Abstract
The assessment of Burkholderia diversity in agricultural areas is important considering the potential use of this genus for agronomic and environmental applications. Therefore, the aim of this work was to ascertain how plant species and land use management drive the diversity of the genus Burkholderia. In a greenhouse experiment, different crops, i.e., maize, oat, barley, and grass, were planted in pots containing soils with different land use histories, i.e., maize monoculture, crop rotation, and permanent grassland, for three consecutive growth cycles. The diversity of Burkholderia spp. in the rhizosphere soil was assessed by genus-specific PCR-denaturing gradient gel electrophoresis (DGGE) and analyzed by canonical correspondence analysis (CCA). CCA ordination plots showed that previous land use was the main factor affecting the composition of the Burkholderia community. Although most variation in the Burkholderia community structure was observed between the permanent grassland and agricultural areas, differences between the crop rotation and maize monoculture groups were also observed. Plant species affected Burkholderia community structure to a lesser extent than did land use history. Similarities were observed between Burkholderia populations associated with maize and grass, on the one hand, and between those associated with barley and oat, on the other hand. Additionally, CCA ordination plots demonstrated that these two groups (maize/grass versus barley/oat) had a negative correlation. The identification of bands from the DGGE patterns demonstrated that the species correlated with the environmental variables were mainly affiliated with Burkholderia species that are commonly isolated from soil, in particular Burkholderia glathei, B. caledonica, B. hospita, and B. caribiensis.
The genus Burkholderia was created in 1992, when Yabuuchi et al. (41) reclassified Pseudomonas species belonging to rRNA group II. Since then, this genus has undergone many changes. It now comprises over 30 species, including the Burkholderia cepacia complex, which consists of nine so-called genomovars (7). Many species have potential for agricultural or environmental purposes, such as biological control, bioremediation, atmospheric nitrogen fixation, and plant growth stimulation (11, 12, 16, 38). Moreover, the ability of these microorganisms to colonize the rhizosphere of plants such as maize, wheat, rice, grass, oat, lupine, and coffee at high population densities might expand their potential applications (3, 8, 12, 39). Despite the great agronomic potential of Burkholderia spp., there is general concern about the environment functioning as a source of human-pathogenic organisms, mainly after B. cenocepacia (genomovar III), which is associated with cepacia syndrome in cystic fibrosis patients, was found as a common plant-associated bacterium (3). Although this finding highlighted the importance of assessing the diversity of Burkholderia species in the rhizosphere, most current ecological knowledge is based on B. cepacia populations isolated from the rhizosphere of just one plant species, maize (5, 10, 13). Recently, the diversity of the Burkholderia community associated with woodland rhizospheres was also assessed (27). However, these reports were based on culture-dependent techniques, which are likely to underestimate the natural bacterial population (23). In order to overcome this problem, we developed a method, based on PCR-denaturing gradient gel electrophoresis (PCR-DGGE), which allows the direct analysis of the diversity of Burkholderia species in environmental samples (30).
Plant roots play important roles in shaping microbial communities in soil by releasing a wide range of compounds. Although root-released products comprise an important pool of organic compounds for soil microorganisms, their composition and quality can vary according to the plant species, the soil type, and the plant developmental stage (33, 40). Due to this variation in exudation, different plant species growing in the same soil type are known to select divergent bacterial communities (19, 20, 40). However, when the microbial communities associated with the same plant species growing in different soil types are analyzed, the soil type may exert a large influence on microbial diversity (9, 40).
In view of the fact that plants have a large impact on microbial diversity, one might expect agricultural management to play an important role as well. Indeed, many agricultural practices, such as crop rotation, continuous cropping, and tillage, induce changes in microbial communities in soil (2, 20, 39) which may persist long after the management practice took place (4). Although agricultural practices induce general changes in soil microbial communities, specific microbial groups may respond differently. Clegg et al. (6) showed that the application of inorganic nitrogen had a significant impact on eubacterial and actinomycete community structures, whereas soil drainage significantly affected the community structures of actinomycetes and pseudomonads. In addition, continuous wheat cropping affected the community structure of pseudomonads, such that an increase in the population of antibiotic-producing Pseudomonas spp. induced the natural suppression of Take-all disease in wheat (26). Similarly, the establishment of apple orchards in a field where wheat had previously been grown led to an increase in suppressiveness against Rhizoctonia solani which was correlated with an increase in the B. cepacia and Pseudomonas putida populations (22).
Considering the fact that agricultural management and plant species affect soil microbial communities, the main objective of this work was to gain a better understanding of how land use and crop species, specifically maize, oat, grass, and barley, affect the diversity of the genus Burkholderia. In addition, this study aimed to address which Burkholderia species are selected by specific crops and which factor (plant species or land use) has a larger influence on soilborne populations. To assess the diversity of Burkholderia species in soil, we applied a PCR-DGGE system with primers specific for this genus (30), which allowed for the evaluation of the total Burkholderia population, including the nonculturable fraction.
MATERIALS AND METHODS
Microcosm experiment.
In order to evaluate the effect of different plant species on the diversity of Burkholderia species, we designed a pot experiment in the greenhouse. The treatments consisted of four plant species, namely maize (Zea mays L.), oat (Avena sativa L.), barley (Hordeum vulgare L.), and grass (a commercial mix containing Lolium perenne as the main species), planted in replicate pots containing soils with three different land use histories.
Soil.
The soils used for this experiment were collected from different locations (according to land use) in a field (Wildekamp) located in Wageningen, The Netherlands. The soil was a loamy sand (3% clay, 10% silt, and 87% sand) with about 2% organic matter and a pH of 4.8. The site was composed originally of a long-term (>50 years) permanent grassland (G) field that was partially converted to agricultural land (A) about 20 years ago, with the latter being used mainly for crop rotation. In 2000, different treatments were established in both areas (G and A) before the growing season, using triplicate (10 by 10 m) plots for each treatment in a randomized block design. The treatments comprised 4-year crop rotation (oat, maize, barley, and potato), a monoculture of maize, and grassland. For the greenhouse experiment, we decided to focus mainly on the differences between permanent grassland (G), arable land under crop rotation (A-R), and arable land under maize monoculture (A-M). Therefore, at the end of the growing season of 2001, soil was collected from each triplicate plot of treatments G, A-R, and A-M. At that time, the plots undergoing the crop rotation treatment had only had the first two crops (oats and maize). From this point on, the treatments mentioned above will be referred to as land use histories.
Experimental design.
Soil collected from each triplicate plot of the three different land use histories (G, A-R, and A-M) was sieved and homogenized separately, and approximately 600 g (500 ml) was then transferred to a pot in the greenhouse. Pregerminated seeds (three) of each plant species (oat, maize, and barley) were transferred to the pots, in three replicates per crop. For the pots containing grass, approximately 300 mg of nongerminated seeds were used. As controls, two pots per land use history were kept fallow.
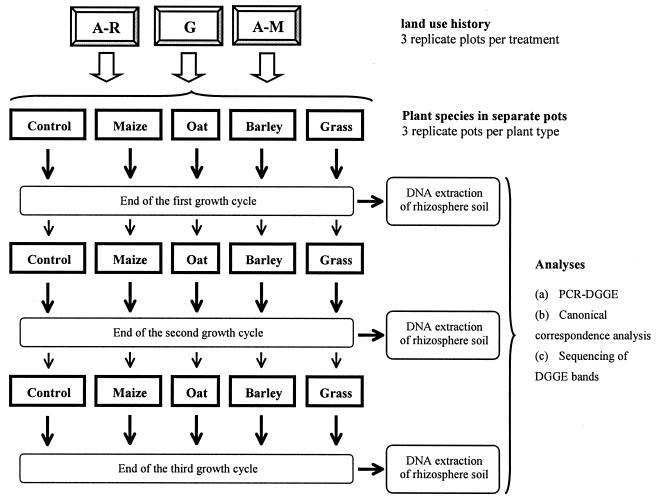
In order to enhance the rhizosphere effect of each plant species, at the end of the first growth cycle we removed the plants, homogenized the soil contained in each pot separately, and transferred new seedlings (corresponding to the previous plant species) into the soil in the pots. This procedure was repeated one more time, thus producing samples from the second and third growth cycles (Fig. 1).
FIG. 1.
Schematic representation of the microcosm experiment. Soils collected from field plots with different cropping histories were distributed in pots, and four plant species were planted. At the end of the growth cycles, the plants were removed from the pots and rhizosphere soil was collected for further DNA extraction. Subsequently, the bulk soil from each pot was mixed separately and new seedlings from the previous plant species were transferred to the pot. A-R, arable land under crop rotation; A-M, arable land under maize monoculture; G, permanent grassland.
Rhizosphere soil sampling.
At the end of each growth cycle (approximately 4 months after sowing), plants were removed from the pots and the roots were shaken gently, removing the loosely adhering soil. Twenty grams of roots containing tightly adhering soil (rhizosphere soil) was transferred to an Erlenmeyer flask containing 90 ml of sterile sodium pyrophosphate (0.1%) and gravel (10 g). After the flasks were shaken for 30 min at 180 rpm, 2 ml of solution containing the rhizosphere soil was used for DNA extraction.
DNA extraction.
DNA from rhizosphere soil was extracted by use of an UltraClean soil DNA isolation kit (Mo Bio Laboratories, BIOzymTC, Landgraaf, The Netherlands). Briefly, 0.5 ml of sodium pyrophosphate solution containing rhizosphere soil and 50 mg of glass beads (≤106-μm diameter) were added to microcentrifuge tubes, and the cells were lysed by bead beating for 60 s in a cell disrupter (Ribolyser; Hybaid, Middlesex, United Kingdom) in order to achieve maximal cell lysis. After the bead-beating step, DNA was extracted according to the protocol described by the supplier.
PCR amplification of partial 16S ribosomal RNA genes of Burkholderia.
The amplification of 16S rRNA genes from rhizosphere soil DNA was done by using primers specific for the genus Burkholderia in a seminested PCR performed according to the methodology described by Salles et al. (30). Briefly, the PCR procedure consisted of a first PCR with primer Burk3 used in combination with the universal eubacterial primer R1378 (17) under the conditions described by Rosado et al. (28). The products from the first PCR were diluted 1:1,000 and used as the template in a second PCR, which was performed with primers Burk3 (GC clamped) and BurkR, as follows. Fifty-microliter reaction mixtures contained 1 μl of DNA (5 to 10 ng), 200 μmol of each deoxyribonucleoside triphosphate per liter, 400 nmol of each primer per liter, 1× TaqPlus Precision buffer (Stratagene, Leusden, The Netherlands), and 2 U of TaqPlus Precision polymerase mixture (Stratagene). Amplification was performed in a PTC-100 thermal cycler (MJ Research, Inc., Tilburg, The Netherlands) (30), and the PCR products, expected to be approximately 500 bp long, were analyzed by electrophoresis in a 1.5% (wt/vol) agarose gel in 0.5× Tris-borate-EDTA buffer (31). When necessary, products were stored at −20°C before they were used for DGGE analysis.
DGGE.
DGGE analysis was performed by using the phorU2 system (Ingeny, Goes, The Netherlands) and the method described by Salles et al. (30). After electrophoresis, the gels were stained with the SYBR Gold I nucleic acid gel stain (Molecular Probes Europe, Leiden, The Netherlands), photographed, and digitized with an Imago compact apparatus (B&L Systems, Maarssen, The Netherlands).
Banding pattern analysis and statistics.
DGGE banding patterns were analyzed with Molecular Analyst software (version 1.61; Bio-Rad, Veenendaal, The Netherlands). In order to compensate for internal distortions during electrophoresis, we aligned the gels by using an external reference pattern. The pattern was composed of pooled PCR products from four Burkholderia species loaded in at least four different lanes distributed along the gel. Subsequently, subtraction of the nonlinear background was achieved by using the rolling disk mechanism with an intensity of 8. For the completion of the gel analysis, identification and quantification of the bands present in each lane were performed by setting the tolerance and optimization at 0.75%. A table containing the calculated surface and position of each band was then exported to Excel, in which the band surface was converted to the relative intensity of the band per lane, with a value between 0 and 1. The relative intensity was obtained by dividing the surface of the band by the sum of the surfaces of all the bands within the lane, thus eliminating the variation in band intensity caused by differences in the amounts of PCR products loaded in the gel.
To perform a statistical analysis of the DGGE profiles versus the environmental variables, we chose canonical correspondence analysis (CCA), as it explains the structure of a “species” data table (in this case, band intensities) by using environmental variables, assuming a unimodal distribution of species (35). For that purpose, community structures based on the relative intensity of each band were analyzed by performing a CCA (CANOCO 4.5; Biometris, Wageningen, The Netherlands). Community similarities were graphed by using ordination plots with scaling focused on intersample differences (21). The ordination plots for species and environmental variables were characterized by biplots that approximated the weighted averages of each species with respect to each of the environmental variables. Thus, the ordination diagram represents not only a pattern of community distribution, but also the main features of the distribution of species along the environmental variables (35). Although classes of nominal environmental variables are more often symbolized by a point at the centroid (the weighted average), they can also be represented by arrows (35). Hence, to facilitate the interpretation of the ordination plots, we used arrows to represent the nominal environmental variable “plant species.” Both the length and the slope of the vector are significant parameters, as long vectors forming smaller angles with an ordination axis are more strongly correlated with that ordination axis (34). In addition, the angle between the vectors provides an approximation of the correlation. Consequently, vectors pointing in the same direction are positively correlated and those pointing in opposite directions are negatively correlated (35). The nominal variables “land use history” and “growth cycle” were represented by centroids, whose positions determine the relationship of these variables with either of the ordination axes (34). In order to investigate statistical significance, we used a Monte Carlo permutation test based on 499 random permutations, assuming the null hypothesis that species data are unrelated to environmental data and the alternative hypothesis that the species respond to the environment.
Soil clones and sequence analyses.
DGGE bands, which were correlated with environmental variables, were selected for sequence analysis. After the inner part of the DGGE band was cut, the DNA was eluted in 20 μl of sterile Milli-Q water and subsequently used as a template for PCR (with primers Burk3 and BurkR), as described above. The reamplified PCR products were purified with a High Pure PCR product purification kit (Boehringer, Mannheim, Germany) and cloned into the pGEM-T easy vector, which was used to transform Escherichia coli strain JM109 according to the procedure recommended by the manufacturer (Promega Benelux, Leiden, The Netherlands). Plasmid extraction from randomly selected colonies was done with the Wizard Plus SV miniprep DNA purification system (Promega Benelux). Plasmids containing the inserts corresponding to the cut DGGE bands were sequenced in an ABI Prism automatic sequencer (BaseClear B.V., Leiden, The Netherlands), and the identities of the sequences were determined by BLAST analyses (1).
Sequence alignment.
The sequences generated in this study or recovered from the GenBank/EMBL database were aligned by using Clustal_X (37), considering only the 16S rRNA gene partial sequence covered by the Burkholderia-specific primers (30). Phylogenetic trees were constructed by the neighbor-joining method (29) based on distance estimations calculated by the method of Jukes and Cantor (18). This analysis was performed with the TREECON program, version 1.3b (Yves van de Peer, Department of Biochemistry, University of Antwerp, Antwerp, Belgium).
Nucleotide sequence accession numbers.
The sequences generated in this study have been deposited in the GenBank database under accession numbers AY571292 to AY571305.
RESULTS
DGGE analysis and identification of bands.
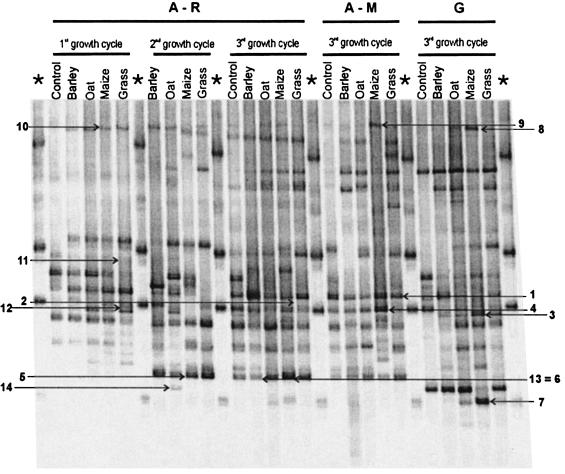
The numbers of bands in the DGGE profiles varied mainly along growth cycles. Figure 2 shows that the number of bands for A-R increased from an average of 14.2 for the first growth cycle to 18.4 for the third growth cycle. A-M and G had averages of 18.4 and 19 bands per lane (Fig. 2) for the third growth cycle, while the averages for the first growth cycle were 14.7 and 16.6 bands, respectively (data not shown). For all samples, the DGGE patterns after the first growth cycle comprised bands which were limited to an area in the middle of the gel (around 55% denaturant), whereas for the last growth cycle the bands were distributed over the whole gel (between denaturant concentrations of roughly 50 and 60%), indicating a change in the structure of the Burkholderia communities (Fig. 2). Additionally, the numbers of bands in the patterns for fallow pots (control) were lower than those in the patterns for pots containing plants, irrespective of the land use history or growth cycle (Fig. 2). Although no difference was observed in the average numbers of bands among plant species or land use histories for the same growth cycle, the Burkholderia communities associated with G were more even than those associated with A-M and A-R, containing fewer dominant bands (Fig. 2). Moreover, an effect of plant species could be observed by differences in the intensities of the bands.
FIG. 2.
DGGE patterns of Burkholderia communities associated with the rhizospheres of barley, oat, maize, and grass grown in soils with different land use histories. Control lanes represent pots without plants (bulk soil). *, Burkholderia marker containing (from top to bottom) B. andropogonis LMG6872, B. multivorans LMG13010, B. cepacia ATCC 25416, and B. dolosa LMG18941. The arrows indicate bands identified by sequencing (Table 1).
CCA not only allows the interpretation of DGGE profiles in relation to environmental variables but also correlates species (band positions) to those environmental variables. Therefore, after analyzing the ordination plots, we selected 15 band positions on the basis of their association with some of the treatments. Bands corresponding with these positions were thus identified in different samples, excised from the gel, and cloned. Bands corresponding to 5 of the 15 band positions could not be amplified or cloned and therefore could not be identified. From the remaining 10 band positions, 43 clones were obtained (around four per band position) and identified by sequencing. Five of the 43 sequences were considered chimeric and were removed from the analyses. All clones were affiliated with Burkholderia species, with identities varying from 97 to 100%, confirming the specificity of the PCR system. DGGE analysis of the clones revealed that 32% migrated to a different position on the gel than the band position that they originated from. Although these clones were affiliated with Burkholderia species, they were discarded from the analysis. Table 1 shows a list of the 10 band positions and respective clones considered in the analysis. Only one sequence per band position is shown in Table 1, since the similarities between them (sequences per band position) varied from 99.8 to 100%. Bands that were present at the same position but obtained from different samples reassuringly showed sequence similarity of >99.2% (30) but surprisingly did not always show the same bacterial sequence as the closest hit in the database (bands 8, 9, and 10) (Table 1; Fig. 2). On the other hand, some bands located in different positions in the gel were identified by the database as being affiliated with the same organism; this held true for bands present at the bottom of the gel (Table 1; Fig. 2). The percentages of similarity between these latter sequences ranged from 97.4 to 99.2%, which might indicate that organisms that possess several rRNA operons with sequence microheterogeneity were present.
TABLE 1.
Sequence analysis of bands excised from DGGE gels and their relationship to environmental variables
| Band no. (GenBank accession no.) | Position in CCA plota | Environ- mental variable | Most closely related bacterial sequenceb | % Identity | Accession no. of related sequence | Reference |
|---|---|---|---|---|---|---|
| 1 (AY571292) | A27 | G | Uncultured eubacterium WD232 | 99 | AJ292667 | 29 |
| Burkholderia sp. strain OY715 | 99 | AJ300696 | 32 | |||
| 2 (AY571302) | A29 | A-R | Burkholderia sp. strain OY715 | 99 | AJ300696 | 32 |
| 3 (AY571296) | A30 | G | Uncultured bacterial clone F2-41 | 97 | AY096172 | Y. Ding, W. B. Whitman, K. Das, and J. R. Kastner, unpublished data |
| B. terricola | 97 | AY040362 | 17 | |||
| 4 (AY571294) | A31 | A-M | Burkholderia sp. strain OY715 | 100 | AJ300696 | 32 |
| 5 (AY571293) | A36 | A-R | Uncultured earthworm cast bacterium | 99 | AY154615 | D. R. Singleton, P. F. Hendrix, D. C. Coleman, and W. B. Whitman, unpublished data |
| 6 (AY571303) | A36 | A-R | Uncultured earthworm cast bacterium | 99 | AY154615 | Singleton et al., unpublished data |
| 7 (AY571295) | A40 | G | Uncultured earthworm cast bacterium | 98 | AY154615 | Singleton et al., unpublished data |
| 8 (AY571300) | B4 | Maize | Burkholderia sp. strain NF23 | 97 | AJ300698 | 32 |
| 9 (AY571304) | B4 | Maize | Burkholderia sp. strain P18G1120 | 97 | AF214131 | 37 |
| 10 (AY571299) | B4 | Maize | Burkholderia sp. strain NF23 | 98 | AJ300698 | 32 |
| 11 (AY571298) | B26 | Grass | Uncultured Burkholderia sp. clone Ba04 | 97 | AF407355 | 35 |
| Burkholderia sp. strain NF23 | 97 | AJ300698 | 32 | |||
| 12 (AY571297) | B32 | Grass | B. hospita | 98 | AY040365 | 17 |
| 13 (AY571301) | B40 | Barley | Uncultured earthworm cast bacterium | 99 | AY154615 | Singleton et al., unpublished data |
| 14 (AY571305) | B47 | Oat | Uncultured earthworm cast bacterium | 97 | AY154615 | Singleton et al., unpublished data |
Species (band) position in CCA ordination plots; species preceded by the same letter were analyzed in the same gel.
Only one clone per band is listed. Clones for which the closest hit was an unculturable organism have the first hit with a species identification also mentioned, except for AY154615, which showed a low identity (<90%) to any culturable organism.
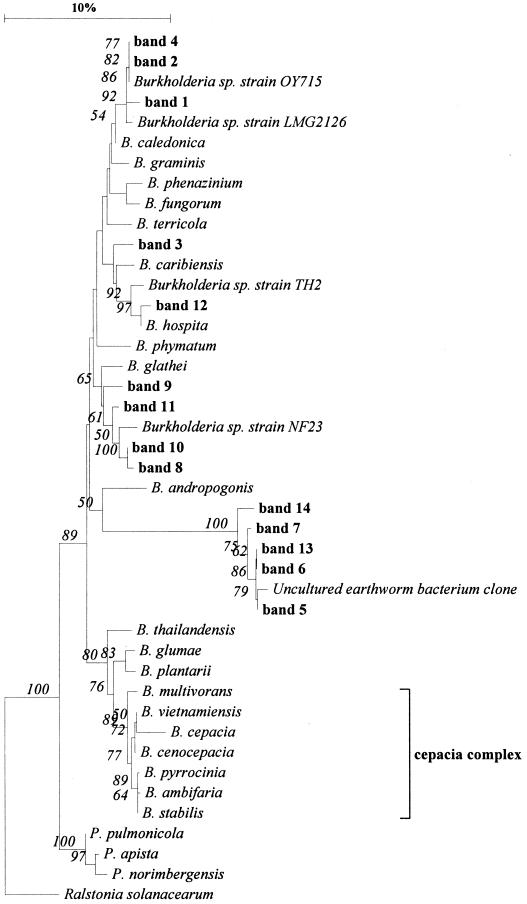
In order to determine the distribution of these clones within the genus Burkholderia, we constructed a phylogenetic tree based on the region amplified by the primers (Fig. 3). Clones representing the 10 band positions were distributed within three branches of the Burkholderia phylogenetic tree, clustering apart from those belonging to the cepacia complex (Fig. 3). Two main groups of discriminating clones were distributed close to B. glathei (four clones) and B. caledonica (three clones). Another main group of clones was affiliated with an unculturable (or uncultured) earthworm cast bacterium (five clones). The remaining two clones were affiliated with B. hospita or B. caribiensis (Fig. 3). All species displaying high levels of similarity with the clones were originally isolated from soil.
FIG. 3.
Phylogenetic tree showing the relationship between some Burkholderia species and bands excised from DGGE gels. The tree was constructed based on the fragment amplified by the Burkholderia-specific primers (35) by using the neighbor-joining method (34). A bootstrap analysis was performed with 100 repetitions, and only values above 50 are shown. A description of the bands is given in Table 1.
Plant species effect.
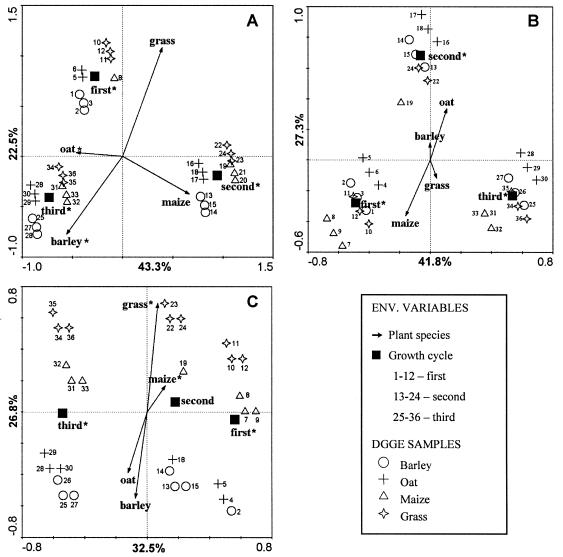
To eliminate artifacts associated with DGGE image analysis on the basis of different gels, we analyzed the effect of plant species separately with each land use history treatment, giving rise to three different ordination plots (Fig. 4). In addition, we used soil that was originally from one field plot which was representative of the triplicate plots. In all ordination plots, the control samples (pots without plants) clearly clustered distantly from the other (planted) samples (data not shown). The controls were then excluded from the analyses to facilitate the visualization of the treatment effects. For the same reason, we opted for ordination plots consisting of biplots of environmental variables and samples (DGGE lanes) instead of species (DGGE bands).
FIG. 4.
Ordination plots of Burkholderia communities associated with the rhizospheres of maize, barley, oat, and grass grown in soils with different land use histories, either A-M (A), A-R (B), or G (C). The plots were generated by CCA of the DGGE profiles. All environmental variables are shown, but only those marked with asterisks were significant (P < 0.05). Values on the axes indicate the percentages of total variation explained by each axis.
Figure 4 shows that samples generally clustered according to growth cycle rather than plant species. The ordination plots corresponding to arable land (A-M and A-R) (Fig. 4A and B) show that the growth cycle was a highly significant explanatory variable (P ≤ 0.05), consistently separating the samples along the first (most important) ordination axis. In A-M, oat and barley were the only plant species that were significant as explanatory variables, being correlated with the first and second axes, respectively (Fig. 4A). Even though maize was not significant as an explanatory variable (P > 0.05), maize and oat were negatively correlated, as inferred by their opposition in the ordination plots. The same configuration was detected for barley and grass (Fig. 4A). A similar pattern for plant species distribution was observed for A-R (Fig. 4B), for which a positive correlation between maize and grass or between oat and barley was observed (Fig. 4B). However, in A-R, none of these ordinations showed a significant P value (P < 0.05%), with all of them clustering close to the origin of the plot. In addition, the explanatory variable growth cycle was mainly separated along the first axis, whereas plant species were distributed along the second axis (Fig. 4B). In the ordination plot representing land use history G (Fig. 4C), the growth cycle was again the main explanatory variable, being spread along the first axis. However, growth cycle 2 was not significant according to a P value estimation. In addition, it is noteworthy that the distribution of the samples followed a different pattern for G than for A-R or A-M, being scattered for the former and concentrated around the centroids of growth cycle for the latter (Fig. 4). Maize and grass were significant as explanatory variables, as explained by the second axis (Fig. 4C). Again, these two explanatory variables were positively correlated with each other and negatively correlated with oat and barley (Fig. 4C). The positive correlation between maize and grass and between oat and barley was also observed when the DGGE profiles were compared by a cluster analysis considering only the presence or absence of bands (data not shown).
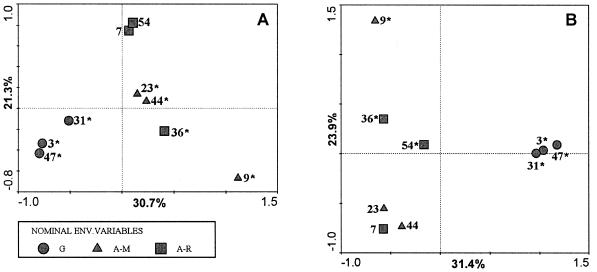
Land use history effect.
To evaluate the effect of land use history on the Burkholderia communities, we chose samples collected after the last growth cycle for the analyses. Thus, we generated biplots of environmental variables (land use history) and of species (DGGE bands). For the rhizosphere soil samples taken from maize (Fig. 5A), we observed that the three replicate G plots clustered together, separating from the A-M and A-R plots along the first axis. The second axis, though, explained the difference between the two treatments of arable land, i.e., crop rotation (A-R) and maize monoculture (A-M). However, although the replicate samples belonging to the same land use history tended to group together, for the A-R and A-M replicate plots there was always one outlier. Land use history was a significant explanatory variable, with all treatments being significant except for two A-R replicate plots (Fig. 5A). In order to determine if a similar pattern would be observed for other plant species, we performed the same analysis with oat plants. Mostly similar results were obtained, but the distinction between the A-M and A-R treatments was not clear (Fig. 5B). Oat was selected instead of the other plants because its effect was negatively correlated with that of maize. This analysis was not performed for grass and barley due to their similarity to maize and oat, respectively.
FIG. 5.
Ordination plots of Burkholderia communities associated with the rhizospheres of maize (A) and oat (B) grown in soils with different land use histories. The plots were generated by CCA of the DGGE profiles. The numbers after the nominal environmental variables correspond to the identification of each one of the three replicate plots from which the soil originated. All environmental variables are shown, but only those marked with asterisks were significant (P < 0.05). Values on the axes indicate the percentages of total variation explained by each axis.
To verify which environmental variable (plant species versus previous land use) had a more significant effect on the community structure of Burkholderia species in soil, we analyzed samples obtained from all of the plant species after the third growth cycle in one representative plot for each land use history treatment. The results showed that land use history had a greater effect on the Burkholderia community structure than did plant species, being largely responsible for the distribution of the samples in the ordination plot (data not shown). In addition, all land use history treatments and the plant species barley were significant as explanatory variables. The distribution of plant species along the ordination plot followed the same pattern as that described above for the plant species effect (data not shown). Similar results were obtained when all plant growth cycles were included in the analysis, confirming that land use history had the greatest influence on the Burkholderia community structure (data not shown).
DISCUSSION
Due to the relevance of the genus Burkholderia with respect to its application for agronomic purposes and its pathogenicity towards cystic fibrosis patients, it is important to assess the Burkholderia diversity in agricultural areas. Therefore, the aim of this work was to ascertain which Burkholderia species in rhizosphere soil are selected by specific crops and which factor (current plant species or previous land use) has the largest influence on Burkholderia populations in soil. To assess the total Burkholderia community structure in the rhizosphere samples, we used a PCR-DGGE system that is specific for determining the diversity of types within this genus (30).
PCR-DGGE has been used in molecular microbial ecology for about a decade (25), and its efficacy in analyzing microbial communities has greatly improved, as (i) primer systems for narrow taxonomic groups have been, and are still being, developed (17, 30) and (ii) different statistical strategies have been applied for analyzing DGGE fingerprinting data (for a review, see reference 14). The most common way to perform analyses of DGGE profiles is by UPGMA (unweighted pair group method with mathematical averages)-based clustering, which identifies samples with similar patterns based on the presence or absence of bands (30) but does not take into account the band intensities or the correlation between banding patterns and environmental variables. This type of correlation can be achieved by using ordination methods, which are vastly used in macroecology (36) and have been recently applied to DGGE fingerprinting analysis (21, 24). However, care should be taken in selecting the most appropriate statistical procedure for the ordination of the molecular profiles, and the underlying theoretical model should be carefully assessed (14). Multivariate analyses such as CCA can be applied to link changes in communities to changes in the environment, correlating the community structure with explanatory variables which can be evaluated by statistical tests. The applicability of multivariate analyses of DGGE patterns was recently confirmed by Muylaert et al. (24), who monitored the bacterial community compositions in four eutrophic lakes. By using relative band intensities instead of presence/absence data matrices, they were able to find a stronger correlation between the bacterial community composition and explanatory variables than by using the latter method. In addition, by using an artificial data set to which potential sources of error associated with PCR-DGGE analysis were introduced, they obtained similar results (24). After applying CCA to analyze the Burkholderia community structure revealed by PCR-DGGE, we were able to ascertain that land use history (long-term) had a larger effect on this community than plant species (short-term), even after three sequential growth cycles in pots.
Grassland and agricultural land subjected to crop rotation represent two types of land use, with each one having a distinct effect on both microbial communities and soil properties. Indeed, our results showed that the soil collected from the arable land plot (which was turned into arable land by the cultivation of a part of the permanent grassland about 20 years ago) had a Burkholderia community structure that differed from that in the permanent grassland plot. Most likely, this was related to the clear differences between both soil management regimens. A comparison of different land use (soil management) regimens revealed that factors such as microbial biomass, pH, and management factors were highly correlated with differences in microbial community composition (34). These results supported the hypothesis that soil disturbance as a result of cultivation, rather than the plant species alone, distinguishes the microbial communities of arable fields from those of grasslands (34). Furthermore, the influence of soil agricultural history might persist long after changes in land management have been made (4). Although the time necessary to overcome the persisting effects of land use history may vary depending on the type of conversion (A to G or G to A), we observed changes due to agricultural practices, mainly in the arable land, as A-R and A-M only partially clustered together. However, the distribution of these two clusters depended on the plant species from which the rhizosphere soil samples were taken, and maize exerted an extra effect in separating the land use history treatments. This could be explained by the fact that the continuous growth of maize for three cycles in the A-M pots represented a prolongation of the land use management (monoculture of maize).
The effect of plant species could be observed by analyzing land use history treatments separately. Although these environmental variables did not always explain the distribution of the samples, the ordination plots indicated two groups of positively correlated crops, with one composed of maize and grass and the other composed of barley and oat. Moreover, these two groups showed a negative correlation between each other. In addition, the difference observed between samples obtained from control (fallow) and planted pots by CCA indicated that the plant species did indeed have an effect on Burkholderia populations, either by increasing the diversity or by affecting the evenness of the samples.
Interestingly, there was a distinct growth cycle effect which could be observed by a rise in the numbers of bands from the first to the third growth cycle, regardless of the plant species or land use history. This increase in diversity with growth cycle number could be explained by the fact that the pots were kept for a total of 1 year under constant greenhouse conditions, which may have been optimally selective for specific organisms. Since the bands that started to appear or became more intense were mainly present at the bottoms of the gels, Burkholderia species with higher G+C% values were apparently stimulated. Some of these bands were identified and showed a high level of similarity to an unculturable or uncultured 16S rRNA gene clone isolated from a cast of Lumbricus rubellus (32). However, although earthworm casts are composed primarily of soil organisms (15), it is not clear whether this clone is specifically associated with earthworms, since it was isolated only once (D. R. Singleton, personal communication).
When comparing different land use histories within a growth cycle, we observed a more striking effect in the arable land treatments (A-M and A-R) than in the grassland (G), from which samples did not show the same tendency to cluster. A plausible explanation could be that the permanent grassland, due to its long land use history, has a more stable and even community that is recalcitrant to (drastic) changes. In the treatments originating from arable land (A-R and A-M), shifts in the Burkholderia community structure after the three plant growth cycles occurred in a more drastic manner, indicating that areas under this agricultural management regimen are more amenable to changes.
The use of CCA proved to be an effective tool for evaluating how Burkholderia communities respond to changes in land use management and how the communities change in response to different plant types. Furthermore, by plotting species (DGGE bands) and environmental variables, we were able to identify band species that were correlated with certain treatments. However, as each DGGE band may harbor more sequence species, we cannot provide substantial data on the actual diversity of the community. Based on this study, we concluded that although species belonging to the cepacia complex might be present in the rhizosphere soil, they seem to be less influenced by agricultural practices, since for 10 selected bands responding to changes in crop and land use management, only typical soil Burkholderia species were found. These results were in agreement with previous work (30), in which the analysis of randomly selected soil clones showed that most clustered close to species with biocontrol and bioremediation abilities. This trend does not seem to be correlated with the origin of the soil, since similar results were found by Richardson et al. (27), who assessed the diversity of Burkholderia isolates from woodland rhizosphere environments.
Acknowledgments
We thank P. Garbeva and K. Voesenek for their assistance with the greenhouse experiment, J. Postma for her comments on the manuscript, and A. Speksnijder for inspiring discussions.
This work was supported by the National Council for Scientific and Technological Development (CNPq) of Brazil, project 20.0849/98-0, and by grant DKW352 (Ministry of Agriculture, Nature Conservation and Fisheries, The Netherlands).
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. H. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alvey, S., C.-H. Yang, A. Buerkert, and D. E. Crowley. 2003. Cereal/legume rotation effects on rhizosphere bacterial community structure in west African soils. Biol. Fertil. Soils 37:73-82. [Google Scholar]
- 3.Balandreau, J., V. Viallard, B. Cournoyer, T. Coenye, S. Laevens, and P. Vandamme. 2001. Burkholderia cepacia genomovar III is a common plant-associated bacterium. Appl. Environ. Microbiol. 67:982-985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buckley, D. H., and T. M. Schmidt. 2003. Diversity and dynamics of microbial communities in soils from agro-ecosystems. Environ. Microbiol. 5:441-452. [DOI] [PubMed] [Google Scholar]
- 5.Chiarini, L., V. Giovannelli, A. Bevivino, C. Dalmastri, and S. Tabacchioni. 2000. Different portions of the maize root system host Burkholderia cepacia populations with different degrees of genetic polymorphism. Environ. Microbiol. 2:111-118. [DOI] [PubMed] [Google Scholar]
- 6.Clegg, C. D., R. D. L. Lovell, and P. J. Hobbs. 2003. The impact of grassland management regime on the community structure of selected bacterial groups in soils. FEMS Microbiol. Ecol. 43:263-270. [DOI] [PubMed] [Google Scholar]
- 7.Coenye, T., and P. Vandamme. 2003. Diversity and significance of Burkholderia species occupying diverse ecological niches. Environ. Microbiol. 5:719-729. [DOI] [PubMed] [Google Scholar]
- 8.Dalmastri, C., L. Chiarini, C. Cantale, A. Bevivino, and S. Tabacchioni. 1999. Soil type and maize cultivar affect the genetic diversity of maize root-associated Burkholderia cepacia populations. Microb. Ecol. 38:273-284. [DOI] [PubMed] [Google Scholar]
- 9.Da Silva, K. R. S., J. F. Salles, L. Seldin, and J. D. van Elsas. 2003. Application of a novel Paenibacillus-specific PCR-DGGE method and sequence analysis to assess the diversity of Paenibacillus spp. in the maize rhizosphere. J. Microbiol. Methods 54:213-231. [DOI] [PubMed] [Google Scholar]
- 10.DiCello, F., A. Bevivino, L. Chiarini, R. Fani, D. Paffetti, S. Tabacchioni, and C. Dalmastri. 1997. Biodiversity of a Burkholderia cepacia population isolated from the maize rhizosphere at different plant growth stages. Appl. Environ. Microbiol. 63:4485-4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.El Banna, N., and G. Winkelmann. 1998. Pyrrolnitrin from Burkholderia cepacia: antibiotic activity against fungi and novel activities against streptomycetes. J. Appl. Microbiol. 85:69-78. [DOI] [PubMed] [Google Scholar]
- 12.Estrada-De los Santos, P., R. Bustillos-Cristales, and J. Caballero-Mellado. 2001. Burkholderia, a genus rich in plant-associated nitrogen fixers with wide environmental and geographic distribution. Appl. Environ. Microbiol. 67:2790-2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fiore, A., S. Laevens, A. Bevivino, C. Dalmastri, S. Tabacchioni, P. Vandamme, and L. Chiarini. 2001. Burkholderia cepacia complex: distribution of genomovars among isolates from the maize rhizosphere in Italy. Environ. Microbiol. 3:137-143. [DOI] [PubMed] [Google Scholar]
- 14.Fromin, N., J. Hamelin, S. Tarnawski, D. Roesti, K. Jourdain-Miserez, N. Forestier, S. Teyssier-Cuvelle, F. Gillet, M. Aragno, and P. Rossi. 2002. Statistical analysis of denaturing gel electrophoresis (DGE) fingerprinting patterns. Environ. Microbiol. 4:634-643. [DOI] [PubMed] [Google Scholar]
- 15.Furlong, M. A., D. R. Singleton, D. C. Coleman, and W. B. Whitman. 2002. Molecular and culture-based analyses of prokaryotic communities from an agricultural soil and the burrows and casts of the earthworm Lumbricus rubellus. Appl. Environ. Microbiol. 68:1265-1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gillis, M., V. Tran Van, R. Bardin, M. Goor, P. Hebbar, A. Willems, P. Segers, K. Kerstens, T. Heulin, and M. P. Fernandez. 1995. Polyphasic taxonomy in the genus Burkholderia leading to an amended description of the genus and proposition of Burkholderia vietnamiensis sp. nov. for N2-fixing isolates from rice in Vietnam. Int. J. Syst. Bacteriol. 45:274-289. [Google Scholar]
- 17.Heuer, H., M. Krsek, P. Baker, K. Smalla, and E. M. H. Wellington. 1997. Analysis of actinomycete communities by specific amplification of genes encoding 16S rRNA and gel-electrophoretic separation in denaturing gradients. Appl. Environ. Microbiol. 63:3233-3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jukes, T. H., and C. R. Cantor. 1969. Evolution of protein molecules, p. 21-132. In H. H. Munro (ed.), Mammalian protein metabolism. Academic Press, New York, N.Y.
- 19.Kowalchuk, G. A., D. S. Buma, W. de Boer, P. G. L. Klinkhamer, and J. A. van Veen. 2002. Effects of above-ground plant species composition and diversity on the diversity of soil-borne microorganisms. Antonie Leeuwenhoek 81:509-521. [DOI] [PubMed] [Google Scholar]
- 20.Lupwayi, N. Z., W. A. Rice, and G. W. Clayton. 1998. Soil microbial diversity and community structure under wheat as influenced by tillage and crop rotation. Soil Biol. Biochem. 30:1733-1741. [Google Scholar]
- 21.Marschner, P., and K. Baumann. 2003. Changes in bacterial community structure induced by mycorrhizal colonisation in split-root maize. Plant Soil 251:279-289. [Google Scholar]
- 22.Mazzola, M. 1999. Transformation of soil microbial community structure and Rhizoctonia-suppressive potential in response to apple roots. Phytopathology 89:920-927. [DOI] [PubMed] [Google Scholar]
- 23.Miller, S. C. M., J. J. LiPuma, and J. L. Parke. 2002. Culture-based and non-growth-dependent detection of the Burkholderia cepacia complex in soil environments. Appl. Environ. Microbiol. 68:3750-3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muylaert, K., K. Van der Gucht, N. Vloemans, L. D. Meester, M. Gillis, and W. Vyverman. 2002. Relationship between bacterial community composition and bottom-up versus top-down variables in four eutrophic shallow lakes. Appl. Environ. Microbiol. 68:4740-4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muyzer, G., E. C. de Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Raaijmakers, J. M., and D. M. Weller. 1998. Natural plant protection by 2,4-diacetylphloroglucinol-producing Pseudomonas spp. in take-all decline soils. Mol. Plant-Microbe Interact. 11:144-152. [Google Scholar]
- 27.Richardson, J., D. E. Stead, J. G. Elphinstone, and R. H. A. Coutts. 2002. Diversity of Burkholderia isolates from woodland rhizosphere environments. J. Appl. Microbiol. 93:616-630. [DOI] [PubMed] [Google Scholar]
- 28.Rosado, A. S., G. F. Duarte, L. Seldin, and J. D. van Elsas. 1998. Genetic diversity of nifH sequences in Paenibacillus azotofixans strains and soil samples analyzed by denaturing gradient gel electrophoresis of PCR-amplified gene fragments. Appl. Environ. Microbiol. 64:2770-2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 30.Salles, J. F., F. A. De Souza, and J. D. van Elsas. 2002. Molecular method to assess the diversity of Burkholderia species in environmental samples. Appl. Environ. Microbiol. 68:1595-1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 32.Singleton, D. R., P. F. Hendrix, D. C. Coleman, and W. B. Whitman. 2003. Identification of uncultured bacteria tightly associated with the intestine of the earthworm Lumbricus rubellus (Lumbricidae; Oligochaeta). Soil Biol. Biochem. 35:1547-1555. [Google Scholar]
- 33.Sorensen, J. 1997. The rhizosphere as a habitat for soil microorganisms, p. 21-46. In J. D. V. Elsas, J. T. Trevors, and E. M. H. Wellington (ed.), Modern soil microbiology. Marcel Dekker Inc., New York, N.Y.
- 34.Steenwerth, K. L., L. E. Jackson, F. J. Calderon, M. R. Stromberg, and K. M. Scow. 2002. Soil microbial community composition and land use history in cultivated and grassland ecosystems of coastal California. Soil Biol. Biochem. 34:1599-1611. [Google Scholar]
- 35.ter Braak, C. J. F. 1987. The analysis of vegetation-environment relationships by canonical correspondence analysis. Vegetation 69:69-77. [Google Scholar]
- 36.ter Braak, C. J. F. 1995. Ordination, p. 91-173. In R. H. G. Jongman, C. J. F. T. Braak, and O. F. R. V. Tongeren (ed.), Data analysis in community and landscape ecology, 2nd ed. Cambridge University Press, Cambridge, United Kingdom.
- 37.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.TrÂn Van, V., O. Berge, S. N. Ke, J. Balandreau, and T. Heulin. 2000. Repeated beneficial effects of rice inoculation with a strain of Burkholderia vietnamiensis on early and late yield components in low fertility sulphate acid soils of Vietnam. Plant Soil 218:273-284. [Google Scholar]
- 39.van Elsas, J. D., P. Garbeva, and J. F. Salles. 2002. Effects of agronomical measures on the microbial diversity of soils as related to the suppression of soil-borne plant pathogens. Biodegradation 13:29-40. [DOI] [PubMed] [Google Scholar]
- 40.Wieland, G., R. Neumann, and H. Backhaus. 2001. Variation of microbial communities in soil, rhizosphere, and rhizoplane in response to crop species, soil type, and crop development. Appl. Environ. Microbiol. 67:5849-5854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yabuuchi, E., Y. Kosako, H. Oyaizu, I. Yano, H. Hotta, Y. Hashimoto, T. Ezaki, and M. Arakawa. 1992. Proposal of Burkholderia gen. nov. and transfer of seven species of the genus Pseudomonas homology group II to the new genus, with the type species Burkholderia cepacia (Palleroni and Holmes 1981) comb. nov. Microbiol. Immunol. 36:1251-1275. [DOI] [PubMed] [Google Scholar]