Abstract
A longitudinal study of bacteriophages and their hosts was carried out at a broiler house that had been identified as having a population of Campylobacter-specific bacteriophages. Cloacal and excreta samples were collected from three successive broiler flocks reared in the same barn. Campylobacter jejuni was isolated from each flock, whereas bacteriophages could be isolated from flocks 1 and 2 but were not isolated from flock 3. The bacteriophages isolated from flocks 1 and 2 were closely related to each other in terms of host range, morphology, genome size, and genetic content. All Campylobacter isolates from flock 1 were genotypically indistinguishable by pulsed-field gel electrophoresis (PFGE). PFGE and multilocus sequence typing indicated that this C. jejuni type was maintained from flock 1 to flock 2 but was largely superseded by three genetically distinct C. jejuni types insensitive to the resident bacteriophages. All isolates from the third batch of birds were insensitive to bacteriophages and genotypically distinct. These results are significant because this is the first study of an environmental population of C. jejuni bacteriophages and their influence on the Campylobacter populations of broiler house chickens. The role of developing bacteriophage resistance was investigated as this is a possible obstacle to the use of bacteriophage therapy to reduce the numbers of campylobacters in chickens. In this broiler house succession was largely due to incursion of new genotypes rather than to de novo development of resistance.
The food-borne pathogen Campylobacter jejuni causes acute enteritis in humans and is a major concern to the poultry industry in the United Kingdom, where more than 80% of birds harbor the organism (5). The reason for this high incidence is that campylobacters are commensal organisms in many mammals and birds, including poultry (19). They are excreted into the environment in large numbers and are efficient colonizers of poultry (20). Campylobacters cannot usually be detected in broiler house chickens until the birds are around 3 weeks old despite the efficient colonization of mature birds (19). Whether the campylobacters are simply undetectable up to this age or whether other factors, such as maternal antibodies or inhibitory cecal microflora, play a part has still to be established (10, 24). The origin of infection also remains elusive. Infection probably results from a combination of horizontal factors; for example, inadequate hygiene barriers, contaminated drinking water, and transfer by farm personnel on boots and equipment have all been suggested (10, 14, 17, 21). Another possibility that has been considered is that it occurs by vertical transfer from breeder hen stock (6, 7). The reasons for the difficulties in tracing the origins of infection include the high degree of diversity of campylobacters and the wide variety of typing methods used. It is now acknowledged (32) that a combination of genotypic methods, including fla typing, pulsed-field gel electrophoresis (PFGE), multilocus sequence typing (MLST), and amplified fragment length polymorphism, used in conjunction with phenotypic methods, such as phage typing and serotyping, gives far more information than any single technique used in isolation. The inherent genetic instability of campylobacters complicates the use of genetic methods (33), which is why combinations of techniques produce the most useful discrimination. The choice of techniques depends on the number of strains to be analyzed and the information sought (12).
Bacteriophages occur naturally in any environment where their specific hosts proliferate. Campylobacter-specific bacteriophages have been isolated from broiler chickens (13), from retail poultry (3), and from other sources, including pig manure, abattoir effluents, and sewage (18, 26). Some of these bacteriophages have been characterized and form the basis of the United Kingdom phage typing scheme (12). The dynamics of the Campylobacter bacteriophage population in relation to their prey has never been established. It is likely that Campylobacter bacteriophages flourish where their hosts are actively growing, including the gut of a broiler chicken. As a consequence, they are excreted into the broiler house environment, where we assume they survive until the right host is available to them under conditions that support growth of the host (for example, after ingestion by a Campylobacter-colonized chicken). Bacteriophages are well adapted to survive for long periods of time in the absence of suitable host bacteria (2). Cleaning procedures in a broiler house that are adequate to remove bacteria may not be sufficient to remove all bacteriophages (there is no specific reason why bacteriophages need to be inactivated). Moreover, the methods of detection for bacteriophages are simply not sensitive enough to detect low numbers in large areas, such as broiler houses.
The use of naturally occurring bacteriophages to reduce the numbers of campylobacters entering the food chain is a promising area of research (4, 29). However, concerns have been raised that campylobacters will simply become resistant to bacteriophages, rendering this strategy ineffective (20). Bacteria can rapidly mutate to become resistant to bacteriophages in vitro on laboratory media (2). This is often associated with antigenic changes that block absorption of the bacteriophage or, alternatively, block replication of the bacteriophage by various means. Bacteriophages may also adapt to overcome host resistance by host-induced modification of the bacteriophage DNA (2). Clearly, there is a balance that allows both host and prey to proliferate.
Here we describe a study of a broiler house naturally infected with both Campylobacter and bacteriophages. We examined the relationships between Campylobacter and bacteriophages over three rearing cycles with particular reference to acquisition of bacteriophage resistance by the campylobacters and the phenotypic and genotypic characteristics of the host bacteria.
MATERIALS AND METHODS
Samples.
A United Kingdom broiler house used for rearing 25,000 broiler chickens was selected for this study, as bacteriophages had already been isolated from this site. Samples were collected from three consecutive flocks. Ten cloacal swab samples were taken by using charcoal transport swabs (Transweb swabs; MW171; Medical Wire and Equipment Co. Ltd., Corsham, United Kingdom), and 10 freshly voided excreta samples were collected from individual birds located in different sectors of the broiler house to obtain maximum coverage of the rearing area. Samples were collected on day 32 of a 41-day rearing cycle, in November 2001, January 2002, and February 2002. Samples were transported to the laboratory and processed on the day of receipt. The broiler house was thoroughly cleaned and disinfected after each batch of birds was removed according to the United Kingdom Assured Chicken Production Scheme.
Isolation of campylobacters from chicken excreta and cloacal swabs.
Campylobacters were isolated from chickens by direct plating of excreta or cloacal swabs by using modified cefoperazone charcoal deoxycholate agar (CM739; selective supplement code SR155; Oxoid, Basingstoke, United Kingdom). The plates were incubated at 42°C for 48 h under microaerobic conditions (5% O2, 5% H2, 10% CO2, 80% N2). Suspect colonies were confirmed as Campylobacter spp. colonies by having typical colony morphology and typical cell morphology as determined by Gram staining and by being oxidase positive. Catalase and hippurate hydrolysis tests were also performed. Strains of Campylobacter were stored at −80°C in Microbank storage beads (Pro Lab Diagnostics, Neston, United Kingdom).
Isolation of phage from excreta and cloacal swabs by filtration.
A 10% suspension of excreta was prepared in SM buffer (50 mM Tris-HCl [pH 7.5], 0.1 M NaCl, 8 mM MgSO40 · 7H2O, 0.01% gelatin; Sigma). Cloacal swabs were emulsified in 2 ml of SM buffer. After 24 h of incubation at 4°C (to allow bacteriophages to dissociate), 1 ml of each suspension was centrifuged at 3,000 × g for 5 min in an Eppendorf tube to remove debris. The supernatant was transferred to a new tube and centrifuged at 13,000 × g for 10 min to remove bacteria. The resulting supernatant was filtered through a 0.2-μm-pore-size disposable filter (Minisart; Sartorius, Goettingen, Germany) to remove any remaining bacteria. The filtrate was then applied to lawns of C. jejuni NCTC 12662 (phage type 14 [PT 14]), which is known to be sensitive to most C. jejuni and Campylobacter coli phage isolates. Lawns of Campylobacter isolates from the same source were also used to propagate phages. The lawns were prepared from overnight blood agar plate cultures (CM0271 [Oxoid] with 5% defibrinated horse blood [TCS, Buckingham, United Kingdom]). Bacteria were scrapped off with a sterile swab and suspended in ice-cold 10 mM Mg SO4 to obtain a suspension containing approximately 1010 CFU/ml as determined by using McFarland standards. Bacterial lawns were prepared by using the method described by Sambrook et al. (27). Briefly, for each lawn, 100 μl of cells was added to 5 ml of molten NZCYM overlay agar (NZCYM broth [Difco, Oxford, United Kingdom] with 0.6% Select agar [Sigma Aldrich, Gillingham, United Kingdom]), and the preparation was poured immediately onto a prewarmed NZCYM agar plate (NZCYM broth with 1.5% Select agar). The plates were allowed to set for 10 min and then dried at 42°C for 30 min. Each sample suspension was applied as a 10-μl spot to a prepared lawn and allowed to absorb into the overlay agar. The plates were then incubated for 24 h at 42°C under microaerobic conditions. Plaques were collected from the overlays by using a pipette tip and were suspended in 100 μl of SM medium. Single plaques were propagated in this way a total of three times to ensure that the isolates represented a single clone; this procedure resulted in 25 individual bacteriophage suspensions designated Cφ1 to Cφ25.
Characterization of Campylobacter strains by serotyping and phage typing.
All strains isolated were phage typed and serotyped by heat-stable antigen agglutination by using standard protocols (11, 12).
PFGE of digested genomic DNA from Campylobacter isolates.
Genomic DNA for PFGE was prepared from Campylobacter isolates by using a standardized protocol (23) used by PulseNet (http://www.cdc.gov/pulsenet/) to allow comparisons of strain fingerprints between different laboratories. Briefly, restriction fragments produced by SmaI (Roche, Lewes, United Kingdom) digestion of genomic DNA from each strain isolated from three consecutive flocks were separated by PFGE by using a CHEF DRII apparatus (Bio-Rad, Hemelhemstead, United Kingdom) at 6 V/cm, ramped from 6 to 35 s over 18 h, at 14°C in 0.5× TBE (45 mM Tris HCl, 45 mM borate, 1 mM EDTA, pH 8.3; Sigma Aldrich). Ethidium bromide-stained images were aligned and compared by using concatenated lambda DNA markers (Sigma Aldrich). The criteria of Tenover et al. (30) were used to interpret the PFGE patterns. Macro restriction patterns (MRPs) were given arbitrary numbers if more than three fragments were different from the fragments of other isolates. MRPs with one to three fragment differences were considered to be probably closely related to each other and were included within a single MRP. MRPs with no fragment differences were interpreted as being genetically indistinguishable.
The pulsed-field gels were also treated with 0.25 M HCl and Southern blotted by capillary transfer onto Hybond-N+ (Amersham Pharmacia Biotech, Little Chalfont, United Kingdom) nylon membranes before fixation with UV light. The blots were probed with phage Cφ1 and Cφ6 DNAs labeled by random priming by using the DIG system (Boehringer Mannheim, Lewes, United Kingdom). Colorimetric detection was performed in the presence of the nitroblue tetrazolium and 5-bromo-4-chloro-3-indolylphosphate (BCIP) substrates according to the manufacturer's instructions (Boehringer Mannheim).
MLST of Campylobacter isolates.
Selected isolates from each flock showing different MRPs following PFGE separation of SmaI-digested DNA (when the defined criteria were used) were analyzed by MLST (9) with reference to the database at http://mlst.zoo.ox.ac.uk.
Lysis of Campylobacter isolates by bacteriophages (lytic spectra).
Bacterial lawns were prepared by using each Campylobacter isolate as described above. Each of the 25 plaque-purified test bacteriophage suspensions (adjusted to contain approximately 106 PFU/ml) was applied as a 10-μl spot to a prepared lawn and allowed to absorb into the top agar layer. The plates were then incubated for 24 h at 42°C under microaerobic conditions. If lysis of the strain (defined as 20 or more plaques visible) was observed with all 25 bacteriophages, the result was recorded as sensitive; otherwise the ratio of sensitive phages to resistant phages was recorded.
Characterization of C. jejuni bacteriophages by their lytic spectra.
Four representative bacteriophages (Cφ1, Cφ5, Cφ6, and Cφ7), two from each bacteriophage-positive flock, were propagated on C. jejuni 12662 (PT 14). These bacteriophages were screened for lytic activity at a routine test dilution (defined as the concentration at which a bacteriophage causes just less than complete lysis of the propagating strain) by using 80 Campylobacter strains whose phage type had been previously defined on the basis of the 16 bacteriophages currently in use in the United Kingdom phage typing scheme (12). The bacteriophages were applied to lawns prepared from these strains and incubated as previously described (12).
Examination of C. jejuni bacteriophage morphology by electron microscopy.
Bacteriophage isolates Cφ1, Cφ5, Cφ6, and Cφ7 were examined by electron microscopy as previously described (3). Briefly, glutaraldehyde-fixed phage suspensions on Pioloform grids were negatively stained with 0.5% uranyl acetate. The specimens were observed with a JEOL 100CX transmission electron microscope at an acceleration voltage of 80 kV.
Phage genome size determination by PFGE.
Genomic DNAs from bacteriophages Cφ1, Cφ5, Cφ6, and Cφ7 were prepared and run by using a CHEF DRII (Bio-Rad) as previously described (3). A 2-mm slice of each agarose plug was inserted into the wells of a 1% agarose gel. The gel was run by using a Bio-Rad CHEF DRII system in 0.5× TBE for 18 h at 6 V/cm with a switch time of 30 to 60 s. Lambda concatemers (Sigma Aldrich) were used as markers.
Bacteriophage DNA restriction digests.
Genomic DNA was prepared from each bacteriophage by using a standard protocol described for lambda phage (27) by using proteinase K digestion, followed by phenol-chloroform extraction and precipitation. DNAs were digested with restriction enzymes (DraI, EcoRI, EcoRV, HhaI, HindIII, PstI, and SspI; Promega Ltd., Southampton, United Kingdom), and the enzyme digests were run on a standard 0.6% Tris-acetate-EDTA agarose gel and stained with ethidium bromide following electrophoresis (27).
RESULTS
Isolation of campylobacters and bacteriophages.
Campylobacters were isolated from each of the three flocks, each isolate was identified to the species level, serotyped, and phage typed, and SmaI genomic DNA digests were subjected to PFGE. The overall Campylobacter isolation rate from the cloacal and excreta samples was 77% (46 of 60 samples), and the percentage of positive samples varied from 40 to 100% (Table 1). Cloacal swabs were just as likely to harbor campylobacters as excreta samples (Table 2). Bacteriophages were isolated from flock 1 and flock 2 but not from flock 3 (Table 1). The overall phage isolation rate for all three flocks was 42% (26 of 60 samples), but the range was 0 to 100% depending on the batch. There was no discernible difference in the bacteriophage isolation rate between the two types of samples (results not shown). In flock 2, bacteriophages were isolated from two excreta samples that did not yield campylobacters.
TABLE 1.
Numbers of bacteriophages and campylobacters isolated from 20 samples taken from each of three successive flocksa
| Flock | No. of bacteriophage isolates | No. of Campylobacter isolates |
|---|---|---|
| 1 | 5 | 8 |
| 2 | 20 | 18 |
| 3 | 0 | 20 |
| Total | 25 | 46 |
Isolation of bacteriophages and Campylobacter was attempted from 20 samples taken from each successive flock.
TABLE 2.
Characteristics of campylobacters isolated from the three flocks
| Flock | Isolatea | Sero- type | PT | Lytic activityb | MRPc | MLST typed |
|---|---|---|---|---|---|---|
| 1 | F1C4 | UTe | 14 | S | 01 | |
| F1C5 | UT | RDNCf | S | 01 | ||
| F1C6 | UT | 35 | S | 01 | ||
| F1C8 | UT | 1 | S2/R23 | 01 | 661 | |
| F1E3 | HS2 | RDNC | S | 01 | 661 | |
| F1E4 | HS8 | UT | S2/R23 | 01 | ||
| F1E6 | HS8 | RDNC | S | 01 | ||
| F1E9 | UT | 35 | S | 01 | ||
| 2 | F2C1 | HS13 | RDNC | R | 02 | Unique within ST21 complex |
| F2C2 | UT | RDNC | S | 01 | 661 | |
| F2C3 | HS13 | RDNC | R | 02 | ||
| F2C4 | HS13 | RDNC | R | 02 | ||
| F2C5 | HS13 | RDNC | R | 02 | ||
| F2C6 | UT | 68 | R | 04 | ||
| F2C7 | UT | RDNC | S | 01 | 661 | |
| F2C8 | HS13 | RDNC | R | 02 | ||
| F2C9 | HS13 | 33 | R | 03 | Unique within ST21 complex | |
| F2C10 | HS37 | 35 | S | 01 | 661 | |
| F2E1 | HS13 | 1 | R | 03 | Unique within ST21 complex | |
| F2E2 | UT | 68 | R | 04 | Unique within ST21 complex | |
| F2E3 | HS13 | 33 | S6/R19 | 03 | Unique within ST21 complex | |
| F2E6 | HS13 | 1 | R | 03 | Unique within ST21 complex | |
| F2E7 | HS13 | RDNC | R | 02 | ||
| F2E8 | HS13 | 14 | R | 02 | ||
| F2E9 | HS13 | 14 | R | 02 | ||
| F2E10 | HS13 | RDNC | R | 02 | ||
| 3 | F3C1 | HS9 | UT | R | 05 | |
| F3C2 | HS9 | UT | R | 05 | 814 | |
| F3C3 | HS9 | UT | R | 05 | 814 | |
| F3C4 | HS9 | UT | R | 05 | ||
| F3C5 | HS9 | UT | R | 05 | ||
| F3C6 | HS9 | UT | R | 05 | ||
| F3C7 | HS9 | UT | R | 05 | ||
| F3C8 | HS37 | UT | R | 05 | 814 | |
| F3C9 | HS9 | UT | R | 05 | ||
| F3C10 | HS9 | UT | R | 05 | ||
| F3E1 | HS9 | UT | R | 05 | ||
| F3E2 | HS9 | UT | R | 05 | ||
| F3E3 | HS9 | UT | R | 05 | ||
| F3E4 | HS9 | UT | R | 05 | ||
| F3E5 | HS9 | UT | R | 05 | ||
| F3E6 | HS9 | UT | R | 05 | ||
| F3E7 | HS9 | UT | R | 05 | ||
| F3E8 | UT | UT | R | 05 | ||
| F3E9 | HS9 | UT | R | 05 | ||
| F3E10 | HS9 | UT | R | 05 |
In the strain designations C indicates that the strain was obtained from a cloacal sample, and E indicates that the strain was obtained from an excreta sample.
Lytic activity with all 25 bacteriophage isolates in this study. S, sensitive to all bacteriophages; R, resistant to all bacteriophages; S2/R23, sensitive to 2 bacteriophages and resistant to 23 bacteriophages.
MRP determined by PFGE of Smal-digested genomic DNA phage type.
MLST (9) was carried out with selected strains.
UT, untypeable.
RDNC, reacts with phage but does not conform to a recognized phage type.
Characterization of the campylobacters.
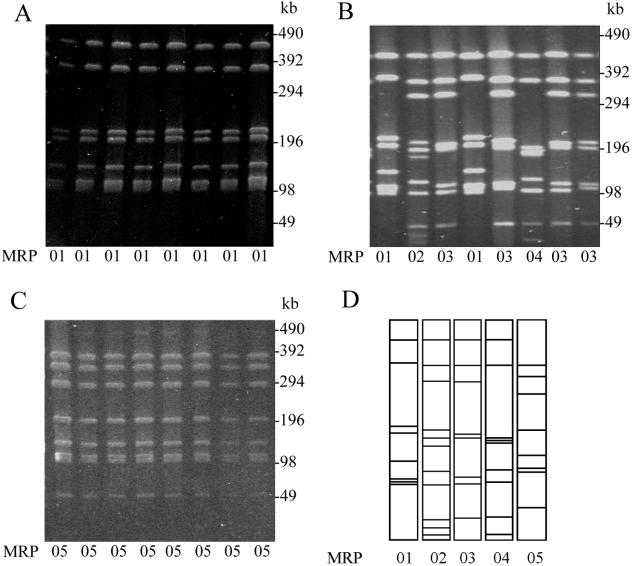
A total of 46 campylobacter isolates were characterized from the three flocks (Table 2). All isolates were identified as C. jejuni based on the fact that they were hippurate positive. Phage typing was not a useful method for distinguishing between Campylobacter isolates from this particular broiler house. Twenty-one isolates (46%) were untypeable, and the remainder were very heterogeneous in terms of phage type. Serotyping indicated that for flock 1 most strains were untypeable (five of eight strains), and the remaining strains were serotypes HS2 (n = 1) and HS8 (n = 2). Most isolates in flock 2 (13 of 18 isolates) were serotype HS13, while four strains were untypeable. In flock 3 the most common serotype was HS9 (18 of 20 strains); one isolate was untypeable, and one was serotype HS37. PFGE of SmaI digests of genomic DNA provided profiles for each of the strains. Five different MRPs were identified and designated 01 to 05. All eight isolates from flock 1 possessed a common MRP (MRP 01) following SmaI digestion (Fig. 1), indicating that all the corresponding birds were colonized by genetically indistinguishable strains despite the observed variation in their serotypes and phage types. All 20 of the isolates from flock 3 produced the same MRP (MRP 05), indicating that it was likely that this flock was also colonized by genetically indistinguishable strains that were different from those observed in flock 1. Four different MRPs were identified among the 18 isolates from flock 2 by using the criteria defined by Tenover et al. (30). Three MRPs (MRPs 02, 03, and 04) showed some similarity to each other (although they differed by more than three fragments), indicating that the strains could possibly be related. Several isolates from this flock differed in their assigned MRPs by between one and three fragments (e.g., F2E1 and F2E6) (Fig. 1), indicating that these strains were probably closely related (30). Three isolates from flock 2 had the MRP 01 present in flock 1. In order to further understand the relationships among these isolates, MLST was carried out with representative strains for each MRP type (Table 2) and a group of isolates in the MRP 03 group that possessed variant fragment patterns. The allelic profiles of these isolates are presented in Table 3. All MRP 01 isolates possessed the same allelic profile recognized as sequence type 661 (ST 661), irrespective of whether they arose from flock 1 or flock 2. The MRP 02, 03, and 04 isolates from flock 2 all belonged to the ST 21 complex. The representative MRP 02, 03, and 04 isolates were distinct strains as determined by MLST, sharing some alleles but not all seven alleles. However, MRPs 02 and 04 differed by only one allele, and this represented a change in the sequence between the two alleles of one nucleotide. Three of four of the MRP 03 isolates examined had identical allelic profiles despite slight variations in their MRP fragment profiles, but a fourth isolate (F2E3) differed at one allele (pgm) from the other isolates. The MRP 05 isolates from flock 3 were all ST 814.
FIG. 1.
PFGE profiles of genomic DNAs, cut with the SmaI restriction endonuclease, from representative isolates of C. jejuni from each of the three flocks. (A) Flock 1. Lanes 1 to 8, isolates C4, C5, C6, C8, E3, E4, E6, and E9, respectively. (B) Flock 2. Lanes 1 to 8, isolates C7, C8, C9, C10, E1, E2, E3, and E6, respectively. (C) Flock 3. Lanes 1 to 8, isolates C1, C3, C5, C6, C7, C9, E5, and E6, respectively. (D) Graphic interpretation of the different patterns produced by each flock with their assigned MRPs.
TABLE 3.
MLST allelic profiles of selected isolates representing the different PFGE MRPs
| Flock | Isolate | PFGE MRP | Allelic profilea
|
STa | Com- plexa | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| aspA | glnA | gltA | glyA | pgm | tkt | uncA | |||||
| 1 | F1C8 | 01 | 02 | 75 | 04 | 48 | 103 | 34 | 01 | 661 | UNb |
| F1E3 | 01 | 02 | 75 | 04 | 48 | 103 | 34 | 01 | 661 | UN | |
| 2 | F2C2 | 01 | 02 | 75 | 04 | 48 | 103 | 34 | 01 | 661 | UN |
| F2C7 | 01 | 02 | 75 | 04 | 48 | 103 | 34 | 01 | 661 | UN | |
| F2C10 | 01 | 02 | 75 | 04 | 48 | 103 | 34 | 01 | 661 | UN | |
| F2C1 | 02 | 02 | 75 | 12 | 03 | 02 | 62 | 25 | NDc | ST21 | |
| F2C9 | 03 | 02 | 01 | 04 | 03 | 02 | 34 | 01 | ND | ST21 | |
| F2E1 | 03 | 02 | 01 | 04 | 03 | 02 | 34 | 01 | ND | ST21 | |
| F2E3 | 03 | 02 | 01 | 04 | 03 | 103 | 34 | 01 | ND | ST21 | |
| F2E6 | 03 | 02 | 01 | 04 | 03 | 02 | 34 | 01 | ND | ST21 | |
| F2E2 | 04 | 02 | 75 | 12 | 03 | 02 | 62 | 05 | ND | ST21 | |
| 3 | F3C2 | 05 | 02 | 75 | 04 | 48 | 141 | 34 | 01 | 814 | UN |
| F3C3 | 05 | 02 | 75 | 04 | 48 | 141 | 34 | 01 | 814 | UN | |
| F3C8 | 05 | 02 | 75 | 04 | 48 | 141 | 34 | 01 | 814 | UN | |
Allelic profiles, STs, and complexes were obtained from the MLST website (http://mlst.zoo.ox.ac.uk) (9).
UN, currently not assigned to a lineage.
ND, not defined (between five and eight close matches).
Each of the C. jejuni strains was tested for lysis with each bacteriophage isolate (Table 2) to distinguish bacteriophage-sensitive strains from bacteriophage-resistant strains. The results correlated well with the MRP typing data. MRP 01 was associated with sensitivity of the strains to bacteriophages isolated from both flock 1 and flock 2, although two isolates from flock 1 were resistant to 23 of the 25 bacteriophages. Most of the alternate strains having different genotypes (MRPs, MLST) were associated with resistance to all the bacteriophages; the only exception was one strain (F2E3, which had the MRP 03 profile) for which sensitivity to some but not all bacteriophages was observed.
Characterization of C. jejuni bacteriophages.
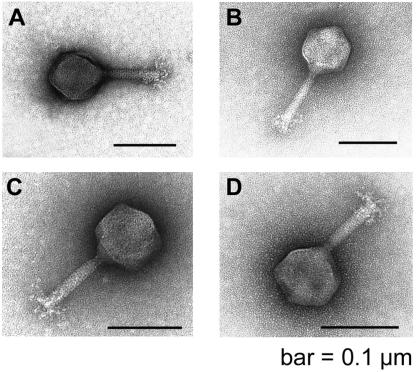
Twenty-five bacteriophages were isolated and characterized. The relationship with regard to host range was tested for each phage with each of the C. jejuni isolates. The host ranges of the bacteriophages proved to be identical except for three C. jejuni isolates, as noted above. Specifically, bacteriophages Cφ1 and Cφ2 from flock 1 were able to lyse two C. jejuni strains from flock 1 which were resistant to all the remaining bacteriophages (F1C8 and F1E4) (Table 2). One Campylobacter strain (F2E3) from flock 2 was sensitive to 6 bacteriophages (including Cφ1 and Cφ2) but was resistant to the remaining 19 bacteriophages (Table 2). The MRP of this strain (MRP 03) indicates that it is typical of the bacteriophage-resistant types present in the majority of the birds from this flock. Two representative bacteriophages from flock 1 (Cφ1 and Cφ5) and two representative bacteriophages from flock 2 (Cφ6 and Cφ7) were selected for extensive lytic characterization with campylobacters representing the 80 different phage types in the bacteriophage typing scheme. Each of these strains has by definition a unique lytic profile with the 16 bacteriophages in the United Kingdom phage typing scheme (12). Of the 80 strains, 46 were resistant to all four representative bacteriophages, while 31 were sensitive to them. There were only three minor differences in the susceptibilities of these strains, indicating their close relationship. The differences were that PT 46 and PT 61 were both lysed by Cφ1 but were resistant to the other representatives. PT 43 was resistant to Cφ6 but was lysed by Cφ1, Cφ5, and Cφ7. The same bacteriophages were examined by electron microscopy (Fig. 2) and were found to be identical in structure and size, possessing icosahedral heads (diameter, 87 ± 5.6 nm [mean ± standard deviation]; n = 19) and contractile tails (length, 113 ± 1.1 nm [mean ± standard deviation]; n = 19). These characteristics are typical of the Myoviridae family of bacteriophages. The genomic DNA sizes of these phages were indistinguishable by PFGE; the DNAs migrated at 140 kb. Restriction enzyme HhaI has previously been reported to be one of the few useful enzymes for cutting Campylobacter bacteriophage DNA and distinguishing between the phages (3, 25). DNAs of isolates Cφ1, Cφ5, Cφ6, and Cφ7 produced similar HhaI profiles consisting of four fragments of similar sizes (range, 7 kb to approximately 60 kb). The phage DNAs were not cut with the enzymes EcoRI, EcoRV, HindIII, PstI, and SspI, whereas the DraI enzyme cut the DNAs into many fragments that were less than 2 kb long. Genomic SmaI digests of DNAs representing all five Campylobacter MRPs were separated by PFGE and Southern blotted before they were probed with digoxigenin-labeled DNAs from representative bacteriophages Cφ1 (isolate from flock 1) and Cφ6 (isolated from flock 2). No hybridization signals were observed compared with single-copy DNA control probes, suggesting that the Campylobacter genomes did not contain DNA related to phage Cφ1 or Cφ6.
FIG. 2.
Electron micrographs of bacteriophages Cφ1 (A), Cφ5 (B), Cφ6 (C), and Cφ7 (D).
DISCUSSION
The high isolation rate of Campylobacter from broiler house chickens (77% overall) was expected as most United Kingdom poultry is colonized by this bacterium (5). The significance of the fact that 42% of the samples contained bacteriophage is not known as there are no published data on the rates of carriage of bacteriophages in poultry except for the data obtained in this study, which focused on a single barn.
Characterization of the C. jejuni strains and the bacteriophages isolated revealed some interesting features of this ecological niche. In common with epidemiological studies of campylobacters in broiler houses, the dynamics of phage predation are complicated by the general lack of understanding regarding the origin of most Campylobacter strains isolated from chickens. We aimed to define the host strains on the basis of phenotypic (phage typing and serotyping) and genotypic (PFGE and MLST) methods, as a combination of the two is generally regarded as the best approach. In this study we encountered a few strains that were genetically indistinguishable by PFGE but were untypeable or had variable (between-isolate) serotypes. It has previously been reported that C. jejuni isolates having the same serotype do not always have similar genotypes (as determined by PFGE or MLST) and that strains with the same defined genotype may have different serotypes (9, 15). MLST was employed to confirm that the PFGE MRP 01 isolates from flock 1 and flock 2 were genetically related to each other and genetically distinct from the other isolates. It was also used to explore the relationships between isolates from flock 2 that had similar PFGE MRPs (MRPs 02, 03, and 04) to confirm that they were indeed genetically distinguishable but notably shared some alleles and fell within the same clonal complex, the ST21 complex. This finding was similar to that reported by Wassenaar et al. (31), who found that 21 single-colony isolates had 14 different PFGE genotypes. These authors alluded to the possibility that phages were involved in the apparent genomic instability, possibly in combination with environmental pressure (other possible causes were also discussed). In contrast to flock 2, there was no heterogeneity in the MRPs of the C. jejuni isolates from flock 1 and flock 3 despite the presence of bacteriophages in flock 1. All of the flock 3 isolates possessed MRP 05, and representative isolates shared the MLST ST 814 alleles. ST 814 differs by one allele from ST 661 of the MRP 01 isolates found in flock 1. It is possible that these isolates originated from a common environmental reservoir.
The C. jejuni isolates recovered from flock 3 did not support replication of any of the bacteriophages of flocks 1 and 2 and contained no bacteriophage that could be propagated on C. jejuni NCTC 12662 or the previously described C. jejuni isolates. However, it is possible that the bacteriophages remained in the broiler house environment until new sensitive strains were introduced.
Phage typing was not found to be a useful tool for studying the relationships among these groups of strains. This may be because the presence of native phages may influence the susceptibility to the bacteriophages used in the phage typing scheme, altering the underlying type (31). While no firm conclusions could be drawn regarding the influence of native bacteriophages on the resultant phage types for this limited set of strains, it would be surprising if they had no effect.
Using genetic typing data, we showed that similar phage-sensitive C. jejuni strains were present in consecutive flocks, likely due to transfer from one to flock to the next either by persistence in the broiler house or by reintroduction from a common environmental source. These data confirm the reports of others (22, 28) who have demonstrated carryover of Campylobacter strains from flock to flock within a broiler house. In our study it was particularly interesting that the same bacteriophages (as far as could be determined) were present in conjunction with these strains. Although the concepts of species and strain cannot readily be applied to bacteriophages in the same way that they are applied to higher organisms (1, 16), it appeared that the bacteriophages in the broiler house represented a population of very highly related organisms. The lytic profiles of the representative bacteriophages were remarkably similar when they were tested with 80 distinct strains of Campylobacter. Bacteriophages of campylobacters are extremely difficult to characterize by other methods often applied to bacteria or phages from other species, such as restriction fragment length polymorphism. This is due the refractory nature of their DNAs to digestion by restriction endonucleases (25). However, the similarity of the HhaI digests of genomic bacteriophage DNAs indicated that the chromosomes of the four selected bacteriophages were similar in size and structure. The characteristics of these bacteriophages indicated that they were members of the Myoviridae, whose members have lytic lifestyles and are not known to form lysogens. The absence of hybridization between Campylobacter genomic DNAs and bacteriophage Cφ1 and Cφ6 DNAs verifies this presumption. Therefore, although a bacteriophage and its host had been transferred from one flock to the next, it is unlikely that the intermediate vector included bacteria carrying an integrated bacteriophage genome which could be later excised and propagated. It was also interesting that in flock 2 the bacteriophages appeared to flourish despite the fact that at least one new bacteriophage-resistant strain had been introduced. Bacteriophages were present in every sample examined from flock 2, even in the absence of Campylobacter. The most likely explanation is that although campylobacters were in fact originally present in the samples, they had not remained viable long enough to be cultured, possibly due to bacteriophage activity.
Partial resistance to bacteriophages occurred in two Campylobacter variants from flock 1, but the resistant types were not apparent in flock 2. While this may indicate that Campylobacter acquired resistance to the bacteriophages, the bacteriophages may also have adapted in response, maintaining the ability to infect the host. However, bacteriophages Cφ1 and Cφ5 of flock 1 were indistinguishable in terms of host profile from the bacteriophages isolated from flock 2. Flock 2 contained multiple genotypes (MRPs 01, 02, 03, and 04), and notably microheterogeneity was observed among the MRP 03 isolates. The multiplicity of related strains has been argued to be a result of genetic exchange between campylobacters in the avian intestinal tract (8). It is of interest to consider whether the frequency of these events may be provoked if two or more strains are in the presence of bacteriophage. Examination of the allelic profiles determined for MLST of variable MRP 03 isolates indicated in general that they retained common alleles. However, a single isolate from flock 2, F2E3, was found to possess a typical MRP 03 profile but had an allelic difference at the pgm locus, one of the seven loci that constitute the MLST scheme. This isolate was also notable in that it was the only member of the MRP 03 group that was found to be partially sensitive to some but not all of the bacteriophage isolates. The pgm 103 allele was found in all the MRP 01 phage-sensitive isolates from flock 1 and flock 2. It is tempting to speculate that F2E3 may have arisen through genetic recombination between MRP 01- and MRP 03-derived genomes that yielded phage-sensitive MRP 03. How this isolate was maintained in the chicken gut in the presence of phage while it was in competition with related phage-insensitive MRP 03 types is an intriguing question. Indeed, bacteriophages could be isolated from the majority of samples that yielded bacteriophage-resistant campylobacters. Either bacteriophages were present in these samples because large numbers had been ingested by the birds, or mixed populations of resistant and sensitive campylobacters were present in the same birds. In a situation where mixed populations occur, the resistant strains may be the predominant isolates either because they represent a greater proportion of the population than their sensitive bacteriophage-predated competitors or because the isolation procedures somehow facilitate phage infection.
With regard to bacteriophage therapy, long-term development of phage resistance is not indicated by this study. Although some resistant C. jejuni strains were isolated, these strains did not dominate and outgrow the sensitive types as one might expect. Instead, they coexisted, implying that sensitive and resistant states were maintained in the chicken gut in the presence of phage.
Before bacteriophage therapy is adopted as a means of controlling campylobacters in broiler chickens, it is important to understand the ecology of bacteriophages in a naturally infected flock. In conclusion, this study indicates that the ecology of campylobacters and their bacteriophages in the broiler house is complex and may play a significant role in influencing which strains of Campylobacter enter the human food chain. This may occur by positive selection pressure for phage-resistant strains through succession but probably not through de novo acquisition of resistance.
Acknowledgments
This work was supported by the United Kingdom Department for the Environment, Food, and Rural Affairs through its Food Quality and Safety LINK research program.
We thank Stefan Hyman at the University of Leicester for his expert assistance with the phage electron microscopy.
REFERENCES
- 1.Ackermann, H.-W., M. S. DuBow, A. W. Jarvis, L. A. Jones, V. N. Krylov, J. Maniloff, J. Rocourt, R. S. Saferman, J. Schneider, L. Seldin, T. Sozzi, P. R. Stewart, M. Werquin, and L. Wunsche. 1992. The species concept and its application to tailed phages. Arch. Virol. 124:69-82. [DOI] [PubMed] [Google Scholar]
- 2.Adams, M. H. (ed.). 1959. Bacteriophages. Interscience Publishers Inc., New York, N.Y.
- 3.Atterbury, R. J., P. L. Connerton, C. E. R., Dodd, C. E. D. Rees, and I. F. Connerton. 2003. Isolation and characterization of Campylobacter bacteriophages from retail poultry. Appl. Environ. Microbiol. 69:4511-4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Atterbury, R. J., P. L. Connerton, C. E. R. Dodd, C. E. D. Rees, and I. F. Connerton. 2003. Application of host-specific bacteriophages to the surface of chicken skin leads to a reduction in recovery of Campylobacter jejuni. Appl. Environ. Microbiol. 69:6302-6306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Corry, J. E. L., and H. I. Atabay. 2001. Poultry as a source of Campylobacter and related organisms. J. Appl. Microbiol. 90:96S-114S. [DOI] [PubMed] [Google Scholar]
- 6.Cox, N. A., N. J. Stern, K. L. Hiett, and M. E. Berrang. 2002. Identification of a new source of Campylobacter contamination in poultry: transmission from breeder hens to broiler chickens. Avian Dis. 46:535-541. [DOI] [PubMed] [Google Scholar]
- 7.Cox, N. A., N. J. Stern, J. L. Wilson, M. T. Musgrove, R. J. Buhr, and K. L. Hiett. 2002. Isolation of Campylobacter spp. from semen samples of commercial broiler breeder roosters. Avian Dis. 46:717-720. [DOI] [PubMed] [Google Scholar]
- 8.de Boer, P., J. A. Wagenaar, R. P. Achterberg, J. P. M. van Putten, L. M. Schouls, and B. Dium. 2002. Generation of Campylobacter jejuni genetic diversity in vivo. Mol. Microbiol. 44:351-359. [DOI] [PubMed] [Google Scholar]
- 9.Dingle, K. E., F. M. Colles, D. R. A. Wareing, R. Ure, A. F. Fox, F. E. Bolton, H. J. Bootsma, R. J. L. Willems, R. Urwin, and M. C. J. Maiden. 2001. Multilocus sequence typing system for Campylobacter jejuni. J. Clin. Microbiol. 39:14-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Evans, S. J., and A. R. Sayers. 2000. A longitudinal study of campylobacter infection of broiler flocks in Great Britain. Prev. Vet. Med. 46:209-223. [DOI] [PubMed] [Google Scholar]
- 11.Frost, J. A., A. N. Oza, R. T. Thwaites, and B. Rowe. 1998. Serotyping scheme for Campylobacter jejuni and Campylobacter coli based on direct agglutination of heat-stable antigens. J. Clin. Microbiol. 36:335-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frost, J. A., J. M. Kramer, and S. A. Gillanders. 1999. Phage typing of Campylobacter jejuni and Campylobacter coli and its use as an adjunct to serotyping. Epidemiol. Infect. 123:47-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grawjewski, B. A., J. W. Kusek, and H. M. Gelfand. 1985. Development of a bacteriophage typing scheme for Campylobacter jejuni and Campylobacter coli. Epidemiol. Infect. 104:403-414. [Google Scholar]
- 14.Gregory, E., H. Barnhart, D. W. Dreesen, N. J. Stern, and J. L. Corn. 1997. Epidemiological study of Campylobacter spp. in broilers: source, time of colonization and prevalence. Avian Dis. 41:890-898. [PubMed] [Google Scholar]
- 15.Hänninen, M. L., P. Perko-Makela, H. Rautelin, B. Duim, and J. A. Wagenaar. 2001. Genomic relatedness within five common Finnish Campylobacter jejuni pulsed-field gel electrophoresis genotypes studied by amplified fragment length polymorphism analysis, ribotyping, and serotyping. Appl. Environ. Microbiol. 67:1581-1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hendrix, R. W., M. C. M. Smith, R. N. Burns, M. E. Ford, and G. F. Hatfull. 1999. Evolutionary relationships among diverse bacteriophages and prophages: all the world's a phage. Proc. Natl. Acad. Sci. USA 96:2192-2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hiett, K. L., N. J. Stern, P. Fedorka-Cray, N. A. Cox, M. T. Musgrove, and S. Ladely. 2002. Molecular subtype analyses of Campylobacter spp. from Arkansas and California poultry operations. Appl. Environ. Microbiol. 68:6220-6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khakhria, R., and H. Lior. 1992. Extended phage-typing scheme for Campylobacter jejuni and Campylobacter coli. Epidemiol. Infect. 108:403-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Newell, D. G., and C. Fearnley. 2003. Sources of Campylobacter colonization in broiler chickens. Appl. Environ. Microbiol. 69:4343-4351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Newell, D. G., and J. A. Wagenaar. 2000. Poultry infections and their control at farm level, p. 497-510. In I. Nachamkin and M. J. Blaser (ed.), Campylobacter, 2nd ed. ASM Press, Washington, D.C.
- 21.Pearson, A. D., M. Greenwood, T. D. Healing, D. Rollins, M. Shahamat, J. Donaldson, and R. R. Colwell. 1993. Colonization of broiler chickens by waterborne Campylobacter jejuni. Appl. Environ. Microbiol. 59:987-996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Petersen, L., and A. Wedderkopp. 2001. Evidence that certain clones of Campylobacter jejuni persist during successive broiler flock rotations. Appl. Environ. Microbiol. 67:2739-2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ribot, E. M., C. Fitzgerald, K. Kubota, B. Swaminathan, and T. J. Barrett. 2001. Rapid pulsed-field gel electrophoresis protocol for subtyping of Campylobacter jejuni. J. Clin. Microbiol. 39:1889-1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sahin, O., N. Luo, S. Huang, and Q. Zhang. 2003. Effect of Campylobacter-specific maternal antibodies on Campylobacter jejuni colonization in young chickens. Appl. Environ. Microbiol. 69:5372-5379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sails, A. D., D. R. A. Wareing, F. J. Bolton, A. J. Fox, and A. Curry. 1998. Characterisation of 16 Campylobacter jejuni and C. coli typing bacteriophages. J. Med. Microbiol. 47:123-128. [DOI] [PubMed] [Google Scholar]
- 26.Salama, S. M., F. J. Bolton, and D. N. Hutchinson. 1989. Improved method for the isolation of Campylobacter jejuni and Campylobacter coli bacteriophages. Lett. Appl. Microbiol. 8:5-7. [Google Scholar]
- 27.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 28.Shreeve, J. E., M. Toszeghy, A. Ridley, and D. G. Newell. 2002. The carry-over of Campylobacter isolates between sequential poultry flocks. Avian Dis. 46:378-385. [DOI] [PubMed] [Google Scholar]
- 29.Stone, R. 2002. Stalin's forgotten cure. Science 298:728-731. [DOI] [PubMed] [Google Scholar]
- 30.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. A. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wassenaar, T. M., B. Geilhausen, and D. G. Newell. 1998. Evidence of genomic instability in Campylobacter jejuni isolated from poultry. Appl. Environ. Microbiol. 64:1816-1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wassenaar, T. M., and D. G. Newell. 2000. Genotyping of Campylobacter spp. Appl. Environ. Microbiol. 66:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wassenaar, T. M., S. L. W. On, and R. J. Meinersmann. 2000. Genotyping and the consequences of genetic instability, p. 369-380. In I. Nachamkin and M. J. Blaser (ed.), Campylobacter, 2nd ed. ASM Press, Washington, D.C.