Abstract
To obtain a mannitol-producing Lactococcus lactis strain, the mannitol 1-phosphate dehydrogenase gene (mtlD) from Lactobacillus plantarum was overexpressed in a wild-type strain, a lactate dehydrogenase(LDH)-deficient strain, and a strain with reduced phosphofructokinase activity. High-performance liquid chromatography and 13C nuclear magnetic resonance analysis revealed that small amounts (<1%) of mannitol were formed by growing cells of mtlD-overexpressing LDH-deficient and phosphofructokinase-reduced strains, whereas resting cells of the LDH-deficient transformant converted 25% of glucose into mannitol. Moreover, the formed mannitol was not reutilized upon glucose depletion. Of the metabolic-engineering strategies investigated in this work, mtlD-overexpressing LDH-deficient L. lactis seemed to be the most promising strain for mannitol production.
Mannitol is a sugar alcohol that is produced by a wide variety of organisms. It is assumed to have several beneficial effects as a food additive. It can serve as an antioxidant (5, 6, 24, 25) and as a low-calorie sweetener, which can replace sucrose (7, 10). Efiuvwevwere and coworkers (11) showed that mannitol has an osmoprotectant and antioxidant effect on the dairy lactic acid bacterium Lactococcus lactis subjected to decreased water activity and that mannitol enhances survival during drying of starter cells. The viability of starter cultures of L. lactis, which are extensively used in the dairy industry, may thus be enhanced by mannitol production in these strains. In addition, the use of mannitol-producing L. lactis strains may result in fermented products with extra nutritional value.
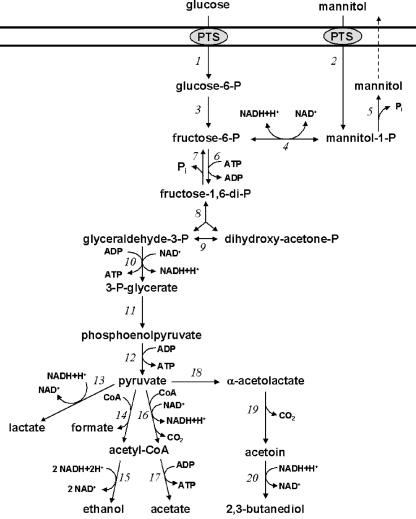
Mannitol biosynthesis in homofermentative lactic acid bacteria, such as L. lactis, starts with the glycolysis intermediate fructose 6-phosphate (Fig. 1). Mannitol 1-phosphate dehydrogenase (MPDH) (EC 1.1.1.17) catalyzes the reduction of fructose 6-phosphate, and also the reverse reaction, the oxidation of mannitol 1-phosphate (4, 12). Mannitol 1-phosphate is dephosphorylated to mannitol by mannitol phosphatase activity. Although the gene encoding MPDH (mtlD) has been reported for L. lactis IL-1403 (2), mannitol production by L. lactis and other homofermentative lactic acid bacteria is not very likely. Presumably, the mtlD gene, which is located in a mannitol operon, is not transcribed due to catabolite repression, as described for Bacillus stearothermophilus (14, 15), and no mannitol production can take place during growth on certain sugar substrates. Therefore, overexpression of MPDH might be an important step for mannitol synthesis in L. lactis.
FIG. 1.
Proposed pathway for hexose metabolism of homofermentative lactic acid bacteria. (1) Phosphoenolpyruvate-dependent sugar phosphotransferase system (PTS). (2) Mannitol-specific PTS. (3) Phosphoglucose isomerase. (4) Mannitol 1-phosphate dehydrogenase. (5) Mannitol 1-phosphatase. (6) 6-Phosphofructokinase. (7) Fructose-diphosphatase. (8) Fructose 1,6-diphosphate aldolase. (9) Triosephosphate isomerase. (10) Glyceraldehyde 3-phosphate dehydrogenase and phosphoglycerate kinase. (11) Phosphoglyceromutase and enolase. (12) Pyruvate kinase. (13) Lactate dehydrogenase. (14) Pyruvate-formate lyase. (15) Acetaldehyde dehydrogenase and alcohol dehydrogenase. (16) Pyruvate dehydrogenase. (17) Acetate kinase. (18) α-Acetolactate synthase. (19) α-Acetolactate decarboxylase. (20) 2,3-Butanediol dehydrogenase.
In contrast to some heterofermentative lactic acid bacteria, mannitol production by homofermentative lactic acid bacteria is not very common. However, mannitol production by L. lactis was observed by Neves and coworkers (22). In resting high-density cell suspensions of a lactate dehydrogenase (LDH)-deficient L. lactis strain, high levels of intracellular mannitol were produced. Upon glucose depletion, mannitol was remetabolized. Also an LDH-negative mutant of Lactobacillus plantarum produced small amounts of mannitol (12). In these cases, mannitol production was believed to be an alternative pathway to regenerate NAD+ instead of lactate formation.
Metabolic engineering can be a helpful tool to achieve mannitol production in L. lactis. In this report, mtlD from L. plantarum was cloned and overexpressed in L. lactis. The involvement of fructose 6-phosphate as a substrate of MPDH could imply that the accumulation of fructose 6-phosphate, such as was reported in L. lactis with reduced phosphofructokinase (PFK) activity (1), could coincide with mannitol production. Also, alternative NAD+ regeneration via MPDH, as reported in an LDH-deficient L. lactis strain (22), could contribute to mannitol production. Therefore, mtlD was overexpressed in different genetic backgrounds: the parental L. lactis strain, NZ9000; an LDH-deficient strain, NZ9010; and a strain with reduced PFK activity, HWA217. Also, a comparison was made of mannitol production by growing cells and by high-density resting cells.
MATERIALS AND METHODS
L. lactis strains, plasmids, and media.
The L. lactis strains and plasmids used in this study are listed in Table 1. The L. lactis strains were grown at 30°C in M17 broth (Oxoid) supplemented with 0.5% glucose. For (semi)anaerobic cultivations, cells were grown in batch cultures in 50-ml tubes without aeration. When cells were grown aerobically, shaking flasks with baffles were used. When applicable, chloramphenicol and erythromycin were supplemented at 10 and 5 μg/ml, respectively. Growth was followed by measuring the optical density at 600 nm (OD600) with an Ultrospec 2000 spectrophotometer (Pharmacia Biotech). For inducing MPDH activity, 1 ng of nisin/ml was added to a growing culture at an OD600 of 0.5.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strains and plasmids | Characteristics | Reference(s) or source |
|---|---|---|
| Strains | ||
| L. lactis NZ9000 | MG1363 pepN::nisRK | 9, 20a |
| L. lactis NZ9010 | NZ9000 ldh::ery Eryr | 3, 17 |
| L. lactis HWA217 | Reduced PFK activity | 1 |
| Plasmids | ||
| pNZ8148 | pNZ8048 derivative; Cmr; lactococcal cloning and expression vector with nisA promoter upstream of a multi- ple cloning site | 9, 20a |
| pNZmt1D | pNZ8148 carrying L. plantarum mtlD gene | This work |
| pNZ9530 | EryrnisRK | 18 |
Construction of plasmid pNZmtlD.
The gene encoding mannitol 1-phosphate dehydrogenase (mtlD) from L. plantarum was cloned into the nisin-inducible expression vector pNZ8148 (Table 1). For this, mtlD was amplified by PCR from L. plantarum genomic DNA (accession no. NP 784055) (19) using theprimers MPDHLP-1FW (5′-TCGTACCATGGTAGACGTACATTTTG-3′)and MPDHLP-3RV (5′-GTCAGTCTAGACTACTTTGCTGCAGCTAAG-3′), with introduced NcoI and XbaI digestion sites, respectively (underlined). NcoI-XbaI-digested mtlD was cloned into pNZ8148, resulting in pNZmtlD containing mtlD fused to the nisA promoter. The sequence of mtlD was verified by sequencing the cloned PCR product (Eurogentec, Seraing, Belgium). pNZmtlD was cloned into the L. lactis strains NZ9000, NZ9010, and HWA217 (Table 1). Plasmid pNZ9530, containing nisR and nisK genes, was cotransformed in L. lactis HWA217 to make nisin induction of mtlD possible. The nisRK genes, coding for the histidine protein kinase NisK and the response regulator NisR, are the only nis genes required for nisA promoter activation on the pNZmtlD plasmid (18).
Analysis of fermentation products and glucose consumption.
During the growth experiments, samples were taken from the L. lactis cultures and centrifuged for 1 min at 10,000 × g, and the supernatants were stored at −20°C until they were analyzed. In the supernatant, lactate, acetate, formate, glucose, mannitol, ethanol, 2,3-butanediol, and acetoin were analyzed by high-performance liquid chromatography (HPLC). Separation was performed with a 30-cm-long ION-300 ion exclusion column (Alltech, Breda, The Netherlands) at a flow rate of 0.4 ml/min and a temperature of 90°C. The eluent consisted of 3 mM sulfuric acid. The products were detected on a refractive-index detector (Waters 410). The distribution of glucose to different fermentation products was calculated as the slope of the product concentration versus the consumed glucose.
Preparation of cell extracts.
Cell extracts were prepared by disruption of cells by glass beads. A cell culture (50 ml) was centrifuged (4°C; 20 min at 2,000 × g) and washed with 50 mM morpholineethanesulfonic acid (MES) buffer (pH 7.0). The cells were resuspended in 2 ml of 50 mM MES buffer (pH 7.0). For cell disruption, 1 ml of cell suspension was added to 1.0 g of 0.1-mm-diameter zirconium-silica beads (BioSpec Products, Inc.) in a 2-ml Eppendorf cup, and the cells were disrupted by vigorous shaking at 4°C for 5 min. Cell debris was removed by centrifugation (4°C; 2 min at 10,000 × g), and the supernatant was used for all enzyme assays. The protein contents of the extracts were determined by the bicinchoninic acid protein assay (Pierce) with bovine serum albumin as the standard.
Enzyme assays.
Cell cultures (50 ml) were harvested for enzyme assays at an OD600 of ±1.3 or 2 h after induction with nisin, and cell extracts were prepared as described above. LDH activity was determined by the method of Hillier and Jago (16). PFK activity was assayed according to the method of Grobben et al. (13), with the modification that 5 mM fructose 6-phosphate was used to initiate the reaction. The PFK activities of extracts with high MPDH activity could not be determined, as fructose 6-phosphate is a substrate for both PFK and MPDH. Mannitol 1-phosphate oxidation by MPDH was determined in a reaction mixture containing 25 mM Tris-HCl, pH 8.0, and 1.5 mM NAD. The reaction was started with the addition of 1 mM mannitol 1-phosphate. The reduction of fructose 6-phosphate was assayed in 25 mM sodium phosphate buffer, pH 6, with 0.15 mM NADH. Fructose 6-phosphate (1 mM) was used to initiate the reaction. LDH, PFK, and MPDH activities were determined from the rate of NADH oxidation or formation at 30°C by measuring the absorbance at 340 nm (Ultrospec 2000).
Mannitol 1-phosphatase activity was determined in a 1-ml reaction mixture containing 0.1 to 0.2 mg of protein enzyme extract/ml, 50 mM MES buffer (pH 7.0), 10 mM MgCl2, and 3 mM mannitol 1-phosphate. The amounts of inorganic phosphate formed were determined after 0, 60, 120, and 180 min of incubation at 30°C by a modified protocol of the Sigma inorganic phosphate kit. At the above time points, 200-μl samples were taken, and the reaction was stopped with the addition of 40 μl of acid molybdate solution. Ten microliters of Fiske & SubbaRow reducer solution was added to 200 μl of the clear centrifuged solution in a 96-well microplate, and the absorbance at 655 nm was measured with a microplate reader (3550-UV; Bio-Rad).
NMR experiments.
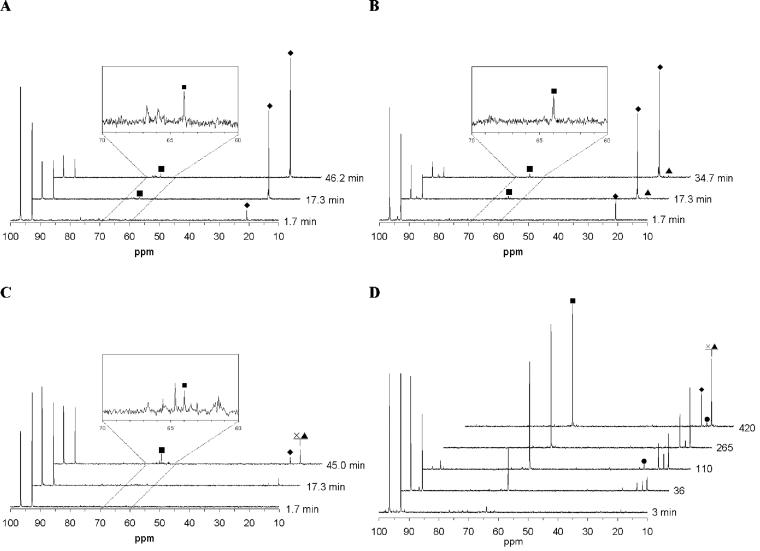
Cells grown on M17 broth were harvested at an OD600 of ∼1.5, centrifuged, washed, and resuspended in 50 mM potassium phosphate buffer (pH 6.5) to an OD600 of ±35. 13C nuclear magnetic resonance (NMR) spectra were taken using a Bruker AMX-400Wb spectrometer at 100.62 MHz. All experiments were carried out at 30°C in a 10-mm NMR tube. Cell suspension (4 ml) was placed in the NMR tube, and an initial spectrum was acquired. At time zero, 20 mM [1-13C]glucose (Campro Scientific, Veenendaal, The Netherlands) was supplied in the NMR tube. 13C-NMR spectra were acquired for 104 s (64 scans). Chemical shifts were referred to the β-C1 of d-glucose (96.6 ppm). Resonances in the spectra (Fig. 2) were identified by spiking with the pure (unlabeled) materials.
FIG. 2.
13C-NMR spectra during metabolism of [1-13C]glucose by high-density suspensions of nongrowing cells of the L. lactis strains NZ9000/pNZmtlD (A), HWA217/pNZmtlD (B), NZ9010 (C), and NZ9010/pNZmtlD (D). The spectra were recorded until no changes could be detected. The resonances of α and β C1 of d-glucose were detected at 92.8 and 96.6 ppm. ⧫, 13C-labeled lactate; •, acetoin; ×, 2,3-butanediol; ▴, ethanol; ▪, mannitol.
Quantification of products by 13C-NMR.
Glucose, lactate, acetoin, 2,3-butanediol, ethanol, and mannitol were quantified during the consumption of [1-13C]glucose. Due to the fast pulsing conditions and short repetition times, the in vivo NMR spectra were not fully relaxed, and therefore there was not a direct correlation between peak intensities and concentrations. To correct for saturation, the saturation recovery method was used by performing relaxation measurements.
RESULTS
In this work, we used different metabolic-engineering approaches to induce mannitol production in L. lactis. Basically, we used the glycolytic model of L. lactis (17; http://jjj.biochem.sun.ac.za) to predict the most efficient metabolic-engineering strategy. The model predicted that knocking out the LDH gene and decreasing the PFK activity could play a significant role in mannitol production in L. lactis by increasing the intracellular NADH and fructose 6-phosphate levels. Mannitol production in three L. lactis strains was determined: the wild-type strain, NZ9000, the LDH-deficient strain NZ9010 (17), and the PFK-reduced strain HWA217 (1). Enzyme activities in mid-log-phase cultures were measured, fermentation products in supernatants of batch grown cultures were analyzed, and 13C-NMR spectra were recorded during [1-13C]glucose consumption by cell suspensions.
MPDH enzyme activity in L. lactis.
Both L. lactis strains NZ9000 and HWA217 that were grown on glucose did not show any MPDH activity, while cell extracts of L. lactis NZ9010 contained very low MPDH activity (Table 2). Since MPDH activity is essential for mannitol production, the corresponding gene of L. plantarum (mtlD) was overexpressed in all three L. lactis strains using pNZmtlD (Table 1). The transformants were grown on glucose and induced with nisin, and the cells were harvested after 2 h of induction. In all three hosts, the introduction of pNZmtlD led to a large increase in MPDH activity (Table 2). The low MPDH activity exhibited by the noninduced culture can be explained by the very low residual activity of the nisin promoter in the pNZ8148 expression vector under noninduced conditions (8, 9).
TABLE 2.
Enzyme activities determined in crude cell extracts of the various L. lactis strains grown on glucose
| L. lactis strain | Nisin (ng/ml) | Activitya
|
||||
|---|---|---|---|---|---|---|
| LDH | PFK | MPDH
|
MP | |||
| (F6P → M1P) | (M1P → F6P) | |||||
| NZ9000 | 0 | 13.5 ± 3.5 | 1.6 ± 0.6 | ND | ND | 4.2 ± 1.5 |
| NZ9000/pNZmt1D | 0 | 15.8 ± 2.7 | 1.8 ± 0.6 | ND | 0.03 ± 0.01 | 5.2 ± 1.0 |
| NZ9000/pNZmt1D | 1 | 16.1 ± 2.8 | −b | 6.2 ± 0.4 | 20.9 ± 0.3 | 3.5 ± 0.3 |
| NZ9010 | 0 | ND | 1.5 ± 0.5 | 0.02 ± 0.02 | ≤0.01 | 4.9 ± 0.4 |
| NZ9010/pNZmt1D | 0 | ND | 1.2 ± 0.6 | 0.03 ± 0.02 | 0.03 ± 0.02 | 5.6 ± 0.3 |
| NZ9010/pNZmt1D | 1 | ND | − | 9.8 ± 1.6 | 30.3 ± 8.3 | 3.6 ± 0.2 |
| HWA217/pNZ9530 | 0 | 17.0 ± 5.2 | 0.6 ± 0.4 | ND | ND | 8.6 ± 1.9 |
| HWA217/pNZ9530/pNZmt1D | 0 | 19.2 ± 1.2 | 0.3 ± 0.2 | 0.04 ± 0.01 | 0.12 ± 0.09 | 7.5 ± 0.8 |
| HWA217/pNZ9530/pNZmt1D | 1 | 17.7 ± 6.0 | − | 2.5 ± 0.9 | 8.4 ± 2.3 | 6.7 ± 2.4 |
In micromoles per minute per milligram of protein (nanomoles per minute per milligram of protein for mannitol 1-phosphatase [MP]). ND, not detected. Cell extracts were made from cultures during exponential growth at an OD600 of 1.3 to 1.6 or after 2 h of induction with nisin (mtlD overexpression strains).
−, not determined.
Mannitol production by mtlD constructs.
The effect of overexpression of mtlD on the formation of fermentation products was determined by cultivating the strains in batch cultures on M17 medium under oxic or anoxic conditions. To induce MPDH activity, 1 ng of nisin/ml was added to growing cultures of the strains containing the pNZmtlD plasmid, and the fermentation products were determined over time in the culture supernatant (Table 3). Furthermore, in vivo 13C-NMR measurements were performed with suspensions of glucose-grown cells (Fig. 2 and Table 4). The consumption of [1-13C]glucose by the suspensions was monitored until no changes in the spectra were observed. Resonances in the spectra of these measurements (Fig. 2) were identified as lactate (20.7 ppm), acetoin (18.8 ppm), ethanol (17.5 ppm), 2,3-butanediol (17.4 ppm), and mannitol (63.9 ppm) by spiking with the pure (unlabeled) materials.
TABLE 3.
Amounts of fermentation products formed per mole of consumed glucose during growth on 0.5% glucose
| L. lactis strain | ox | Nisin (ng/ml) | Amt of product (mol mol of glucose−1)
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Lactate | Formate | Acetate | Ethanol | Acetoin | 2,3-Butanediol | Pyruvate | Mannitol | C recovery | |||
| NZ9000 | 1.71 | 0.86 | |||||||||
| NZ9000/pNZmt1D | 1.74 | 0.87 | |||||||||
| NZ9000/pNZmt1D | 1 | 1.74 | 0.87 | ||||||||
| NZ9010 | 0.13 | 1.1 | 0.15 | 1.1 | 0.28 | 0.07 | 1.04 | ||||
| NZ9010 | +a | 0.02 | 0.09 | 0.12 | 0.57 | 0.03 | 0.26 | 0.85 | |||
| NZ9010/pNZmt1D | 0.06 | 1.0 | 0.37 | 0.77 | 0.17 | 0.16 | 0.006 | 0.94 | |||
| NZ9010/pNZmt1D | 1 | 0.08 | 0.93 | 0.15 | 0.87 | 0.24 | 0.15 | 0.011 | 0.95 | ||
| NZ9010/pNZmt1D | + | 0.02 | 0.22 | 0.58 | 0.16 | 0.78 | |||||
| NZ9010/pNZmt1D | + | 1 | 0.02 | 0.21 | 0.60 | 0.15 | 0.79 | ||||
| HWA217/pNZ9530 | 1.86 | 0.12 | 0.01 | 0.94 | |||||||
| HWA217/pNZ9530/pNZmt1D | 1.83 | 0.01 | 0.005 | 0.93 | |||||||
| HWA217/pNZ9530/pNZmt1D | 1 | 1.87 | 0.01 | 0.008 | 0.95 | ||||||
ox +, oxic growth.
TABLE 4.
Formation of 13C-labeled fermentation products during [1-13C]glucose consumption at 30°C by high-density suspensions of nongrowing L. lactis cells
| L. lactis strain | [13C]glucose consumed (%) | Amt of product (mol mol of [1-13C]glucose−1)a
|
||||||
|---|---|---|---|---|---|---|---|---|
| Lactate | Acetate | Ethanol | Acetoin | 2,3-Butanediol | Mannitol | 13C recovery | ||
| NZ9000 | 70 | 0.68 | 0.68 | |||||
| NZ9000/pNZmt1D | 85 | 0.62 | 0.02 | 0.64 | ||||
| NZ9010 | 44 | 0.17 | 0.22 | 0.24 | 0.02 | 0.65 | ||
| NZ9010/pNZmt1D | 100 | 0.07 (0.3) | (0.14) | 0.03 (0.11) | 0.14 (0.22) | 0.07 (0.15) | 0.15 (0.25) | 0.46 (0.90) |
| HWA217/pNZ9530 | 65 | 0.49 | 0.49 | |||||
| HWA217/pNZ9530/pNZmt1D | 88 | 0.53 | 0.02 | 0.01 | 0.56 | |||
Spectra were recorded for 46.2 min, except for the spectrum of NZ9010/pNZmt1D, which was recorded for 420 min. The values in parentheses are concentrations measured in the supernatants by HPLC.
Overexpression of mtlD in parental L. lactis.
Wild-type L. lactis NZ9000 showed a typical homolactic fermentation pattern, with lactate as the main fermentation product (Table 3). Overexpression of mtlD in NZ9000 resulted in the same homolactic pattern (Table 3), and no mannitol was detected during growth. However, 13C-NMR analysis of [1-13C]glucose-consuming cell suspensions of nisin-induced L. lactis NZ9000/pNZmtlD revealed the accumulation of a small amount of mannitol (Fig. 2A). The conversion rate of glucose into mannitol was 0.02 mol of mannitol per mol of glucose (Table 4).
MtlD overexpression in an L. lactis strain with reduced PFK activity.
Growth of L. lactis with reduced PFK activity (strain HWA217) resulted in a mainly homolactic fermentation pattern with small amounts of acetate and formate as side products, as described by Andersen et al. (1). Introduction of pNZmtlD in HWA217 resulted in mannitol production during growth of 0.005 and 0.008 mol of mannitol per mol of consumed glucose (Table 3). The mannitol production was not dramatically improved when the MPDH activity was increased by nisin induction (Table 2). Analysis of glucose consumption by nongrowing cell suspensions confirmed the homolactic pattern of HWA217 (spectra not shown). 13C-NMR analysis of suspensions of L. lactis HWA217/pNZ9530/pNZmtlD showed that mannitol and ethanol accumulated (spectra not shown). In this strain, 1% of the [1-13C]glucose is converted into [1-13C]mannitol by cell suspensions (Table 4).
Overexpression of mtlD in LDH-deficient L. lactis.
HPLC analysis of the LDH-deficient strain L. lactis NZ9010 showed a mixed-type fermentation pattern when the bacteria were grown on glucose under aerobic and (semi)anaerobic conditions (Table 3), similar to the fermentation patterns of the LDH-deficient strains described by Bongers et al. (3). No mannitol could be detected during growth. The introduction of pNZmtlD in strain NZ9010 resulted in conversion rates of 0.6 and 1.1%, for the uninduced and the induced growing cultures, respectively (Table 3). Production of mannitol was not observed during growth under aerobic conditions (Table 3).
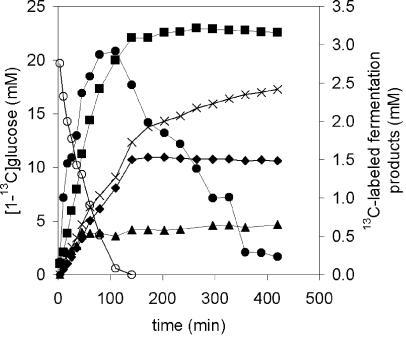
13C-NMR analysis of the glucose metabolism of L. lactis NZ9010 suspensions did not show large amounts of mannitol (Fig. 2C); only 2% of the 13C-labeled glucose was converted into mannitol (Table 4). Furthermore, glucose consumption by the cell suspension of L. lactis NZ9010 was much slower than that of the other strains, since only 44% of the glucose was metabolized during 45 min, whereas the others strains converted 65 to 88% of the glucose in the same time or less (Table 4). Much larger amounts of mannitol were produced by the nisin-induced cell suspensions of L. lactis NZ9010/pNZmtlD (Fig. 2D). A conversion rate as high as 15% (0.15 mol of 13C-labeled-mannitol per mol of [1-13C]glucose) was obtained (Table 4). During glucose consumption, a rapid increase in mannitol was observed, and upon glucose exhaustion, the mannitol produced was not reutilized (Fig. 3).
FIG. 3.
Formation of 13C-labeled fermentation products during [1-13C]glucose consumption by high-density suspensions of nongrowing cells of L. lactis NZ9010/pNZmtlD. The starting concentration of [1-13C]glucose is 20 mM. ○, glucose; ⧫, lactate; •, acetoin; ×, 2,3-butanediol; ▴, ethanol; ▪, mannitol.
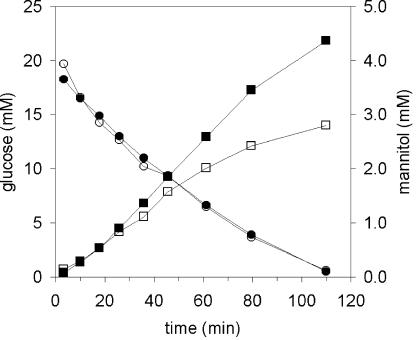
The large difference in mannitol production between growing and nongrowing cells might be explained by intracellular accumulation of mannitol in the resting cells. To determine this, supernatant samples from cell suspensions converting [1-13C]glucose were analyzed by HPLC. The measured concentrations of all fermentation products in the supernatants were higher than the concentrations that were measured with 13C-NMR. The extracellular mannitol production was corrected to 25% (Fig. 4 and Table 4). This suggested the loss of 13C label during the experiment, for example, via CO2 formation. HPLC analysis of cell extracts of glucose-consuming cell suspensions confirmed that mannitol was excreted and not accumulated in the cells (data not shown).
FIG. 4.
Mannitol production and glucose consumption by high-density suspensions of nongrowing cells of L. lactis NZ9010/pNZmtlD. ▪, mannitol, and •, glucose measured by HPLC in the supernatant; □, 13C-labeled mannitol, and ○, glucose measured by 13C-NMR analysis.
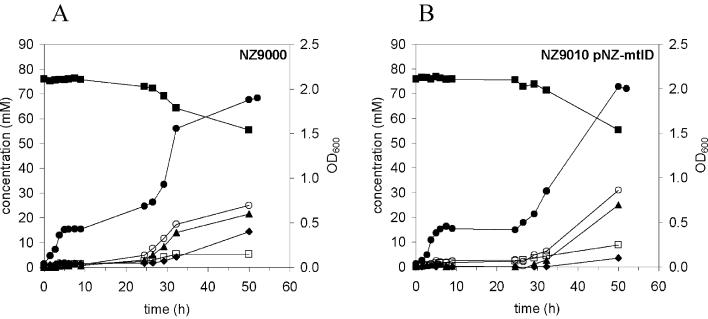
To investigate the mannitol-utilizing abilities of L. lactis NZ9010/pNZmtlD, glucose-precultured NZ9000 and NZ9010/pNZmtlD cells were grown anaerobically on M17 broth supplemented with mannitol. Both NZ9000 and NZ9010/pNZmtlD strains showed a lag phase of ∼24 h before growth on mannitol was observed (Fig. 5).
FIG. 5.
Growth, mannitol consumption, and product formation by L. lactis NZ9000 (A) and NZ9010/pNZ-mtlD (B) cultured under anaerobic conditions. M17 broth supplemented with 75 mM mannitol was inoculated with 2% (vol/vol) glucose-grown overnight culture. ▪, mannitol; ⧫, lactate; ▴, ethanol; ○, formate; □, acetate; •, OD600.
DISCUSSION
We investigated the effect of overexpression of the L. plantarum mtlD gene in three L. lactis hosts: the parental strain NZ9000, the LDH-deficient strain NZ9010, and the PFK-reduced strain HWA217. The mannitol-producing capacities of the constructed transformants were determined by HPLC analysis of supernatants of growing cultures, in vivo 13C-NMR analysis of glucose-consuming cell suspensions, and enzyme activity measurements. By combining these data, the effect of overexpression of mtlD in different genetic backgrounds on the mannitol-producing capacity of L. lactis could be determined.
When mtlD was overexpressed in wild-type L. lactis NZ9000, no mannitol was detected in supernatants of growing cells (Table 3). Although both MPDH and mannitol 1-phosphatase activities were present in cell extracts (Table 2), no mannitol production could be observed. Apparently, the glycolytic flux in the growing cells is too high, and subsequently, the fructose 6-phosphate concentration is too low to enable a flux to mannitol. Also, NAD is regenerated by LDH, so the organism has no need to regenerate NAD by an alternative pathway, such as mannitol synthesis via MPDH. Under nongrowing conditions, mtlD overexpression resulted in a small amount of mannitol (Fig. 2A). About 2% of the glucose was converted to mannitol by this transformant (Table 4). Due to the low glycolytic flux in the nongrowing cells (20), the fructose 6-phosphate concentration might have been just high enough for MPDH to convert fructose 6-phosphate into mannitol 1-phosphate.
Increasing the intracellular fructose 6-phosphate concentration by lowering the PFK activity might be a strategy to enable mannitol production in L. lactis. Andersen and coworkers (1) showed that glycolytic fluxes were reduced in growing cells of an L. lactis mutant with 40% of the PFK activity of the MG1363 wild type. In this strain, sugar phosphates, such as glucose 6-phosphate and fructose 6-phosphate, accumulated. The increase in the fructose 6-phosphate concentration might contribute to a flux toward mannitol 1-phosphate via MPDH. Indeed, when mtlD is overexpressed in the PFK-reduced L. lactis strain HWA217, mannitol production is observed in the supernatants of growing cultures (Table 3). Since no mannitol was produced by the mtlD-overexpressing parental strain NZ9000, the results may imply that the reduction in PFK has resulted in the increased flux toward mannitol. However, nongrowing cells of MPDH-overexpressing HWA217 produced amounts of mannitol similar to those produced by the parental strain with high MPDH overexpression. Since PFK reduction increases the fructose 6-phosphate pool (1), it can be assumed that further increase of the fructose 6-phosphate pool does not increase the conversion of glucose into mannitol by nongrowing cells.
Another strategy to increase the flux to mannitol is to increase the NADH pool by knocking out the LDH activity, which is mainly responsible for the regeneration of NAD+ in L. lactis. Neves et al. (22) showed that nongrowing cells of an LDH-deficient L. lactis strain transiently produced mannitol intracellulary under anaerobic conditions to relieve the pressure to regenerate NAD+. In contrast to the LDH-deficient strain of Neves and coworkers (22), no high (intracellular) mannitol levels were produced by nongrowing cells of NZ9010 (Fig. 2C and Table 4). Moreover, both anaerobic and aerobic growing cultures of NZ9010 produced no mannitol at all. The formation of ethanol and 2,3-butanediol under anaerobic conditions suggests that the LDH-deficient strain has used these pathways to regenerate NAD+. Analysis of the fermentation products of the LDH-deficient strains showed that under anaerobic conditions lactate is formed. This can be attributed to transcriptional activation of the alternative LDH gene ldhB under anaerobic conditions (3).
When mtlD was overexpressed in NZ9010, mannitol production by growing cultures could be observed (Table 3). Both nisin-induced and uninduced cultures produced mannitol in small amounts: ∼1% of the glucose was converted into mannitol. Apparently, the low MPDH activity in the noninduced cultures, due to residual activity of the nisin promoter in pNZ8148, is sufficient to accomplish a flux to mannitol. Nisin-induced overexpression of mtlD did not result in much higher mannitol production. Apparently, MPDH activity is not rate limiting in mannitol production. Noting that mannitol 1-phosphatase activity is rather low in the different L. lactis strains (Table 2), this enzyme might have the highest control of the mannitol synthesis pathway.
Larger amounts of mannitol were detected by 13C-NMR in the NZ9010/pNZmtlD cell suspensions (Fig. 2D and Table 4). About 25% of the glucose was converted into mannitol, and conversion rates of glucose into acetoin, ethanol, and 2,3-butanediol were lower than in the NZ9010 strain. This implies that NAD+ regeneration resorted to mannitol production via the introduced MPDH activity. In theory, up to 66.7% of glucose can be converted into mannitol when no NAD+ is regenerated through lactate, ethanol, or 2,3-butanediol formation. These high conversion rates cannot be expected, considering the regained lactate production and the high ethanol production by the LDH-deficient L. lactis strain. In addition, the fairly low mannitol 1-phosphatase activities in all three strains might be limiting for high glucose-mannitol conversions.
We showed that the mannitol concentration remains constant (Fig. 3), even after glucose exhaustion. This is in contrast with the observations of Neves et al. (22), which clearly demonstrated reutilization of the mannitol produced. Also, in the supernatants of growing cultures used in this work, mannitol was still present after 24 h, when glucose had already been depleted for at least 10 h (data not shown). For L. lactis IL-1403, a putative catabolite-responsive element (CRE) site in the promoter region of the mannitol operon was identified (accession no. AE006241 and AE006242), which suggests a possible involvement of CcpA in the regulation of the mannitol operon (21), and thus transcription of the genes involved in mannitol transport and metabolism will be derepressed when glucose is absent and mannitol is present. Moreover, L. lactis MG1363 and the LDH-deficient variant of Neves et al. are capable of growing on mannitol (23), so one would expect that mannitol is consumed when glucose is depleted. Apparently, the disruption of the LDH gene in the LDH-deficient strain used in this work did not induce the expression of genes coding for mannitol transport and utilization, unlike the mutant of Neves et al. (22, 23). In contrast to the findings of Neves and coworkers (23), the LDH-deficient strain did not grow better than the parental strain (Fig. 5). The 24-h lag phase of L. lactis NZ9010/pNZmtlD growing on mannitol (Fig. 5) emphasizes that mannitol utilization genes have to be induced prior to growth on mannitol. Hence, immediate reutilization of mannitol by high-density resting cell suspensions of NZ9010/pNZmtlD would not be expected.
The great difference in mannitol production between the growing and the nongrowing cells may be caused by accumulation of NADH in the resting cells. In the resting cells, where no ATP is needed for biomass production, the ATP demand is low (20), so ATP-generating steps, such as the conversion of phosphoenolpyruvate to pyruvate, are less important. Still, NADH is generated in glycolysis, and hence the nongrowing cells might give priority to regeneration of NAD+ over ATP generation. Because of the LDH deficiency, MPDH takes a major part in NADH oxidation in the NZ9010/pNZmtlD strain.
Our work presented here showed that mtlD overexpression, PFK reduction, and LDH deficiency have contributed to mannitol production in L. lactis. The most promising combination for mannitol production in L. lactis was mtlD overexpression in an LDH-deficient background. Despite the capability of growing on mannitol, no concomitant mannitol reutilization was observed in the LDH-deficient mutant, which is very desirable for the possibilities of mannitol in food products.
Acknowledgments
We thank Peter Ruhdal Jensen for providing the L. lactis HWA217 strain.
REFERENCES
- 1.Andersen, H. W., C. Solem, K. Hammer, and P. R. Jensen. 2001. Twofold reduction of phosphofructokinase activity in Lactococcus lactis results in strong decreases in growth rate and in glycolytic flux. J. Bacteriol. 183:3458-3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bolotin, A., P. Wincker, S. Mauger, O. Jaillon, K. Malarme, J. Weissenbach, S. D. Ehrlich, and A. Sorokin. 2001. The complete genome sequence of the lactic acid bacterium Lactococcus lactis ssp lactis IL1403. Genome Res. 11:731-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bongers, R. S., M. H. N. Hoefnagel, M. J. C. Starrenburg, M. A. J. Siemerink, J. G. A. Arends, J. Hugenholtz, and M. Kleerebezem. 2003. IS981-mediated adaptive evolution recovers lactate production by ldhB transcription activation in a lactate dehydrogenase-deficient strain of Lactococcus lactis. J. Bacteriol. 185:4499-4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chakravorty, M. 1964. Metabolism of mannitol and introduction of mannitol 1-phosphate dehydrogenase in Lactobacillus plantarum. J. Bacteriol. 87:1246-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chaturvedi, V., A. Bartiss, and B. Wong. 1997. Expression of bacterial mtlD in Saccharomyces cerevisiae results in mannitol synthesis and protects a glycerol-defective mutant from high-salt and oxidative stress. J. Bacteriol. 179:157-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chaturvedi, V., B. Wong, and S. L. Newman. 1996. Oxidative killing of Cryptococcus neoformans by human neutrophils. Evidence that fungal mannitol protects by scavenging reactive oxygen intermediates. J. Immunol. 156:3836-3840. [PubMed] [Google Scholar]
- 7.Debord, B., C. Lefebvre, A. M. Guyot-Hermann, J. Hubert, R. Bouche, and J. C. Guyot. 1987. Study of different forms of mannitol: comparative behaviour under compression. Drug Dev. Ind. Pharm. 13:1533-1546. [Google Scholar]
- 8.de Ruyter, P. G. G. A., O. P. Kuipers, M. M. Beerthuyzen, I. van Alen-Boerrigter, and W. M. de Vos. 1996. Functional analysis of promoters in the nisin gene cluster of Lactococcus lactis. J. Bacteriol. 178:3434-3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Ruyter, P. G. G. A., O. P. Kuipers, and W. M. de Vos. 1996. Controlled gene expression systems for Lactococcus lactis with the food-grade inducer nisin. Appl. Environ. Microbiol. 62:3662-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dwivedi, B. K. 1978. Low calorie and special dietary foods. CRC Press, Inc., West Palm Beach, Fla.
- 11.Efiuvwevwere, B. J. O., L. G. M. Gorris, E. J. Smid, and E. P. W. Kets. 1999. Mannitol-enhanced survival of Lactococcus lactis subjected to drying. Appl. Microbiol. Biotechnol. 51:100-104. [Google Scholar]
- 12.Ferain, T., A. N. Schanck, and J. Delcour. 1996. 13C nuclear magnetic resonance analysis of glucose and citrate end products in an ldhL-ldhD double-knockout strain of Lactobacillus plantarum. J. Bacteriol. 178:7311-7315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grobben, G. J., M. R. Smith, J. Sikkema, and J. A. M. De Bont. 1996. Influence of fructose and glucose on the production of exopolysaccharides and the activities of enzymes involved in the sugar metabolism and the synthesis of sugar nucleotides in Lactobacillus delbrueckii subsp. bulgaricus NCFB 2772. Appl. Microbiol. Biotechnol. 46:279-284. [Google Scholar]
- 14.Henstra, S. A., R. H. Duurkens, and G. T. Robillard. 2000. Multiple phosphorylation events regulate the activity of the mannitol transcriptional regulator MtlR of the Bacillus stearothermophilus phosphoenolpyruvate-dependent mannitol phosphotransferase system. J. Biol. Chem. 275:7037-7044. [DOI] [PubMed] [Google Scholar]
- 15.Henstra, S. A., M. Tuinhof, R. H. Duurkens, and G. T. Robillard. 1999. The Bacillus stearothermophilus mannitol regulator, MtlR, of the phosphotransferase system. J. Biol. Chem. 274:4754-4763. [DOI] [PubMed] [Google Scholar]
- 16.Hillier, A. J., and G. R. Jago. 1982. l-Lactate dehydrogenase, FDP-activated, from Streptococcus cremoris. Methods Enzymol. 89:362-367. [DOI] [PubMed] [Google Scholar]
- 17.Hoefnagel, M. H. N., M. J. C. Starrenburg, D. E. Martens, J. Hugenholtz, M. Kleerebezem, S. Van II, R. Bongers, H. V. Westerhoff, and J. L. Snoep. 2002. Metabolic engineering of lactic acid bacteria, the combined approach: kinetic modelling, metabolic control and experimental analysis. Microbiology 148:1003-1013. [DOI] [PubMed] [Google Scholar]
- 18.Kleerebezem, M., M. M. Beerthuyzen, E. E. Vaughan, W. M. de Vos, and O. P. Kuipers. 1997. Controlled gene expression systems for lactic acid bacteria: transferable nisin-inducible expression cassettes for Lactococcus, Leuconostoc, and Lactobacillus spp. Appl. Environ. Microbiol. 63:4581-4584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kleerebezem, M., J. Boekhorst, R. van Kranenburg, D. Molenaar, O. P. Kuipers, R. Leer, R. Tarchini, S. A. Peters, H. M. Sandbrink, M. W. E. J. Fiers, W. Stiekema, R. M. K. Lankhorst, P. A. Bron, S. M. Hoffer, M. N. N. Groot, R. Kerkhoven, M. de Vries, B. Ursing, W. M. de Vos, and R. J. Siezen. 2003. Complete genome sequence of Lactobacillus plantarum WCFS1. Proc. Natl. Acad. Sci. USA 100:1990-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koebmann, B. J., H. W. Andersen, C. Solem, and P. R. Jensen. 2002. Experimental determination of control of glycolysis in Lactococcus lactis. Antonie Leeuwenhoek 82:237-248. [PubMed] [Google Scholar]
- 20a.Kuipers, O. P., P. G. G. A. de Ruyter, M. Kleerebezem, and W. M. de Vos. 1998. Quorum sensing-controlled gene expression in lactic acid bacteria. J. Biotechnol. 64:15-21. [Google Scholar]
- 21.Luesink, E. J., R. E. M. A. Van Herpen, B. P. Grossiord, O. P. Kuipers, and W. M. De Vos. 1998. Transcriptional activation of the glycolytic las operon and catabolite repression of the gal operon in Lactococcus lactis are mediated by the catabolite control protein CcpA. Mol. Microbiol. 30:789-798. [DOI] [PubMed] [Google Scholar]
- 22.Neves, A. R., A. Ramos, C. Shearman, M. J. Gasson, J. S. Almeida, and H. Santos. 2000. Metabolic characterization of Lactococcus lactis deficient in lactate dehydrogenase using in vivo C-13-NMR. Eur. J. Biochem. 267:3859-3868. [DOI] [PubMed] [Google Scholar]
- 23.Neves, A. R., A. Ramos, C. Shearman, M. J. Gasson, and H. Santos. 2002. Catabolism of mannitol in Lactococcus lactis MG1363 and a mutant defective in lactate dehydrogenase. Microbiology 148:3467-3476. [DOI] [PubMed] [Google Scholar]
- 24.Shen, B., R. G. Jensen, and H. J. Bohnert. 1997. Mannitol protects against oxidation by hydroxyl radicals. Plant Physiol. 115:527-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smirnoff, N., and Q. J. Cumbes. 1989. Hydroxyl radical scavenging activity of compatible solutes. Phytochemistry 28:1057-1060. [Google Scholar]