Abstract
Photorhabdus sp. strain Az29 is symbiotic with an Azorean nematode of the genus Heterorhabditis in a complex that is highly virulent to insects even at low temperatures. The virulence of the bacteria is mainly attributed to toxins and bacterial enzymes secreted during parasitism. The bacteria secrete proteases during growth, with a peak at the end of the exponential growth phase. Protease secretion was higher in cultures growing at lower temperatures. At 10°C the activity was highest and remained constant for over 7 days, whereas at 23 and 28°C it showed a steady decrease. Two proteases, PrtA and PrtS, that are produced in the growth medium were purified by liquid chromatography. PrtA was inhibited by 1,10-phenantroline and by EDTA and had a molecular mass of 56 kDa and an optimal activity at pH 9 and 50°C. Sequences of three peptides of PrtA showed strong homologies with alkaline metalloproteases from Photorhabdus temperata K122 and Photorhabdus luminescens W14. Peptide PrtA-36 contained the residues characteristic of metzincins, known to be involved in bacterial virulence. In vitro, PrtA inhibited antibacterial factors of inoculated Lepidoptera and of cecropins A and B. PrtS had a molecular mass of 38 kDa and was inhibited by 1,10-phenanthroline but not by EDTA. Its activity ranged between 10 and 80°C and was optimal at pH 7 and 50°C. PrtS also destroyed insect antibacterial factors. Three fragments of PrtS showed homology with a putative metalloprotease of P. luminescens TTO1. Polyclonal antibody raised against PrtA did not recognize PrtS, showing they are distinct molecules.
Photorhabdus spp. are non-free-living Enterobacteriaceae (6, 19). The entomopathogen species of Photorhabdus have a symbiotic relation with nematodes of the genus Heterorhabditis for transport with their infective juveniles (IJs). The IJs actively seek for an insect host, penetrating through its natural openings and cuticle. Inside the insect hemocoel, the bacteria are released and actively multiply avoiding the host defenses and causing an acute disease condition that is followed by insect death within 48 h. The bacteria also create the nutritional conditions and protective environment for the development of its nematode symbiont (2, 21).
Bacteria of the Photorhabdus genus produce toxins and other potentially virulent factors (16). The characterized toxins are organized in pathogenic islands (53) and include the Tc complex, which is responsible for oral toxicity (9, 52), and the Mcf toxin, which causes loss of insect body turgor, followed by death (14). Among other potential virulence factors, there is a complex set of extracellular enzymes, including proteases, lipases, lecithinases, chitinases, and phosphatases (5, 12, 20). Proteases represent an important part of these enzymes, although their role on the virulence process is yet unclear. Until now, all extracellular proteases purified and characterized from Photorhabdus were classified as metalloproteases, but in reports on proteases from different strains it has been suggested that there are differences in the number and characteristics of these molecules (7, 8, 40, 44). Recently, the genetic characterization of the major metalloproteases produced by Photorhabdus luminescens W14 and by Photorhabdus temperata K122 showed that they are repeats-in-toxin (RTX)-like metalloproteases belonging to the metzincin clan and similar to those produced by Pseudomonas, Erwinia, Serratia, and Yersinia spp. (7). In characterization studies of the proteases produced by these bacteria, different biochemical characteristics and DNA sequences have been found for metalloproteases that are secreted by different species or even by different strains of the same species (33, 56). The metzincin clan is part of the zincin group of metalloproteases, which includes proteases involved on bacterial virulence (36). Major examples for human pathogenic bacteria include the alkaline protease and elastase produced by Pseudomonas aeruginosa (3, 17, 22, 26), the metalloproteases of Vibrio vulnificus and Vibrio cholerae (27, 37), serralysin from Serratia marcescens (29, 32, 35, 51), and enterotoxin from Bacteroides fragilis (38, 45, 55). Recently, the yrp1 gene of an RTX-like metalloprotease was characterized from the fish pathogen Yersinia ruckeri, and the enzyme's involvement in pathogenicity was demonstrated by insertional mutation (18). The implication on phytopathogenicity of this type of metalloproteases was also demonstrated on a plant pathogen, Erwinia carotovora subsp. carotovora (4, 33).
Photorhabdus sp. strain Az29 was isolated from the Azorean isolate of Heterorhabditis bacteriophora Az29 (43). This complex proved to be the most pathogenic against the lepidopteran Pseudaletia unipuncta (41). In laboratory assays, the Az29 complex was able to infect and to promote disease in insects at temperatures as low as 10°C (unpublished data). Assays comparing the pathogenic activity induced by the nematode-bacteria and by the bacteria alone demonstrated that the virulence was mainly due to the associated bacteria. Photorhabdus sp. strain Az29 proved to be the most pathogenic against sixth-instar larvae of P. unipuncta, with a median lethal time of 40.6 h after injection of 200 CFU (42). These data enhanced our interest in the virulence factors released by Photorhabdus sp. strain Az29. We report here the purification and characterization of two extracellular metalloproteases recovered in the end of the exponential phase of growth and their activity on insect antibacterial factors.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Photorhabdus sp. strain Az29 was isolated from its symbiotic nematode by the method of Akhurst (1). Twenty IJs were surface sterilized for 10 min in 1% sodium hypochlorite, washed in sterile distilled water, transferred to a petri dish containing 5 ml of TSBYE (3% tryptic soy broth [Difco], 0.5% yeast extract [Difco]), and cut into several pieces with a scalpel. The plates were incubated at 30°C for 24 h and streaked on NBTA plates (2.3% nutrient agar [Difco], 0.0025% bromothymol blue [Merck], 0.004% 2,3,5-triphenyltetrazolium [Merck]). The presence of Photorhabdus colonies was confirmed by dye adsorption on NBTA plates, the production of luminescence, and antibiotic activity. The isolated bacteria were maintained on NBTA plates at 10°C and subcultured weekly.
The growth curves of Photorhabdus sp. strain Az29 cultures were obtained by monitoring the optical density at 600 nm at different incubation times. For this purpose, 250-ml flasks with 50 ml of TSBYE were inoculated with 0.2 ml of a preculture in 5 ml of TSBYE grown at 28°C for 18 h. The flasks were incubated at 10, 23, or 28°C in an orbital shaker at 180 rpm. Three replicates were made for each temperature. To monitor the protease activity during growth, 1-ml aliquots were sampled at different time intervals and centrifuged at 12,000 × g for 10 min at 4°C, and the supernatants were stored at −20°C until needed for the assays.
For purification of the protease, cultures were obtained in flasks as described above at 23°C and 180 rpm for 24 h. The cultures were centrifuged at 12,000 × g for 10 min at 4°C, and the supernatants were stored at −20°C until used.
Assay of proteolytic activity and protein determination.
The proteolytic activity during bacterial growth was monitored by a caseinolytic macroassay (49). Aliquots (50 μl) of culture supernatant were obtained at different time intervals, combined with 80 μl of 2% azocasein (Sigma) in 50 mM Tris-HCl (pH 8.5), and incubated at 37°C for 3 h. The undigested substrate was precipitated by adding 400 μl of 10% trichloroacetic acid to the reaction mixture, followed by incubation for 10 min at 4°C, followed in turn by centrifugation at 10,000 × g for 10 min. The supernatant was neutralized by the addition of 600 μl of 1 N NaOH, and the absorbance at 440 nm (A440) was measured. A blank obtained by reacting the substrate with the Tris-HCl buffer was used to correct the absorbance readings, which were then converted to Pronase E (Sigma) equivalent activity values (μg/ml).
The protease activity in the fractions obtained in the purification steps was tested by a microassay for which 20-μl aliquots from each fraction were added to 40 μl of 3% azocasein in Tris plus an additional 40 μl of 50 mM Tris-HCl (pH 8.5), followed by incubation at 37°C for 30 min. The undigested substrate was precipitated by adding 75 μl of 10% trichloroacetic acid to the reaction mixture, followed by incubation for 10 min at 4°C and centrifugation at 10,000 × g for 10 min. Then, 100 μl of supernatant was transferred to a 96-well microtitration plate and neutralized by the addition of an equal volume of 1 N NaOH. The A440 was measured by using a microplate reader (Bio-Rad). One unit of enzyme activity was defined as the amount of enzyme that yielded an absorbance change of 0.01.
The amount of total protein was determined by the Coomassie blue dye binding method (10), with bovine serum albumin (Merck) as a standard protein.
Protease purification.
The supernatant of the 50-ml bacterial culture was fractionated with 80% ammonium sulfate overnight at 4°C with constant stirring. The precipitate was recovered by centrifugation (15,000 × g for 30 min at 4°C), resuspended in 10 ml of 50 mM Tris-HCl (pH 8.5), and dialyzed in a PD10 (Pharmacia-Amersham) against the same buffer. The dialysate was applied on a DEAE-Sepharose column (1.5 by 5 cm) equilibrated with 50 mM Tris-HCl (pH 8.5; buffer A) that was connected to a Pharmacia fast-performance liquid chromatography system (FPLC). The bound proteins were eluted with 50 ml of NaCl on a linear gradient of 0 to 0.5 M in 50 mM Tris-HCl (pH 8.5) in 1-ml fractions at a 1-ml/min flow. The fractions were tested for protease activity, and the positive fractions were pooled, diluted 1:1 in buffer A, and loaded on a HighTrap Q-Sepharose column equilibrated with the same buffer in a FPLC system. The bound proteins were eluted in 1-ml fractions at a 1-ml/min flow in a step of 15 ml of 0.2 M NaCl in buffer A, followed by a linear gradient of 30 ml from 0.2 to 0.3 M NaCl. The fractions were then assayed for protease activity as described above. The positive fractions collected in the gradient were stored at −20°C. Active fractions eluted on the 0.2 M NaCl step were pooled, diluted 1:1 in buffer A, and loaded on a Mono-Q column equilibrated with the same buffer in FPLC. The bound proteins were eluted in 1-ml fractions at a 1-ml/min flow in a linear gradient of 0.1 to 0.2 M NaCl over 20 ml. The active fractions were also stored at −20°C.
SDS-PAGE, zymograms, and immunoblot analysis.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed as described by Laemmli (31) in 10% polyacrylamide gel by using a Mini-Protean II gel system (Bio-Rad) under nondenaturing conditions. Low-range SDS-PAGE markers (Bio-Rad) containing phosphorylase b (97.4 kDa), bovine serum albumin (66.2 kDa), ovalbumin (45.0 kDa), carbonic anhydrase (31.0 kDa), trypsin inhibitor (21.5 kDa), and lysozyme (14.4 kDa) were used as molecular mass standards. The proteins were visualized by the silver staining method of Morrissey (39).
Zymograms were performed as described by Schmidt et al. (44) with minor modifications. Polyacrylamide gels (10%) were copolymerized with 0.05% gelatin or with 0.05% azocasein. The samples were applied in nonreducing Laemmli buffer without denaturation and run at 100 V. The gels were washed twice in 2.5% (wt/vol) Triton X-100 and then incubated in 50 mM Tris-HCl (pH 8.5) with 10 mM CaCl2 and 0.1 M NaCl for 2 h at room temperature. The gels were stained with Coomassie blue and later washed until the zones of substrate hydrolysis were visible.
For immunodetection, the proteins were separated by SDS-PAGE as described above and electroblotted onto a nitrocellulose membrane (Millipore Corp.) with a liquid transfer apparatus (Bio-Rad) in a continuous buffer system (Tris-glycine and 20% methanol; pH 11) applying 0.8 mA/cm2 for 2 h (50). The membrane was blocked in TBS (0.01 M Tris-HCl [pH 7.5], 0.1 M NaCl) containing 0.05% (vol/vol) Tween 20 (TBS-T) and 0.5% (wt/vol) skim milk for 30 min at room temperature. The membrane was then incubated with primary antibody (1:500) in TBS-T containing 0.5% (wt/vol) of skim milk for 2 h at room temperature. The membrane was washed three times for 10 min in TBS-T, followed by 2 h of incubation with goat anti-rabbit immunoglobulin G-peroxidase conjugates (Sigma) diluted 1:4,000 in TBS. The immunoreactivity was detected by incubating the membrane in TBS containing 0.06% (wt/vol) diaminobenzidine, 0.018% NiCl2 (wt/vol), and 0.3% H2O2 (vol/vol).
Inhibition assays.
Portions (20 μl) of the purified proteases were incubated for 30 min at room temperature in the presence of phenylmethylsulfonyl fluoride (1.5, 2, 2.5, and 3 mM), EDTA (1, 2, 5, 8, and 10 mM), and 1,10-phenantroline (0.2, 1, 3, and 8 mM). The remaining proteolytic activity was accessed by using the azocasein microassay described above.
Determination of optimal pH and temperature.
The caseinolytic activity of the purified proteases was assayed at pH values ranging from 6 to 10 by using the azocasein microassay. Azocasein was buffered with 200 mM sodium phosphate buffer (pH 6), 200 mM Tris-HCl (pH 7), 200 mM Tris-HCl (pH 8), 200 mM Tris-glycine (pH 9), and 200 mM sodium carbonate (pH 10).
To determine the optimal temperature for protease activity, the purified proteases were incubated during 30 min with the azocasein substrate, according to the microassay procedure, at 4, 10, 23, 28, 37, 50, 60, 65, 70, 80, and 90°C. A control reaction was prepared for each incubation temperature.
Assays for antibacterial activity inhibition.
The effect of the two purified proteases on insect antibacterial activity was measured by using a modified growth inhibition zone assay according to the method of Hoffmann et al. (25). Hemolymph with antibacterial activity was collected from Galleria mellonella larvae at 24 h postinoculation with an overnight culture of Escherichia coli, and 10-μl portions of hemolymph were incubated with 10 μl of each protease for 1 h at 37°C. Untreated hemolymph and purified proteases were used as controls. After incubation, the reaction mixture was transferred to wells prepared on E. coli pour plates (∼105 CFU/plate). After 24 h of incubation at 37°C, the diameters of the inhibition zones were recorded. The effect of the proteases on cecropin A and cecropin B were also assayed. For that purpose, 6 μl of 1% cecropin A (Sigma) and 6 μl of 1% cecropin B (Sigma) were incubated with 6 μl of each purified protease for 1 h at 37°C. The samples were tested for the maintenance of antibacterial activity, as described above, with untreated cecropins A and B and proteases as controls.
Production of antiserum.
The purified PrtA was subjected to SDS-PAGE and electroblotted onto nitrocellulose membrane (Millipore Corp.) according to the procedure described above. The membranes were treated with 0.5% Ponceau Red, and the stained bands were excised for the preparation of rabbit polyclonal antibodies. The antiserum was prepared at Eurogentec Laboratories (Scraing, Belgium).
Preparation for N-terminal amino acid sequencing and internal sequences.
The purified proteases were subjected to SDS-PAGE and electroblotted onto polyvinylidene fluoride membranes (Millipore Corp.) as described for the nitrocellulose membranes. The membranes were stained with 0.5% Ponceau Red, and the bands were excised and subjected to sequencing. N-terminal amino acid sequence analysis was performed by automated Edman degradation. For the internal sequencing, the proteases were digested with endo-Lys-C, and the peptide fragments were isolated by reversed-phase HPLC. Several peptides of each protease were sequenced at the Emory University Microchemical Facility (Atlanta, Ga.). The peptide sequences were compared to entries at the NCBI database by using BLASTP 2.2.6. and at the PhotoList database (http://genolist.pasteur.fr/PhotoList/index.html) by using FASTA.
RESULTS
Effect of temperature on bacterial growth and on protease production.
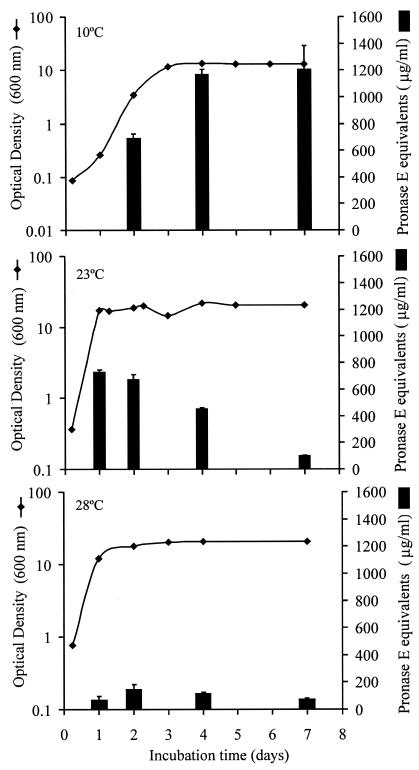
The growth curves of Photorhabdus sp. strain Az29 at 10, 23, and 28°C and the proteolytic activity recovered in the culture media were monitored for 28 days. The optimal growth temperature was 23°C, with a doubling time of 3.2 h, reaching stationary phase after 24 h of incubation (Fig. 1). At 28°C, the stationary phase was also reached after 24 h of incubation, but with a doubling time of 4.3 h. Photorhabdus sp. strain Az29 was able to grow at 10°C with a doubling time of 6 h, but the stationary phase was reached after 72 h of incubation, with an optical density at 600 nm identical to the values reached at 23 and 28°C. The cultures at different temperatures remained in stationary phase during the 28 days of the assay.
FIG. 1.
Caseinolytic activity during growth of Photorhabdus sp. strain Az29 in TSBYE at different incubation temperatures. The bacterial growth was monitored by measuring the culture's optical density at 600 nm. Samples of culture supernatants were collected at different times, and the caseinolytic activity was estimated by using the macroassay described in Materials and Methods. The caseinolytic activity (μg/ml) is expressed in pronase E equivalent activity.
The caseinolytic activities measured in the culture supernatants at each temperature were maximal by the end of the exponential phase (Fig. 1). The highest caseinolytic activity was recovered in the cultures incubated at 10°C; the activity was 1.8 and 8 times higher than the activities at 23 and 28°C, respectively. The caseinolytic activity in cultures at 23 and 28°C decreased rapidly with incubation time. After 7 days the remaining activity was only 15 and 25% at 23 and 28°C, respectively, but at 10°C the activity did not decrease. After 28 days of incubation the caseinolytic activity in the cultures at 10°C remained constant, whereas in the cultures at 23 and 28°C was residual (data not shown).
Purification and characterization of the proteases.
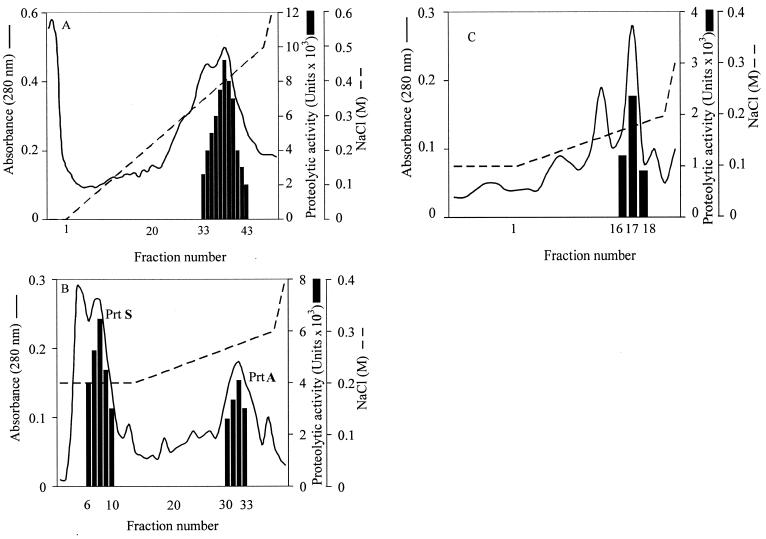
The proteases released by Photorhabdus sp. strain Az29 were purified from 50 ml of TSBYE culture supernatant (Table 1). The ammonium sulfate precipitation resulted in a 1.46-fold increase of specific activity. On DEAE-Sepharose, the proteolytic activity was concentrated in a large peak of protein that was eluted on a gradient at 0.38 M NaCl (Fig. 2A). The active fractions were pooled in a HiTrap Q-Sepharose that separated the fractions with caseinolytic activity into two peaks (Fig. 2B). The first peak was eluted in the continuous 0.2 M NaCl step with a 4.12-fold increase in specific activity, and the second peak was eluted in the gradient at 0.27 M NaCl with a 6.06-fold increase in specific activity. The second peak contained a pure protein with a single band of 56 kDa in SDS-PAGE and a single band of degradation in both gelatin and casein zymograms (Fig. 3), labeled as PrtA. The first peak active fractions from the HiTrap Q-Sepharose were loaded onto a Mono-Q column and eluted on a gradient of NaCl. The caseinolytic activity was recovered in the fractions eluted at 0.18 M NaCl (Fig. 2C), resulting in a 4.8-fold increase of specific activity. This peak had a pure protein with a single band of 38 kDa in SDS-PAGE and a single band of proteolytic activity in the zymogram with casein as the substrate, whereas no band was seen on zymograms with gelatin as substrate (Fig. 3). This protein was labeled as PrtS.
TABLE 1.
Purification of Photorhabdus sp. strain Az29 proteases
| Purification step | Total protein (mg) | Total activity (U) | Sp act (U/mg) | Purification (fold) | Yield (%) |
|---|---|---|---|---|---|
| Supernatant | 33.32 | 184,000 | 5,522 | 11 | 100 |
| Ammonium sulfate | 8.71 | 70,000 | 8,037 | 1.46 | 38 |
| DEAE-Sepharose | 2.38 | 67,340 | 28,306 | 5.13 | 37 |
| HiTrap Q-Sepharose PrtA | 0.39 | 13,076 | 33,442 | 6.06 | 7 |
| HiTrap Q-Sepharose PrtS | 1.0 | 22,950 | 22,723 | 4.12 | 12 |
| Mono-Q PrtS | 0.17 | 4,455 | 26,518 | 4.8 | 2 |
FIG. 2.
Purification of proteases from Photorhabdus sp. strain Az29. (A) Elution of a proteolytic peak in DEAE-Sepharose with a 0 to 0.5 M NaCl linear gradient in 50 mM Tris-HCl; (B) elution on HiTrap Q-Sepharose of purified PrtA at 0.27 M NaCl and of PrtS at 0.2 M NaCl; (C) elution on Mono-Q column of purified PrtS at 0.18 M NaCl.
FIG. 3.
Analysis of PrtA and PrtS by SDS-PAGE, Western blotting, and zymography. Shown are SDS-PAGE analyses for the purified fraction of PrtA (lane A) and PrtS (lane B); Western blot analyses, with polyclonal antibodies raised to PrtA, with PrtA (lane C) and PrtS (lane D); and zymogram analyses of PrtA in gelatin (lane E) and in casein (lane G) and of PrtS in gelatin (lane F) and in casein (lane H).
Polyclonal antibodies raised against PrtA reacted with PrtA but not with PrtS, indicating that two distinct proteases were purified (Fig. 3).
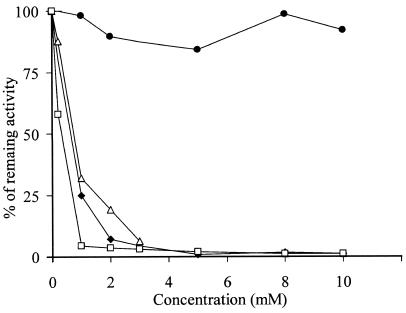
Both proteases were not inhibited by PMSF, a specific inhibitor of serine proteases, even at the highest concentration used (data not shown). The activities of PrtA and PrtS were inhibited by 8 mM 1,10-phenantroline to 1.1 and 1.0% of the initial activity, respectively, thus indicating that they are both metalloproteases. PrtA was also strongly inhibited by 10 mM EDTA, being reduced to 1.0% of the initial activity, whereas PrtS was not (Fig. 4).
FIG. 4.
Inhibition of PrtA and PrtS by protease inhibitors. The inhibited activities of the purified proteases are expressed as a percentage of the uninhibited activity. Symbols: ⧫, PrtA treated with EDTA; •, PrtS treated with EDTA; ▵, PrtA treated with 1,10-phenantroline; □, PrtS treated with 1,10-phenanthroline.
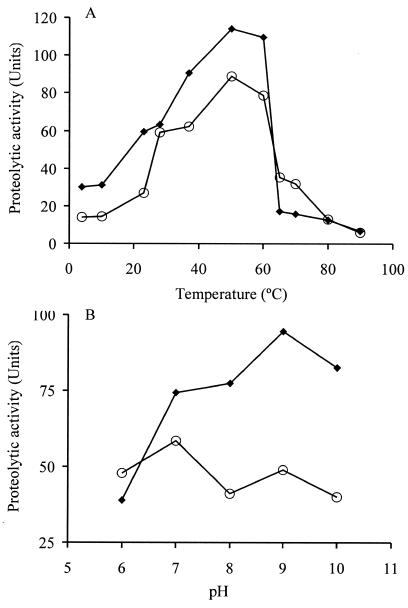
Both proteases remained active on a broad range of temperatures, from 10 to 80°C, showing evident heat stability. The optimal reaction temperature for both proteases was 50°C, resulting in an increase of 26 and 42% for each compared to the activity at 37°C. The activity at 60°C was 20.9 and 26% higher than at 37°C for PrtA and PrtS, respectively. At 65°C, PrtA was reduced to 18.8% of the activity recorded at 37°C, whereas PrtS to only 56.7% (Fig. 5A). The pH effect was more evident on the proteolytic activity of PrtA than in PrtS. Under the assay conditions, the maximal activity was observed at pH 9 for PrtA and at pH 7 for PrtS (Fig. 5B).
FIG. 5.
Effect of temperature (A) and of pH (B) on the caseinolytic activity of PrtA and PrtS. The caseinolytic activity was evaluated by the microassay described in Materials and Methods. Symbols: ⧫, PrtA; ○, PrtS.
Determination of the N-terminal amino acid sequence and internal sequences.
The N-terminal sequencing of PrtA and PrtS did not produce an amino acid sequence, indicating a possible blockage. From the digested peptides of PrtA the following sequences were obtained: PrtA-23, TDDTVYGFNSNTDR; PrtA-27, TFSENDIGVNGYGR, and PrtA-36, QTFTHEIGHTLGLEHPAA.
Four internal sequences were obtained for PrtS: PrtS-31, NYDDEVPSDTLH; PrtS-32, LGGYAXEK (the “X” means that no residue could be identified); PrtS-41, YLEATYNFYK; and PrtS-46, DTTFLSFAK.
A database search for PrtA peptides showed several matches with extracellular metalloproteases secreted by bacteria of the genera Photorhabdus, Erwinia, and Pseudomonas. PrtS-41 and PrtS-46 showed homologies to a putative metalloprotease from P. luminescens subsp. laumondii TTO1 (Table 2).
TABLE 2.
Comparison of PrtA and PrtS peptides sequences with entries at the NCBI and PhotoList databases
| Fragment | Organism | Protein | Identity (%) | Score (bits) | ɛ value | Accession no. |
|---|---|---|---|---|---|---|
| PrtA-23 | P. temperata K122 | Secreted alkaline metalloprotease | 100 | 48.6 | 1e−05 | AAO39136.1 |
| P. luminescens W14 | Secreted alkaline metalloprotease | 100 | 45.6 | 8e−05 | AAO39138.1 | |
| E. chrysanthemi | Secreted protease B precursor | 92 | 43.9 | 3e−04 | P16316 | |
| E. chrysanthemi | Secreted protease A precursor | 92 | 43.9 | 3e−04 | Q07295 | |
| P. aeruginosa | Alkaline protease | 85 | 41.4 | 1e−03 | 1AKL | |
| P. luminescens subsp. laumondii TTO1 | Alkaline metalloprotease precursor PrtA | 100 | 45.6 | 8e−05 | NP 928000.1 | |
| PrtA-27 | P. temperata K122 | Secreted alkaline metalloprotease | 100 | 47.7 | 2e−05 | AAO39136.1 |
| P. luminescens W14 | Secreted alkaline metalloprotease | 85 | 39.7 | 5e−03 | AAO39138.1 | |
| P. temperata K122 | Secreted alkaline metalloprotease | 100 | 60.4 | 3e−09 | AAO39136.1 | |
| P. luminescens subsp. laumondii TTO1 | Alkaline metalloprotease precursor PrtA | 78 | 33.7 | 0.31 | NP 928000.1 | |
| PrtA-36 | P. temperata K122 | Secreted alkaline metalloprotease | 100 | 60.4 | 3e−09 | AAO39136.1 |
| P. luminescens W14 | Secreted alkaline metalloprotease | 94 | 54.9 | 1e−07 | AAO39138.1 | |
| E. chrysanthemi | Metalloprotease A | 88 | 49.4 | 6e−06 | JN0891 | |
| P. fluorescens | Alkaline protease | 87 | 47.7 | 2e−05 | BAA36461.1 | |
| P. luminescens subsp. laumondii TTO1 | Alkaline metalloprotease precursor PrtA | 94 | 54.9 | 1e−07 | NP 928000.1 | |
| PrtS-36 | P. luminescens subsp. laumondii TTO1 | Unnamed protein product (probable metalloprotease) | 75 | 24 | 0.86 | Plu1382 |
| PrtS-41 | P. luminescens subsp. laumondii TTO1 | Unnamed protein product (probable metalloprotease) | 100 | 38 | 0.017 | BX571863 |
| PrtS-46 | P. luminescens subsp. laumondii TTO1 | Unnamed protein product (probable metalloprotease) | 77 | 24 | 365 | BX571863 |
Effect on insect antibacterial activity.
The antibacterial activity of the hemolymph of G. mellonella was reduced to ca. 50% after incubation in vitro with PrtA and PrtS. Both proteases were also able to reduce the antibacterial activity of cecropin A by 80%. PrtS completely reduced the activity of cecropin B, whereas PrtA caused a reduction of 75%.
DISCUSSION
Photorhabdus sp. strain Az29 produces high amounts of extracellular proteases. The maximum proteolytic activity was detected on supernatants at the end of the exponential growth phase at the three temperatures tested. The activity decreased during the stationary phase at 23 and 28°C, whereas it remained constant at 10°C. The maximum proteolytic activity recovered in supernatants was obtained at 10°C, below the optimal growth temperature of this strain (23°C). We suggest that this activity at low temperatures supports the fact that the complex H. bacteriophora Az29 is pathogenic for insects at temperatures ca. 10°C. Protease production by Y. ruckeri (46), Flavobacterium psychrophilum (47) and lipase production by Pseudomonas fluorescens (11, 54) has been also demonstrated to be higher at temperatures below the optimal growth temperature. Despite the mechanisms of regulation by temperature are poorly understood, Woods et al. (54) suggested that on lipase production by P. fluorescens a posttranscriptional or posttranslational regulation mechanism is involved. In this bacterium, the gene for lipase (lipA) is part of the aprX-lipA operon that includes the genes for a metalloprotease (aprX), for a protease inhibitor and for type I secretion functions. This operon has a strong homology to the prtA operon of P. luminescens W14 (7), suggesting a similar mechanism of temperature regulation.
A variable number of proteases obtained from different strains of Photorhabdus under different growth conditions and different times of incubation have been reported. In most cases, a large protease was evidenced on zymograms, along with other smaller bands also exhibiting proteolytic activity. Bowen et al. (7) purified PrtA from P. temperata K122 with 55 kDa, which is probably related to the 61-kDa protease purified from P. luminescens Hm by Schmidt et al. (44), with the 57-kDa protease reported for P. luminescens Hp (40) and with the 55-kDa protease purified from P. luminescens W14 (8). The small molecules with proteolytic activity on zymograms have been suggested to be degradation products of the major protease (8, 40). Guo et al. (24) reported the purification of two metalloproteases secreted by P. luminescens W14 with molecular masses of 58 and 38 kDa, but no further information on these molecules was available.
In the present study we have accomplished the identification of two distinct proteases, PrtA (56 kDa) and PrtS (38 kDa). Despite the lack of information on N-terminal sequences for these proteases, the fact that polyclonal antibody against PrtA did not recognize PrtS supports the assumption that they are two distinct proteins. These differences were reinforced by other biochemical characterization. Both proteases are inhibited by 1,10-phenantroline, indicating that they are metalloproteases; however, PrtS was not inhibited by EDTA, thus showing it is distinct from PrtA. The proteases showed also differences on zymogram analysis with different substrates. PrtA hydrolyzed casein and gelatin, whereas PrtS hydrolyzed only casein. These enzymes also have differences in optimal pH, with PrtA being more sensitive to pH variations than PrtS. Both enzymes were active over a wide range of temperatures, showing optimal caseinolytic activity at 50°C. The thermal stability was comparable to that of proteases purified from P. luminescens W14 and P. temperata K122 (7), and it has also been exhibited by other psychrophilic bacteria belonging to the genus Pseudomonas (30).
Homology analysis with amino acid sequences from peptides resulting from internal digestion of PrtA from Photorhabdus sp. strain Az29 revealed identity with the sequence of PrtA, a metalloprotease sequenced from P. temperata K122 and homology between 85 and 100% with PrtA from P. luminescens W14. The same fragments also show strong homology with metalloproteases from Pseudomonas and Erwinia spp. This homology supports the classification of PrtA as a zinc-dependent metalloprotease, which was also supported by the analysis of the fragment PrtA-36 that reveals the zinc-binding sequence (HEXXHXXGXXH) characteristic of the metzincin clan of metalloproteases (23). Three of the PrtS peptides show homology with a putative metalloprotease from P. luminescens subsp. laumondii TTO1. This putative metalloprotease contains a conserved domain characteristic of bacterial elastases.
One of the most intriguing aspects about the entomopathogenic nematode-bacterium complex is its ability to escape insect defenses. In initial studies, this ability was attributed to the nematode, but Photorhabdus by itself multiplies rapidly escaping insect defenses (13, 15). In insects infected with the nematode-bacterium complex, soluble immunodepression factors were found, suggesting that proteases could be mediators of this activity (28). In the present study we have shown that PrtA and PrtS inhibited the antibacterial activity, particularly in the in vitro cecropin A and B assays. Recently, a factor of unknown nature involved in the inhibition of hemocyte-mediated phagocytosis was identified in the growth medium of P. luminescens W14 (48). The combined action of metalloproteases and phagocytosis inhibition must be key factors on the Photorhabdus invasion. Moreover, it has been suggested that the proteases from Photorhabdus could be considered as virulence factors. It has been proved that the proteases are not orally toxic (8) but that they participated in the maturation of the Tc toxins (24). Just a small group of Photorhabdus strains, however, carry the three genes in the tca operon that confers the oral toxicity, whereas the non-orally toxic group lacks the tcaA and tcaB genes (34). However, all of them cause disease and insect death, thus suggesting they have other active pathogenic mechanisms.
The fact that the metalloprotease of Photorhabdus is expressed during the initial phase of the installation on the insect host simultaneously with the tcaB toxin (15) and that it is an RTX-like metalloprotease reinforces the assumption that it is implicated on the entire process of infection. Once Photorhabdus is inside the insect's hemocoel, the organisms lodge around the midgut, between the cells of the epithelium and the sheathing extracellular matrix, causing severe damage to the surrounding tissues, before they spread all over the cadaver (48). The metzincin clan of metalloproteases, in which PrtA is included, is known to cause severe damage to host tissues (36). In in vitro assays Bowen et al. (7) showed that PrtA was cytotoxic to mammalian cells in a way similar to the cytotoxic activity reported for a metalloprotease from S. marcescens (35). On the other hand, PrtS is homologous with a putative metalloprotease containing a conserved domain of elastases. The participation of elastases on extracellular matrix degradation is also well documented (36). The degradation of insect tissues being determinant on the pathogenic process is also relevant for the symbiosis because it provides nutrients to the associated nematode, which is not able to grow on insects without a previous bioconversion by the symbiotic bacteria. The role of proteases from Photorhabdus, either in parasitism or in symbiosis, will probably be better understood when encoding genes are mutated or expressed in bacteria other than Photorhabdus. Gene sequencing of PrtA and PrtS of Photorhabdus sp. strain Az29 has been undertaken and will give further insight into this matter.
Acknowledgments
This study was supported by Grants of Fundação para a Ciência e Tecnologia (FCT) of Portugal through POCTI (BSE/41630/01) and PRAXIS (PCNA/C/BIA/109/96). A.C. and A.P. were recipients of a postdoctoral fellowship and of an initiation to research fellowship, respectively, from FCT.
We thank A. S. Martins for critical reading and corrections of the manuscript.
REFERENCES
- 1.Akhurst, R. J. 1982. Antibiotic activity of Xenorhabdus spp., bacteria symbiotically associated with insect pathogenic nematodes of the families Heterorhabditidae and Steinernematidae. J. Gen. Microbiol. 128:3061-3066. [DOI] [PubMed] [Google Scholar]
- 2.Akhurst, R. J., and G. B. Dunphy. 1993. Tripartite interaction between symbiotically associated entomopathogenic bacteria, nematodes and their insect host, p. 1-23. In N. E. Beckage, S. N. Thompson, and B. A. Federici (ed.), Parasites and pathogens of insects, vol. I. Academic Press, Inc., San Diego, Calif. [Google Scholar]
- 3.Azghani A. O., J. W. Baker, S. Shetty, E. J. Miller, and G. J. Bhat. 2002. Pseudomonas aeruginosa elastase stimulates ERK signaling pathway and enhances IL-8 production by alveolar epithelial cells in culture. Inflamm. Res. 51:506-510. [DOI] [PubMed] [Google Scholar]
- 4.Basset, A., R. S. Khush, A. Braun, L. Gardan, F. Boccard, J. A. Hoffmann, and B. Lemaitre. 2000. The phytopathogenic bacteria Erwinia carotovora infects Drosophila and activates an immune response. Proc. Natl. Acad. Sci. USA 97:3376-3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boemare, N. E., and R. J. Akhurst. 1988. Biochemical and physiological characterization of colony form variants in Xenorhabdus spp. (Enterobacteriaceae). J. Gen. Microbiol. 134:751-761. [Google Scholar]
- 6.Boemare, N. E., R. J. Akhurst, and R. G. Mourant. 1993. DNA relatedness between Xenorhabdus spp. (Enterobacteriaceae), symbiotic bacteria of entomopathogenic nematodes, and a proposal to transfer Xenorhabdus luminescens to a new genus, Photorhabdus gen. nov. Int. J. Syst. Bacteriol. 43:249-255. [Google Scholar]
- 7.Bowen D. J., T. A. Rochelau, C. K. Grutzmacher, L. Meslet, M. Vallens, D. Marble, A. Dowling, R. H. Ffrench-Constant, and M. A. Blight. 2003. Genetic and biochemical characterization of PrtA, an RTX-like metalloprotease from Photorhabdus. Microbiology 149:1581-1591. [DOI] [PubMed] [Google Scholar]
- 8.Bowen, D., M. Blackburn, T. A. Rocheleau, C. Grutzmacher, and R. H. Ffrench-Constant. 2000. Secreted proteases from Photorhabdus luminescens: separation of the extracellular proteases from the insecticidal Tc toxin complexes. Insect. Biochem. Mol. Biol. 30:69-74. [DOI] [PubMed] [Google Scholar]
- 9.Bowen, D., T. A. Rocheleau, M. Blackburn, O. Andreev, E. Golubeva, R. Bhartia, and R. H. Ffrench-Constant. 1998. Insecticidal toxins from the bacterium Photorhabdus luminescens. Science 280:2129-2132. [DOI] [PubMed] [Google Scholar]
- 10.Bradford, M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 11.Burger, M., R. G. Woods, C. McCarthy, and I. R. Beacham. 2000. Temperature regulation of protease in Pseudomonas fluorescens LS107d2 by an ECF sigma factor and a transmembrane activator. Microbiology 146:3149-3155. [DOI] [PubMed] [Google Scholar]
- 12.Chen, G., Y. Zhang, J. Li, G. B. Dunphy, Z. K. Punja, and J. M. Webster. 1996. Chitinase activity of Xenorhabdus and Photorhabdus species, bacterial associates of entomopathogenic nematodes. J. Invertebr. Pathol. 68:101-108. [DOI] [PubMed] [Google Scholar]
- 13.Clarke, D. J., and B. C. A. Dowds. 1995. Virulence mechanisms of Photorhabdus sp. strain K122 toward wax moth larvae. J. Invertebr. Pathol. 66:149-155. [Google Scholar]
- 14.Daborn, P. J., N. Waterfield, C. P. Silva, C. P. Y. Au, S. Sharma, and R. H. Ffrench-Constant. 2002. A single Photorhabdus gene makes caterpillars floppy (mcf) allows Escherichia coli to persist within and kill insects. Proc. Natl. Acad. Sci. USA 99:10742-10747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Daborn, P. J., N. Waterfield, M. A. Blight, and R. H. Ffrench-Constant. 2001. Measuring virulence factor expression by the pathogenic bacterium Photorhabdus luminescens in culture and during insect infection. J. Bacteriol. 183:5834-5839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dowds, B. C. A., and A. Peters. 2002. Virulence mechanisms, p. 79-98. In R. Gaugler (ed.), Entomopathogenic nematology. CABI Publishing, Oxford, England.
- 17.Duong, F., A. Lazdunski, B. Cami, and M. Murgier. 1992. Sequence of a cluster of genes controlling synthesis and secretion of alkaline protease in Pseudomonas aeruginosa: relationships to other secretory pathways. Gene 121:47-54. [DOI] [PubMed] [Google Scholar]
- 18.Férnandez, L., P. Secades, J. R. Lopez, I. Márquez, and J. A. Guijarro. 2002. Isolation and analysis of a protease gene with an ABC transport system in the fish pathogen Yersinia ruckeri: insertional mutagenesis and involvement in virulence. Microbiology 148:2233-2243. [DOI] [PubMed] [Google Scholar]
- 19.Fischer-Le Saux, M., V. Viallard, B. Brunel, P. Normand, and N. E. Boemare. 1999. Polyphasic classification of the genus Photorhabdus and proposal of new taxa: P. luminescens subsp. luminescens subsp. nov., P. luminescens subsp. akhurstii subsp. nov., P. luminescens subsp. laumondii subsp. nov., P. temperata sp. nov., P. temperata subsp. temperata subsp. nov., and P. asymbiotica sp. nov. Int. J. Syst. Bacteriol. 49:1645-1656. [DOI] [PubMed] [Google Scholar]
- 20.Forst, S., B. Dowds, N. Boemare, and E. Stackebrandt. 1997. Xenorhabdus and Photorhabdus spp.: bugs that kill bugs. Annu. Rev. Microbiol. 51:47-72. [DOI] [PubMed] [Google Scholar]
- 21.Forst, S., and D. Clarke. 2002. Bacteria-nematode symbiosis, p. 57-77. In R. Gaugler (ed.), Entomopathogenic nematology. CABI Publishing, Oxford, England.
- 22.Galloway, D. R. 1991. Pseudomonas aeruginosa elastase and elastolysis revisited: recent developments. Mol. Microbiol. 5:2315-2321. [DOI] [PubMed] [Google Scholar]
- 23.Gomis-Rüth, F. X. 2003. Structural aspects of the metzicin clan of metalloendopeptidases. Mol. Biotechnol. 24:157-202. [DOI] [PubMed] [Google Scholar]
- 24.Guo, L., R. Fatig III, G. L. Orr, B. W. Shafer, J. A. Strickland, K. Sukhapinda, A. T. Woodsworth, and J. K. Pettel. 1999. Photorhabdus luminescens W-14 insecticidal activity consists of at least two similar but distinct proteins. J. Biol. Chem. 14:9836-9842. [DOI] [PubMed] [Google Scholar]
- 25.Hoffmann, D., D. Hultmark, and H. G. Boman. 1981. Insect immunity: Galleria mellonella and other Lepidoptera have cecropia-P9-like factors active against gram-negative bacteria. Insect. Biochem. 11:537-548. [Google Scholar]
- 26.Holder, I. A., and A. N. Neely. 1989. Pseudomonas elastase acts as a virulence factor in burned hosts by Hageman factor-dependent activation of the host kinin cascade. Infect. Immun. 57:3345-3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ichinose, Y., M. Ehara, T. Honda, and T. Miwatani. 1994. The effect on enterotoxicity of protease purified from Vibrio cholerae O1. FEMS Microbiol. Lett. 115:265-271. [DOI] [PubMed] [Google Scholar]
- 28.Jarosz, J. 1998. Active resistance of entomophagous rhabditid Heterorhabditis bacteriophora to insect immunity. Parasitology 117:201-208. [DOI] [PubMed] [Google Scholar]
- 29.Kamata, R., K. Matsumoto, R. Okamura, T. Yamamoto, and H. Maeda. 1985. The serratial 56K protease as a major pathogenic factor in serratial keratitis: clinical and experimental study. Ophthalmology 92:1452-1459. [DOI] [PubMed] [Google Scholar]
- 30.Koka, R., and B. C. Weimer. 2000. Isolation and characterization of a protease from Pseudomonas fluorescens RO98. J. Appl. Microbiol. 89:280-288. [DOI] [PubMed] [Google Scholar]
- 31.Laemmli, U. K. 1970. Cleavage of structural proteins during assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 32.Maeda, H., A. Molla, T. Oda, and T. Katsuki. 1987. Internalization of serratial protease into cells as an enzyme-inhibitor complex with α2-macroglobulin and regeneration of protease activity and cytotoxicity. J. Biol. Chem. 62:10946-10950. [PubMed] [Google Scholar]
- 33.Marits, R., V. Kõiv, E. Laasik, and A. Mäe. 1999. Isolation off an extracellular protease gene of Erwinia carotovora subsp. carotovora strain SCC3193 by transposon mutagenesis and the role of protease in phytopathogenicity. Microbiology 145:1959-1966. [DOI] [PubMed] [Google Scholar]
- 34.Marokhazi, J., N. Waterfield, G. LeGoff, E. Feil, R. Stabler, J. Hinds, A. Fodor, and R. H. Ffrench-Constant. 2003. Using a DNA microarray to investigate the distribution of insect virulence factors in strains of Photorhabdus bacteria. J. Bacteriol. 185:4648-4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marty, K. B., C. L. Williams, L. J. Guynn, M. J. Benedik, and S. R. Blanke. 2002. Characterization of a cytotoxic factor in culture filtrates of Serratia marcescens. Infect. Immun. 70:1121-1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miyoshi, S., and S. Shinoda. 2000. Microbial metalloproteases and pathogenesis. Microbes Infect. 2:91-98. [DOI] [PubMed] [Google Scholar]
- 37.Miyoshi, S., H. Nakazawa, K. Kawata., K. Tomochika, K. Tobe, and S. Shinoda. 1998. Characterization of the hemorrhagic reaction caused by Vibrio vulnificus metalloprotease, a member of the thermolysin family. Infect. Immun. 66:4851-4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moncrieff, J. S., R. Obiso, L. A. Barroso, J. J. Kling, R. L. Wright, R. L. Van Tassel, D. M. Lyerly, and T. D. Wilkins. 1995. The enterotoxin of Bacteroides fragilis is a metalloprotease. Infect. Immun. 63:175-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morrissey, J. H. 1981. Silver stain for proteins in polyacrylamide gels: a modified procedure with enhanced uniform sensitivity. Anal. Biochem. 117:307-310. [DOI] [PubMed] [Google Scholar]
- 40.Ong, K. L., and F. N. Chang. 1997. Analysis of proteins from different phase variants of the entomopathogenic bacteria Photorhabdus luminescens by two-dimensional zymography. Electrophoresis 18:834-839. [DOI] [PubMed] [Google Scholar]
- 41.Rosa, J. S., and N Simões. 2004. Evaluation of twenty-eight strains of Heterorhabditis bacteriophora isolated in the Azores for biocontrol of the armyworm, Pseudaletia unipuncta (Lepidoptea: Noctuidae). Biol. Control 29:409-417. [Google Scholar]
- 42.Rosa, J. S., C. Cabral, and N. Simões. 2002. Differences between the pathogenic processes induced by Steinernema and Heterorhabditis (Nemata: Rhabditida) in Pseudaletia unipuncta (Insecta: Lepidoptera). J. Invertebr. Pathol. 80:46-54. [DOI] [PubMed] [Google Scholar]
- 43.Rosa, J. S., E. Bonifassi, J. Amaral, L. A. Lacey, N. Simões, and C. Laumond. 2000. Natural occurrence of entomopathogenic nematodes (Rhabditida: Steinernema, heterorhabditis) in the Azores. J. Nematol. 32:215-222. [PMC free article] [PubMed] [Google Scholar]
- 44.Schmidt, T. M., B. Bleakley, and K. H. Nealson. 1988. Characterization of an extracellular protease from the insect pathogen Xenorhabdus luminescens. Appl. Environ. Microbiol. 54:2793-2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sears, C. L. 2001. The toxins of Bacteroides fragilis. Toxicon 39:1737-1746. [DOI] [PubMed] [Google Scholar]
- 46.Secades, P., and J. A. Guijarro. 1999. Purification and characterization of an extracellular protease from the fish pathogen Yersinia ruckeri and effect of culture conditions on production. Appl. Environ. Microbiol. 65:3969-3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Secades, P., B. Alvarez, and J. A. Guijarro. 2001. Purification and characterization of a psychrophilic, calcium-induced, growth-phase-dependent metalloprotease from the fish pathogen Flavobacterium psychrophilum. Appl. Environ. Microbiol. 67:2436-2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Silva, C. P., N. R. Waterfield, P. J. Daborn, P. Dean, T. Chilver, C. P. Y. Au, S. Sharma, U. Potter, S. E. Reynolds, and R. H. Ffrench-Constant. 2002. Bacterial infection of a model insect: Photorhabdus luminescens and Manduca sexta. Cell. Microbiol. 6:329-339. [DOI] [PubMed] [Google Scholar]
- 49.Tomarelli, R. M., J. Charney, and M. L. Harding. 1949. The use of azoalbumin as a substrate in the colorimetric determination of peptic and tryptic activity. J. Lab. Clin. Med. 34:428-433. [PubMed] [Google Scholar]
- 50.Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 76:4350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vollmer, P., I. Walev, S. Rose-John, and S. Bhakdi. 1996. Novel pathogenic mechanism of microbial metalloproteases: liberation of membrane-anchored molecules in biologically active form exemplified by studies with the human interleukin-6 receptor. Infect. Immun. 64:3646-3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Waterfield, N. R., D. J. Bowen, J. D. Fetherston, R. D. Perry, and R. H. Ffrench-Constant. 2001. The toxin complex genes of Photorhabdus: a growing gene family. Trends Microbiol. 9:185-191. [DOI] [PubMed] [Google Scholar]
- 53.Waterfield, N., P. J. Daborn, and R. H. Ffrench-Constant. 2002. Genomic islands in the insect pathogen Photorhabdus. Trends Microbiol. 10:541-545. [DOI] [PubMed] [Google Scholar]
- 54.Woods, R. G., M. Burger, C. A. Beven, and I. R. Beacham. 2001. The aprX-lipA operon of Pseudomonas fluorescens B52: a molecular analysis of metalloprotease and lipase production. Microbiology 147:345-354. [DOI] [PubMed] [Google Scholar]
- 55.Wu, S., L. A. Dreyfus, A. O. Tzianabos, C. Hayashi, and C. L. Sears. 2002. Diversity of the metalloprotease toxin produced by enterotoxigenic Bacteroides fragilis. Infect. Immun. 70:2463-2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang, Y., D. D. Bak, H. Heid, and K. Geider. 1999. Molecular characterization of a protease secreted by Erwinia amylovora. J. Mol. Biol. 289:1239-1251. [DOI] [PubMed] [Google Scholar]