locations with low FHB pressure. However, under conditions that are highly favorable for infection, use of a single management strategy often fails to control the disease and limit myxotoxin production to acceptable levels. To keep DON to manageable levels, a 75% reduction of FHB index must be obtained when the disease pressure is great (Shah et al., 2013). Given that there has been considerable interest in combining the use of fungicides with environmentally friendly methods, biological control has been suggested as one of the integrated management strategies to control these infections. Various of microbial agents are able to inhibit fungal growth. For instance, Shi et al. (2014) reported that some strains of Bacillus amyloliquefaciens antagonized F. graminearum growth, which significantly inhibited DON production in wheat seeds. Besides biological control of the fungus, decontamination of mycotoxins using bacteria is the other attractive strategy of biological control for the management of mycotoxins in food and feed (Shetty et al., 2007).
The employment of microbes as biological control agents (BCA) against F. graminearum and F. culmorum have been reported in several studies under greenhouse conditions (Shi et al., 2014). However, BCAs generally do not perform well enough under uncontrolled conditions in soil and they are not used commercially to control FHB (Walter et al., 2010; Matarese et al., 2012). Our BCAs were isolated from soil in Israel, from a field station called Evolution Canyon (Timmusk et al., 2011, 2014a). Earlier, we demonstrated that isolates from the highly stressed south facing slope (SFS) of Evolution Canyon differ remarkably from isolates derived from the more moderate north facing slope (NFS; Timmusk et al., 2011). SFS isolates were significantly better biofilm producers, phosphorus solubilizers and could produce the enzyme 1-aminocyclopropane-1-carboxylate (ACC deaminase; Timmusk et al., 2011). These isolates were also superior in conferring drought tolerance enhancement to treated plants (Timmusk et al., 2014a). Two Paenibacillus polymyxa isolates, one from the SFS (A26) and one from the NFS (E1) were used in the present study (Timmusk et al., 2014a).
Paenibacillus polymyxa is a plant growth- promoting bacterium (PGPB), with a broad host plant range and is known for its the ability to produce different lipopeptide antibiotics. The first lipopeptide antibiotic was isolated form a rhizosphere isolate in 1947 and since then many antibiotic compounds active against gram-positive and gram-negative bacteria have been reported (Stansly and Schloesser, 1947). P. polymyxa is also known as one of the best rhizobacterial biofilm formers – however, its biofilm formation ability varies in habitat dependent manner (Timmusk et al., 2011). One of the SFS isolates, P. polymyxa A26, has been found, in field studies, to be very efficient against Fusarium sp. (Timmusk et al., 2014b). The genomic sequence of strain A26 reveals that it harbors several giant gene clusters directing the synthesis of bioactive peptides and polyketides (PKs) by modularly organized megaenzymes, i.e., by non-ribosomal peptide synthetases, (NRPS) and PK synthetases, (PKS). Synthesis of both non-ribosomal peptides (NRP) and PKs is dependent on the presence of a functional Sfp-type phosphopantheinyl transferase (PPTase; Mootz et al., 2001). The virulence of various pathogens is dependent on NRP or PK production and therefore their Sfp-type PPTases may be characterized as gate keepers to pathogenicity. For this reason, Sfp-type PPTases have been suggested as a potential target for antibacterial drug development in the medical industry as well as a means of fighting agricultural pathogens (Pieters et al., 2002; Leblanc et al., 2012; Zheng and Burr, 2013).
To study the mode of action of strain A26, we developed a method for A26 mutagenesis (Kim and Timmusk, 2013) and inactivated the A26 Sfp-type PPTase gene resulting in a deletion mutation that is deficient in the production of NRP and PK origin antibiotics and has also lost its antagonistic activity on plate assays. It was confirmed by LC MS/MS that the well-known antibiotic types, fusaricidins and polymyxins, are not produced by the mutant (Timmusk et al., 2015). The mutant is greatly enhanced in biofilm production but is otherwise identical to A26 in other physiological and metabolic parameters (Timmusk et al., 2015). Hence, utilization of this mutant provides an excellent tool to study the mechanism of strain A26 biocontrol (Timmusk et al., 2015). Moreover, strain A26 is particularly attractive for field application; it is a Gram-positive bacterium able to resist pesticides and form endospores that can survive under various stress conditions in the field and has remarkable ability to produce rich biofilms and biologically active compounds.
In order to efficiently utilize strain A26 against F. culmorum and F. graminearum, one has to know the likely mechanisms of action of this BCA including genes and bioactive compounds involved in the biocontrol mechanisms. Based on this knowledge, it is possible to construct efficient cultivation, formulation and delivery systems of this BCA as well as evaluate its performance under specific field conditions. Here we developed a toolbox comprising of dual culture plate assays and wheat kernel assays including quantitative PCR (qPCR) monitoring of pathogens and agents and mycotoxins produced. Using this system we show that, besides generally known antibiotic production by P. polymyxa, biofilm formation ability may play a crucial role in the case of F. culmorum antagonism.
Materials and Methods
Microbial Agents and Culture Conditions
The fungal pathogens F. culmorum strain 42344/1997 and F. graminearum strain A602/1998 were obtained from The National Veterinary Institute, Uppsala, Sweden. Both pathogens were grown on potato dextrose agar (PDA) plates at 22°C. P. polymyxa A26 and E1 were isolated from the rhizosphere of wild barley at EC SFS and NFS (Timmusk et al., 2011, 2014a) and grown on tryptic soya agar (TSA) at 29°C. P. polymyxa A26 Sfp-type- 4′-phosphopantetheinyl transferase deletion mutant strain (A26Δsfp), lacking the ability to produce both NRPs and PKSs was grown under the same conditions.
Bioassay of In Vivo Antagonism
Plate Assay
Inhibitory studies between P. polymyxa A26 and E1 and F. culmorum and F. graminearum were conducted on King’s B plates (Ausubel et al., 1988). The bacterial strains were streaked onto the plates after inoculation with fungal plugs. Plates were incubated at 28°C for 5 days.
Assay on Wheat Grains
Antagonistic Activity of P. polymyxa A26 and A26Δsfp Against F. culmorum and F. graminearum
The antagonistic effect of P. polymyxa A26 and A26Δsfp on F. culmorum and F. graminearum was studied in an experimental setup using sterile wheat grains. Bacterial strains were grown in tryptic soya broth (TSB) overnight at 28°C with shaking at 180 rpm. Cultures were centrifuged for 10 min at 10000 rpm; pellets were washed using sterile water and re-suspended in sterile water. Bacterial cells were counted on Petri plates using the colony forming unit (CFU) method and adjusted to 1 × 107 cells/ml.
Sterile 150 ml conical flasks containing 20 g sterile wheat grains were inoculated with 15 ml 1 × 107 cells/ml P. polymyxa A26 or A26Δsfp overnight cultures. Controls were treated with 15 ml sterile water. Flasks were incubated at room temperature for 8 h, and then inoculated with 1 cm2 agar plugs from 2 week old cultures of either F. graminearum or F. culmorum, and incubated at room temperature. Fungal growth was assessed visually and 1 g samples (≈15 grains) were taken from each flask at four time points; i.e., 0, 5, 10, and 15 days after fungal inoculation, and stored at -20°C.
Antagonistic Activity of P. polymyxa A26 and A26Δsfp Culture Filtrates Against F. culmorum
Bacterial strains were grown in TSB at 28°C on a rotary shaker (180 rpm) for 72 h. Cultures were centrifuged for 10 min at 10000 rpm and supernatants were filter sterilized using 0.22 μm Millipore filters. Wheat grains were prepared as described above. Instead of bacterial solutions, equal aliquots of supernatant were used for wheat grain treatments. Controls were treated with 15 ml sterile water. Experiments were performed three times to confirm reproducibility.
DNA Extraction
The grain samples were freeze dried and ground into a fine powder using a Precellys 24 homogenizer (Bertin Technologies, France). Samples were lysed by incubating 100 mg powder for 15 min in 350 μl of glucose buffer [50 mmol-1 glucose, 25 mmol-l Tris-HCl (pH 8), 10 mmol-l EDTA] with 4 mg ml-1 lysozyme (Timmusk et al., 2009a). DNA was extracted using a hexadecyltrimethylammonium bromide-based method (Nygren et al., 2008). Pure bacterial, fungal, and plant DNA was also extracted from liquid cultures, mycelia and grains, respectively, using the same method. DNA quantity and quality was assessed with spectroscopic methods using a Nanodrop 1000 (Thermo Scientific, Wilmington, DE, USA).
PCR and Quantitative PCR Analyses
The presence and amount of F. culmorum, F. graminearum, and P. polymyxa DNA was monitored by PCR and qPCR. Primers (Table 1) targeting EF1α genes in F. culmorum and F. graminearum as well as wheat were used for PCR as described earlier (Nicolaisen et al., 2009). The bacterial DNA was quantified in the same sample with specific primers designed using Primer3 software (Rozen and Skaletsky, 2000; Table 1) targeting the 16S and Sfp-type PPTase gene in P. polymyxa. The PCR reaction volume (25 μl) included Dream Taq polymerase enzyme (Thermo, USA) supplemented with 10 μM primers, 10 mM dNTPs, and DNA as a template. As a positive control, a reaction mixture with pure bacterial DNA was used and fungal DNA was used as a template for the negative control reactions. PCR reactions were performed using the following parameters: initial denaturation for 2 min at 94°C; followed by 30 cycles of 1 min at 95°C, 50 s at 54°C, and 1 min at 72°C; and finally 10 min at 72°C in a thermal Mastercycler (ABI, USA). qPCR assays were carried out in 25 μl consisting of 12.5 μl Maxima SYBR Green master mix (Thermo, USA), 0.5 μM primers (forward and reverse) and 100 ng DNA using the Bio-Rad iCycler iQ5 (Bio-Rad, USA). Standard curves consisting of 10-fold dilutions starting at 100 ng DNA for each assay were prepared using pure DNA extracted from either the pathogens or the bacteria. Fungal and bacterial DNA amounts were expressed as a relative ratio to the amount of plant DNA.
Table 1.
Primers used for PCR and quantitative PCR (qPCR) analysis.
| Target | Primer’s ID | Sequence (5′–3′) | Reference |
|---|---|---|---|
| Fusarium graminearum | FgramB F FgramB R |
CCATTCCCTGGGCGCT CCTATTGACAGGTGGTTAGTGACTGG |
Nicolaisen et al. (2009) |
| F. culmorum | FculC F FculC R |
CACCGTCATTGGTATGTTGTCACT CGGGAGCGTCTGATAGTCG |
Nicolaisen et al. (2009) |
| Paenibacillus polymyxa | 29Pp F 179Pp R |
GAGCGGGGTTGATTAGAAGC CTTTCCTCCTTCTCCCATGC |
Timmusk et al. (2009a) |
| P. polymyxa1 | 16sA26 F 16sA26 R |
GCATGGGAAAAGGAGGAAAG AGCAGTTACTCTACAAGACGTTC |
This study |
| P. polymyxa2 | Sfpdel F Sfpdel R |
GTTGGTCTGCCGGCAATTGA GGTTGTCTGCATCCTCACGCA |
This study |
| Plant EF1α | Hor1 F Hor2 R |
TCTCTGGGTTTGAGGGTGAC GGCCCTTGTACCAGTCAAGGT |
Nicolaisenet al. (2009) |
1Target both P. polymyxa A26 and A26Δsfp.
2Target only P. polymyxa A26.
Mycotoxins Analysis
The concentrations of DON and ZEA were determined in 1 g freeze dried wheat grains at 5, 10, and 15 days after pathogen infection using a LC-MS/MS (Njobeh et al., 2012; Tevell Åberg et al., 2012). Culture filtrate ZEA was quantified using a commercial ELISA kit for ZEA detection (RIDASCREEN, R-Biopharm AG, Darmstadt, Germany) following the manufacturer’s instructions.
Statistical Analysis and Data Validation:
Data were subjected to analysis of variance (ANOVA) to determine the significance between the different treatments using Costat (CoHort software, CA). Experiments were repeated at least two times.
Results
Paenibacillus polymyxa A26 and A26Δsfp Antagonism Against Fungal Pathogens on Agar Plates
The antagonistic ability of P. polymyxa A26, E1, and A26Δsfp against F. culmorum and F. graminearum was assayed on King’s B plates (Table 2). Both wild-type strains of P. polymyxa significantly inhibited the growth of F. culmorum and F. graminearum where P. polymyxa A26 exhibited higher antagonistic activity than strain E1 (Table 2). A26Δsfp lost its ability to antagonize both pathogens (Table 2).
Table 2.
Paenibacillus polymyxa inhibition of F. graminearum and F. culmorum on plate assays.
| Inhibition zone (mm) |
||
|---|---|---|
| Treatments | F. graminearum | F. culmorum |
| P. polymyxa A26 | 17 ± 1a | 16 ± 2a |
| P. polymyxa E1 | 13 ± 1b | 11 ± 1c |
| P. polymyxa A26Δsfp | 0d | 0d |
| Control | 0d | 0d |
The antagonistic ability of Sfp-type PPTase mutant was compared with two wild-type P. polymyxa strains on King’s B plates. The results are presented as the diameter of the inhibition zone (mm) of fungal growth. Different letters indicate statistically significant differences (P ≤ 0.01). The data represent the mean of four biological replicates.
Paenibacillus polymyxa A26 and A26Δsfp Antagonism Against Fungal Pathogens on Wheat Grains
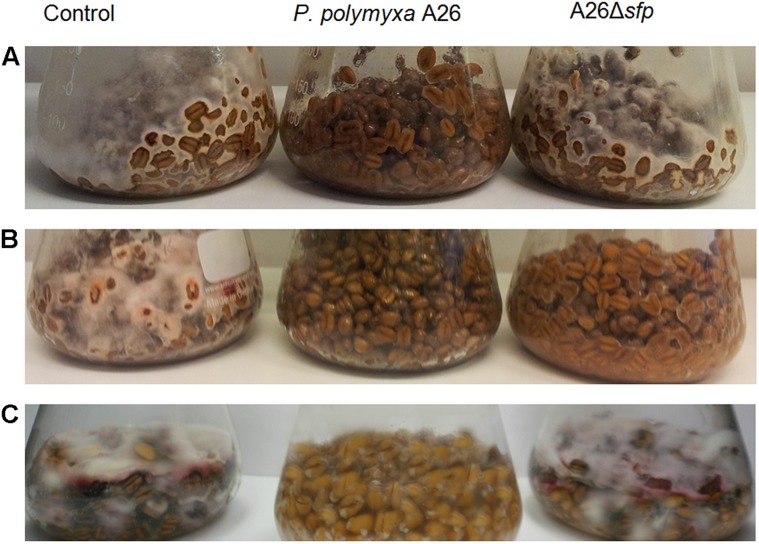
Visual inspection of wheat grains over the experimental period revealed increased amounts of F. culmorum and F. graminearum mycelia in the pathogen control treatment (Figure 1). No fungal mycelia was observed on wheat grains treated with P. polymyxa A26 (Figure 1). The visual observations were followed by pathogen DNA and mycotoxin monitoring. Infection of wheat grains with the pathogens resulted in high DNA and mycotoxin levels (Tables 3 and 4). In the absence of bacteria up to 260 and 382 ng pathogen DNA/ng wheat DNA were detected after 15 days for F. culmorum and F. graminearum, respectively, (Table 3). High levels of DON (up to 6.85 mg/kg) were detected in F. graminearum samples, while F. culmorum strain did not produce any detectable levels of DON (Table 4). Accumulation of high levels (up to 61.2 mg/kg) of ZEA were recorded in grains infected with either F. graminearum or F. culmorum (Table 4). No F. culmorum and F. graminearum DNA nor mycotoxins were detected in P. polymyxa A26 treated wheat grains, respectively, (Tables 3 and 4). The ability of P. polymyxa A26 to inhibit F. graminearum growth in wheat grains was significantly compromised by its Sfp-type PPTase inactivation. As shown in Figure 1, a considerable growth of F. graminearum could be detected in wheat grains treated with A26Δsfp after 15 days. We also detected significant levels of F. graminearum DNA (62.66 ng fungal DNA/ng wheat DNA) after 15 days of fungal infection (Table 3). Despite very low levels of fungal DNA detected after 5 and 10 days, significant levels of both mycotoxins DON and ZEA were detected at all time points.
FIGURE 1.
Fusarium graminearum and F. culmorum antagonism in wheat kernel assay. F. graminearum growth in wheat grains inoculated with Paenibacillus polymyxa A26 or A26Δsfp (A), F. culmorum inoculated with P. polymyxa A26, A26Δsfp (B) and F. culmorum treated with P. polymyxa A26, A26Δsfp culture filtrates (C) after 15 days incubation.
Table 3.
Quantitative PCR (qPCR) analysis for F. culmorum and F. graminearum DNA (ng fungal DNA/ng wheat DNA).
| Treatments | Pathogen DNA (ng Fungal DNA/ng Wheat DNA) |
||
|---|---|---|---|
| 5 days | 10 days | 15 days | |
| F. culmorum | 206.3 ± 28.9a | 197.0 ± 62.6a | 260.7 ± 70.6a |
| A26 + F. culmorum | NDb | NDb | NDb |
| A26Δsfp + F. culmorum | NDb | NDb | NDb |
| F. graminearum | 42.76 ± 10.9a | 82.44 ± 24.4b | 382.38 ± 36.6c |
| A26 + F. graminearum | NDd | NDd | NDd |
| A26Δsfp + F. graminearum | 0.02d | 0.03d | 62.66 ± 17.4e |
Fungal DNA was detected at three time points (5, 10, and15 days) after infection. DNA was extracted from samples collected from A26 and A26Δsfp treated as well as untreated wheat grains. Different letters indicate statistically significant differences (P ≤ 0.01) within all-time points for each pathogen, based on the LSD test.
ND, not detected.
Table 4.
Deoxynivalenol (DON) and zearalenone (ZEA) contents (mg/kg) in wheat grains inoculated with F. culmorum and F. graminearum; Both DON and ZEA were detected at three times points (5, 10, and 15 days) after fungal infection.
| Treatments | Deoxynivalenol (DON) mg/kg |
Zearalenone (ZEA) mg/kg |
||||
|---|---|---|---|---|---|---|
| 5 days | 10 days | 15 days | 5 days | 10 days | 15 days | |
| P. polymyxa A26 + F.graminearum | NDf | NDf | NDf | NDe | NDe | NDe |
| P. polymyxa A26Δsfp + F. graminearum | 1.5e | 0.3e | 0.7e | 0.24de | 0.41d | 0.27de |
| F. graminearum | 6.9d | 2.7b | 1.2c | 5.93b | 1.73c | 10.5a |
| P. polymyxa A26 + F. culmorum | 0 | 0 | 0 | NDc | NDc | NDc |
| P. polymyxa A26Δsfp + F. culmorum | 0 | 0 | 0 | NDc | NDc | NDc |
| F. culmorum | 0 | 0 | 0 | 53.7b | 61.2a | 55b |
| Culture filtrate treatments | ||||||
| P. polymyxa A26 + F. culmorum | 0 | 0 | 0 | NDb | NDb | NDb |
| P. polymyxa A26Δsfp + F. culmorum | 0 | 0 | 0 | 45.5a | 54.1a | 48.0a |
| F. culmorum | 0 | 0 | 0 | 49.4a | 52.8a | 49.7a |
The results represent the means of two biological repeats. Different letters indicate statistically significant differences (P ≤ 0.01) within all-time points for a single mycotoxin, based on the LSD test.
ND, not detected.
Unlike what was seen on the plate assays, A26 Sfp-type PPTase mutant antagonized F. culmorum. Neither pathogen DNA nor mycotoxins were detected F. culmorum DNA in A26Δsfp treated wheat grains (Figure 1; Tables 3 and 4).
Paenibacillus polymyxa A26 and A26Δsfp Culture Filtrate Antagonism Against F. culmorum on Wheat Grains
As A26Δsfp incubation with F. culmorum resulted in significant antagonistic ability, further studies were performed to study the effect of culture filtrates from the wild-type and mutant versions of strain A26. Culture filtrates from wild-type A26 efficiently antagonized the pathogen while the A26Δsfp culture filtrated had no significant effect on pathogen growth and mycotoxin production (Figure 1; Table 4).
Bacterial Growth
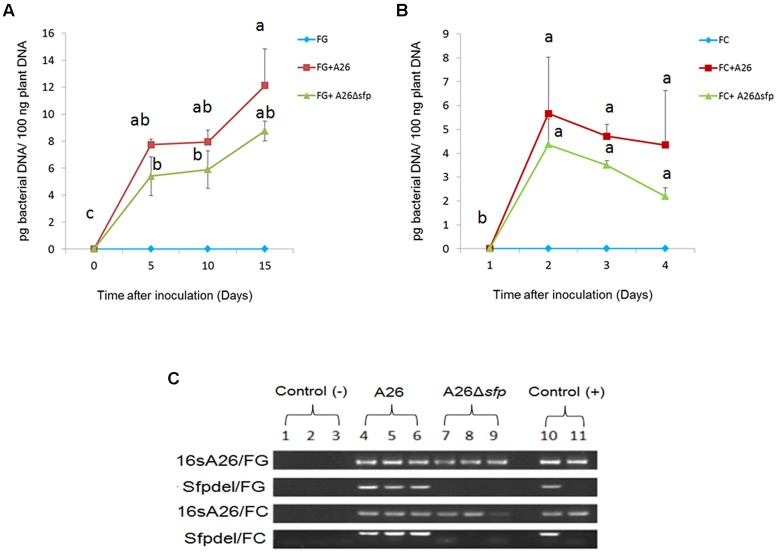
Bacterial DNA in wheat kernel assay was monitored at all three time points using qPCR (Figure 2). Increasing the incubation time did not lead to a significant increase in DNA levels after day 5. The only exception was observed on F. graminearum treated kernels on day 15 then 12.12 pg bacterial DNA/100 ng plant DNA was detected (Figure 2). Using specific PCR primers (Table 1) we confirmed the stability of the Sfp-type PPTase mutation at all-time points (Figure 2B).
FIGURE 2.
A26 and A26Δsfp quantification in wheat kernel assay. QPCR quantification of bacterial DNA extracted from wheat grains inoculated with A26 and A26Δsfp as well as un inoculated wheat grains after 5, 10, and 15 days (A) F. graminearum and (B) F. culmorum; (pg bacterial DNA/100 ng plant DNA). Data shown as a means of two experiments; Bars represents SD; Different letters indicate statistically significant differences (P ≤ 0.01) based on the LSD test. (C) PCR analysis for bacterial DNA using 16S A26 primers (identifying both A26 and A26Δsfp) and sfpdel primers (identifying only A26). DNA extract from pure cultures of A26 and A26Δsfp was used as a positive control while DNA extracted from untreated wheat grains was used as a negative control.
Discussion
Different strategies have been used to manage FHB (Wegulo, 2012). The challenge with using biocontrol bacteria for this purpose is finding and maintaining effective strains for field applications and monitoring the strains active principle(s) under field conditions. The dual culture plate antagonism assay has often been used as a fast method for screening BCAs in vitro. Here we used King’s B medium to compare the antagonistic abilities of a NFS strain E1, SFS strain A26, and A26 Sfp-type PPTase mutant (Table 2). The rationale for using A26 strain is that rhizobacteria from harsh environments were previously shown to be effective at enhancing the drought tolerance of host plants (Timmusk et al., 2014a). The same strains were superior in FHB pathogens biocontrol field assays (Timmusk et al., 2014b). Both abilities are likely a consequence of both ACC deaminase activity and the production of broad range of biologically active compounds by the harsh environment isolates (Glick, 2014 and manuscript in preparation). The results presented here show that SFS strain A26 is highly effective in antagonizing F. graminearum and F. culmorum on the plate assay in comparison to the NFS strain E1 that was tested (Table 2). The A26 strain which shows high antagonistic activity on plate assays, was mutagenized to create a deletion its Sfp-type PPTase gene (Timmusk et al., 2015). The A26 Sfp-type PPTase mutant lost its ability to antagonize both pathogens on the plate assay. This result is expected considering that none of the NRPS PKS lipopeptide antibiotics are produced by the mutant strain, and it confirms the role of the compounds in the antagonism (Timmusk et al., 2015). Then the wild-type and mutant strains were studied further in a gnotobiotic system on wheat kernels (Figure 1). Compared to plate assays, the system provides a surface for colonization as well as nutrition source that might be used by both the pathogen and the BCA under field conditions. It also allows simultaneous quantitative monitoring of pathogen, BCA A26, and A26Δsfp as well as mycotoxin production. In this system, A26 showed full inhibition of F. culmorum and F. graminearum by day 5, which did not change during the course of the 15 day studies (Table 3; Figure 1). The antagonism was followed by qPCR of pathogen, A26 and mycotoxins LC-MS/MS assay which confirm that no pathogens DNA nor mycotoxins were present in the system at any time points (Figure 2; Tables 3 and 4). The mutant, A26Δsfp lost its ability to antagonize F. graminearum, which is the expected result as none of the NRPS PKS lipopeptide antibiotics are produced by the mutant strain (Figure 1; Tables 3 and 4; Timmusk et al., 2015). Direct antagonism of pathogens is widely considered as the most powerful mechanism employed by soil bacteria against pathogens (Cawoy et al., 2014). B. subtilis, the best studied BCA most frequently reported biocontrol mechanisms are connected to non-ribosomally produced cyclic lipopeptides (Ongena and Jacques, 2008; Perez-Garcia et al., 2011; Zeriouh et al., 2011). Lipopeptides which are amphiphilic molecules with an amino or hydroxy-fatty acid integrated into a peptide moiety, interact with the biological membranes of microbial pathogens, including cell leakage and death (Zeriouh et al., 2011). It’s estimated that some Bacillus and Paenibacillus species devote from 4 to 8% of their genomes to synthesize antibiotics (Cawoy et al., 2014). An examination of the A26 genome indicates that polymyxins, fusaricidins as well as quite a number of potentially new non-ribosomal lipopeptides/antibiotics are mediated by its Sfp-type PPTase. Hence, in the future it is necessary to identify the key regions in the respective synthetase gene clusters and perform knockouts of all known and potential antibiotic candidates (Timmusk et al., 2015). While strain A26Δsfp in a great deal lost its antagonistic ability against F. graminearum, the mutant still efficiently antagonized F. culmorum (Figure 1; Tables 3 and 4). On the other hand, a cell free culture supernatant assay showed that the culture filtrates of the mutant were unable to antagonize the pathogen in the kernel assay (Figure 1; Table 4). What mechanism could explain the different effect of the mutant strain in the kernel assay? The Sfp-type PPTase activates peptidyl carrier protein domains (Quadri et al., 1998; Beld et al., 2014; Bunet et al., 2014). During the compound assembly, the biosynthesis intermediates are attached to carrier protein domains of NRPS and PKS via a phosphopantetheinyl arm. The post-translational modification of the domains with 4′-phosphopantetheyinyl as catalyzed by Sfp-type PPTase is crucial for the activation of NRPS and PKS (Beld et al., 2014; Bunet et al., 2014). The mutant A26Δsfp, which is incapable of producing the enzymatically active 4′ phosphopantetheinyl transferase, in turn results in a P. polymyxa A26 mutant strain lacking enzymatically active NRPS and PKS and lipopeptide production (Timmusk et al., 2015). Using specific PCR primers (Table 1) we regularly confirmed the stability of the mutant and wild-type strain, (Figure 2C). These results indicate that fusaricidins, generally believed to be the mechanism of P. polymyxa action against Fusarium sp. may not be the only mechanism functioning on the A26 in biocontrol of F. culmorum. A26Δsfp culture filtrates failed to antagonize F. culmorum in kernel assay in contrast to A26Δsfp cells treatment then we detected the antagonism against the pathogen (Figure 1). This indicates biofilm involvement in the antagonism and is in accordance with our former findings showing that niche exclusion, i.e., antagonist biofilm occupation of the pathogen colonization sites, is responsible for biocontrol (Timmusk et al., 2003, 2009b; Haggag and Timmusk, 2008). Microbial biofilms are comprised of cells and extracellular matrix and can produce a protective layer around infection sites. The dense biofilm matrix limits diffusion of compounds secreted by bacteria and these are therefore concentrated at pathogen infection sites of action. This knowledge is important to be taken into consideration for further selection of biocontrol agents in field conditions where the F. culmorum content is estimated to be high. In parallel with selection of the highest lipopeptide producers, the isolate’s biofilm production should be taken into consideration.
While A26 Sfp-type PPTase mutant fully antagonized F. culmorum and F. graminearum growth was also decreased in the kernel assay first two time points (Table 3). At the same time even if only low levels of the pathogens were detected, significant amounts of both mycotoxins were produced by the pathogen at the times points (Table 4). This confirms the importance of parallel monitoring of pathogen growth and mycotoxins produced.
In the work reported here we established a progressive screening method for FHB BCAs: plate assays combined with a wheat kernel assay. The system allows robust screening of high number of P. polymyxa isolates from harsh environments (Timmusk et al., 2011, 2014a). In the A26 case, plate assays with wild type and its Sfp-type PPTase mutant confirm that NRPS/PKS products have critical importance for the strain antagonistic ability (Table 2). Studies with the more complex system employing wheat kernels show, however, that in case of F. culmorum the BCA-enhanced biofilm formation may be of major importance in antagonizing the pathogen. Simultaneous qPCR monitoring of A26 and pathogens combined with mycotoxin assays supports this finding (Figure 2 2; Tables 3 and 4). The system will be used now to monitor the efficiency of A26 formulation strategies. Moreover, our results confirm that external complementation with A26 metabolite extracts efficiently restores its wild type biofilm formation level indicting the Sfp-type PPTase mediated NRPS/PKS compounds direct involvement in A26 biofilm formation (Timmusk et al., 2015). Dual culture plate and kernel assays with additional knockout mutants of single NRPS/PKS compounds should reveal which of the lipopeptides are active in the observed biofilm formation and antagonistic activity. The qPCR monitoring optimized for the A26 can now be used with the formulated agent monitoring on field application. Furthermore, in parallel with the BCA monitoring the system for the lipopeptide monitoring proven to be involved in A26 biofilm formation and antagonism will be designed. To maintain the BCA active principles during storage is one of the challenges with biocontrol strains. Therefore it is quite important to have a simple system for routine screening of stored A26 and further FHB active BCAs.
It is clear that more studies need to be performed for strain A26 prior to field application. However, the results obtained so far show the importance of combining the strategies described here in order to develop efficient FHB biocontrol agents for field application.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Prof. B. R. Glick is gratefully acknowledged for critically reading the manuscript. The work was performed with financial support from Stiftelsen Oscar och Lili Lamms Minne and FORMAS 222-2014-1326 (to ST).
References
- Ausubel M., Brent R., Kingston E., Moore D. D., Seidman G., Smith J. A., et al. (1988). Current Protocols in Molecular Biology. New York, NY: Wiley Interscience. [Google Scholar]
- Beld J., Sonnenschein E. C., Vickery C. R., Noel J. P., Burkart M. D. (2014). The phosphopantetheinyl transferases: catalysis of a post-translational modification crucial for life. Nat. Prod. Rep. 31 61–108. 10.1039/C3NP70054B [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunet R., Riclea R., Laureti L., Hotel L., Paris C., Girardet J. M., et al. (2014). A single Sfp-type phosphopantetheinyl transferase plays a major role in the biosynthesis of PKS and NRPS derived metabolites in Streptomyces ambofaciens ATCC23877. PLoS ONE 9:e87607 10.1371/journal.pone.0087607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cawoy H., Mariutto M., Henry G., Fisher C., Vasilyeva N., Thonart P., et al. (2014). Plant defense stimulation by natural isolates of bacillus depends on efficient surfactin production. Mol. Plant Microbe Interact. 27 87–100. 10.1094/MPMI-09-13-0262-R [DOI] [PubMed] [Google Scholar]
- Cowger C., Arellano C. (2013). Fusarium graminearum infection and deoxynivalenol concentrations during development of wheat spikes. Phytopathology 103 460–471. 10.1094/PHYTO-03-12-0054-R [DOI] [PubMed] [Google Scholar]
- Fredlund E., Gidlund A., Sulyok M., Borjesson T., Krska R., Olsen M., et al. (2013). Deoxynivalenol and other selected Fusarium toxins in Swedish oats–occurrence and correlation to specific Fusarium species. Int. J. Food Microbiol. 167 276–283. 10.1016/j.ijfoodmicro.2013.06.026 [DOI] [PubMed] [Google Scholar]
- Glick B. R. (2014). Bacteria with ACC deaminase can promote plant growth and help to feed the world. Microbiol. Res. 169 30–39. 10.1016/j.micres.2013.09.009 [DOI] [PubMed] [Google Scholar]
- Haggag W. M., Timmusk S. (2008). Colonization of peanut roots by biofilm-forming Paenibacillus polymyxa initiates biocontrol against crown rot disease. J. Appl. Microbiol. 104 961–969. 10.1111/j.1365-2672.2007.03611.x [DOI] [PubMed] [Google Scholar]
- Kim S.-B., Timmusk S. (2013). A simplified method for Paenibacillus polymyxa gene knockout and insertional screening. PLoS ONE 8:e68092 10.1371/journal.pone.0068092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leblanc C., Prudhomme T., Tabouret G., Ray A., Burbaud S., Cabantous S., et al. (2012). 4’-Phosphopantetheinyl transferase PptT, a new drug target required for Mycobacterium tuberculosis growth and persistence in vivo. PLoS Pathog. 8:e1003097 10.1371/journal.ppat.1003097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindblad M., Gidlund A., Sulyok M., Borjesson T., Krska R., Olsen M., et al. (2013). Deoxynivalenol and other selected Fusarium toxins in Swedish wheat–occurrence and correlation to specific Fusarium species. Int. J. Food Microbiol. 167 284–291. 10.1016/j.ijfoodmicro.2013.07.002 [DOI] [PubMed] [Google Scholar]
- Matarese F., Sarrocco S., Gruber S., Seidl-Seiboth V., Vannacci G. (2012). Biocontrol of Fusarium head blight: interactions between Trichoderma and mycotoxigenic Fusarium. Microbiology 158 98–106. 10.1099/mic.0.052639-0 [DOI] [PubMed] [Google Scholar]
- Mootz H. D., Finking R., Marahiel M. A. (2001). 4’-phosphopantetheine transfer in primary and secondary metabolism of Bacillus subtilis. J. Biol. Chem. 276 37289–37298. 10.1074/jbc.M103556200 [DOI] [PubMed] [Google Scholar]
- Nazari L., Pattori E., Terzi V., Morcia C., Rossi V. (2014). Influence of temperature on infection, growth, and mycotoxin production by Fusarium langsethiae and F. sporotrichioides in durum wheat. Food Microbiol. 39 19–26. 10.1016/j.fm.2013.10.009 [DOI] [PubMed] [Google Scholar]
- Nicolaisen M., Suproniene S., Nielsen L. K., Lazzaro I., Spliid N. H., Justesen A. F. (2009). Real-time PCR for quantification of eleven individual Fusarium species in cereals. J. Microbiol. Methods 76 234–240. 10.1016/j.mimet.2008.10.016 [DOI] [PubMed] [Google Scholar]
- Njobeh P. B., Dutton M., Tevell Åberg A., Haggblom P. (2012). Estimation of multi-mycotoxin contamination in south African compound feeds. Toxins 4 836–848. 10.3390/toxins4100836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nygren C. M., Eberhardt U., Karlsson M., Parrent J. L., Lindahl B. D., Taylor A. F. (2008). Growth on nitrate and occurrence of nitrate reductase-encoding genes in a phylogenetically diverse range of ectomycorrhizal fungi. New Phytol. 180 875–889. 10.1111/j.1469-8137.2008.02618.x [DOI] [PubMed] [Google Scholar]
- Ongena M., Jacques P. (2008). Bacillus lipopeptides: versatile weapons for plant disease biocontrol. Trends Microbiol. 16 115–125. 10.1016/j.tim.2007.12.009 [DOI] [PubMed] [Google Scholar]
- Perez-Garcia A., Romero D., de Vicente A. (2011). Plant protection and growth stimulation by microorganisms: biotechnological applications of Bacilli in agriculture. Curr. Opin. Biotechnol. 22 187–193. 10.1016/j.copbio.2010.12.003 [DOI] [PubMed] [Google Scholar]
- Pieters M. N., Freijer J., Baars B. J., Fiolet D. C., van Klaveren J., Slob W. (2002). Risk assessment of deoxynivalenol in food: concentration limits, exposure and effects. Adv. Exp. Med. Biol. 504 235–248. 10.1007/978-1-4615-0629-4_25 [DOI] [PubMed] [Google Scholar]
- Quadri L. E., Weinreb P. H., Lei M., Nakano M. M., Zuber P., Walsh C. T. (1998). Characterization of Sfp, a Bacillus subtilis phosphopantetheinyl transferase for peptidyl carrier protein domains in peptide synthetases. Biochemistry 37 1585–1595. 10.1021/bi9719861 [DOI] [PubMed] [Google Scholar]
- Rozen S., Skaletsky H. (2000). Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132 365–386. 10.1385/1-59259-192-2:365 [DOI] [PubMed] [Google Scholar]
- Scherm B., Balmas V., Spanu F., Pani G., Delogu G., Pasquali M., et al. (2013). Fusarium culmorum: causal agent of foot and root rot and head blight on wheat. Mol. Plant Pathol. 14 323–341. 10.1111/mpp.12011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah D. A., Molineros J. E., Paul P. A., Willyerd K. T., Madden L. V., De Wolf E. D. (2013). Predicting Fusarium head blight epidemics with weather-driven pre- and post-anthesis logistic regression models. Phytopathology 103 906–919. 10.1094/PHYTO-11-12-0304-R [DOI] [PubMed] [Google Scholar]
- Shetty P. H., Hald B., Jespersen L. (2007). Surface binding of aflatoxin B1 by Saccharomyces cerevisiae strains with potential decontaminating abilities in indigenous fermented foods. Int. J. Food Microbiol. 113 41–46. 10.1016/j.ijfoodmicro.2006.07.013 [DOI] [PubMed] [Google Scholar]
- Shi C., Yan P., Li J., Wu H., Li Q., Guan S. (2014). Biocontrol of Fusarium graminearum growth and deoxynivalenol production in wheat kernels with bacterial antagonists. Int. J. Environ. Res. Public Health 11 1094–1105. 10.3390/ijerph110101094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stansly P. G., Schloesser E. (1947). Studies on polymyxin: isolation and idendification of Bacillus polymyxa and differentiation of polymyxin from certain known antibiotics. J. Bacteriol. 54 549–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tevell Åberg A., Solyakov A., Bondesson U. (2012). Development and in-house validation of an LC-MS/MS method for the simultaneous quantification of the mycotoxins deoxynivalenol, zearalenone, T-2 and HT-2 toxin, ochratoxin A, and fumonisin B1 and B2 in vegetable animal feed. Food Addit. Contam. Part A Chem. Anal. Control Expo. Risk Assess. 30 541–549. 10.1080/19440049.2012.760208 [DOI] [PubMed] [Google Scholar]
- Timmusk S., El Daim I., Copolovici L., Kannaste A., Behers L., Nevo E., et al. (2014a). Drought-tolerance of wheat improved by rhizosphere bacteria from harsh environments: enhanced biomass production and reduced emissions of stress volatiles. PLoS ONE 9:e96086 10.1371/journal.pone.0096086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmusk S., Kim S., Behers L., Nevo E., Häggblom P. (2014b). “Development of a sustainable method for myxotoxin prevention in cereals,” in Poster XIII Meeting of the IOBC Biocontrol of Plant Diseases 2014, Uppsala: (accessed June 15–18, 2014). [Google Scholar]
- Timmusk S., Kim S., Nevo E., Abd El Daim I., Ek B., Bergquist J., et al. (2015). Sfp- type PPTase inactivation promotes bacterial biofilm formation and ability to enhance plant drought tolerance. Front. Microbiol. 6:387 10.3389/fmicb.2015.00387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmusk S., Paalme V., Lagercrantz U., Nevo E. (2009a). Detection and quantification of Paenibacillus polymyxa in the rhizosphere of wild barley (Hordeum spontaneum) with real-time PCR. J. Appl. Microbiol. 107 736–745. 10.1111/j.1365-2672.2009.04265.x [DOI] [PubMed] [Google Scholar]
- Timmusk S., van West P., Gow N. A., Huffstutler R. P. (2009b). Paenibacillus polymyxa antagonizes oomycete plant pathogens Phytophthora palmivora and Pythium aphanidermatum. J. Appl. Microbiol. 106 1473–1481. 10.1111/j.1365-2672.2009.04123.x [DOI] [PubMed] [Google Scholar]
- Timmusk S., Paalme V., Pavlicek T., Bergquist J., Vangala A., Danilas T., et al. (2011). Bacterial distribution in the rhizosphere of wild barley under contrasting microclimates. PLoS ONE 6:e17968 10.1371/journal.pone.0017968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmusk S., van West P., Gow N. A. R., Wagner E. G. H. (2003). “Antagonistic effects of Paenibacillus polymyxa towards the oomycete plant pathogens Phytophthora palmivora and Pythium aphanidermatum,” in Mechanism of Action of the Plant Growth Promoting Bacterium Paenibacillus polymyxa, ed. Timmusk S. (Uppsala: Department of Cell and Moleculer Biology, Uppsala University; ), 21–29. [Google Scholar]
- Walter S., Nicholson P., Doohan F. M. (2010). Action and reaction of host and pathogen during Fusarium head blight disease. New Phytol. 185 54–66. 10.1111/j.1469-8137.2009.03041.x [DOI] [PubMed] [Google Scholar]
- Wegulo S. N. (2012). Factors influencing deoxynivalenol accumulation in small grain cereals. Toxins 4 1157–1180. 10.3390/toxins4111157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J., Jin X., Jia X., Wang H., Cao A., Zhao W., et al. (2013). Transcriptome-based discovery of pathways and genes related to resistance against Fusarium head blight in wheat landrace Wangshuibai. BMC Genomics 14:197 10.1186/1471-2164-14-197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazar S., Omurtag G. Z. (2008). Fumonisins, trichothecenes and zearalenone in cereals. Int. J. Mol. Sci. 9 2062–2090. 10.3390/ijms9112062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeriouh H., Romero D., Garcia-Gutierrez L., Cazorla F. M., de Vicente A., Perez-Garcia A. (2011). The iturin-like lipopeptides are essential components in the biological control arsenal of Bacillus subtilis against bacterial diseases of cucurbits. Mol. Plant Microbe Interact. 24 1540–1552. 10.1094/MPMI-06-11-0162 [DOI] [PubMed] [Google Scholar]
- Zheng D., Burr T. J. (2013). An Sfp-type PPTase and associated polyketide and nonribosomal peptide synthases in Agrobacterium vitis are essential for induction of tobacco hypersensitive response and grape necrosis. Mol. Plant Microbe Interact. 26 812–822. 10.1094/MPMI-12-12-0295-R [DOI] [PubMed] [Google Scholar]