Abstract
Thrombospondin 1 (THBS1) plays an important role in angiogenesis and tumor progression. The aim of the present study was to investigate the effects of single-nucleotide polymorphisms (rs1478605 and rs3743125) in the untranslated regions of the THBS1 gene on the development and progression of gastric cancer. In the case-control study, 275 gastric cancer patients and 275 cancer-free controls were successfully genotyped using polymerase chain reaction-restriction fragment length polymorphism. The data demonstrated that THBS1 rs1478605 genotypic distributions significantly differed between the patient and control groups (P=0.005). Carriers of the CC genotype exhibited a decreased risk of developing gastric cancer compared to the carriers of the CT and TT genotypes [adjusted odd ratio (OR), 0.56; 95% confidence interval (CI), 0.39-0.79; P=0.001]. The CC genotype of rs1478605 was negatively associated with gastric cancer lymph node metastasis (OR, 0.41; 95% CI, 0.23-0.71; P=0.001) and was associated with a reduced risk of lymph node metastasis in male patients (OR, 0.27; 95% CI, 0.14-0.52; P<0.001). The THBS1 CT haplotype was associated with a reduced risk of developing gastric cancer (OR, 0.56; 95% CI, 0.33-0.93; P=0.02). By contrast, no association was observed between THBS1 rs3743125 and the development and progression of gastric cancer. These results suggest that THBS1 rs1478605 represents a potential molecular marker for gastric cancer.
Keywords: gastric cancer, THBS1, single-nucleotide polymorphism
Introduction
Gastric cancer remains the leading cause of cancer-related mortalities worldwide. In China, the incidence and mortality rate of gastric cancer are higher than the international average. Gastric cancer has a complex multi-step etiology, involving environmental and genetic factors (1–5). The individual variations in cancer risk suggest that gene mutations, in addition to genetic polymorphisms, may contribute to the overall risk of gastric cancer (6). Individual genetic susceptibility may represent a critical factor in the development of gastric cancer.
Thrombospondin 1 (THBS1) is a high molecular weight multi-functional glycoprotein that has been shown to be a potent inhibitor of angiogenesis (7). Previous studies have correlated THBS1 expression to tumor angiogenesis, tumor growth and metastasis (8–12). In gastric cancer, THBS1 may have a pro-angiogenic effect, and elevated THBS1 expression levels have been associated with gastric cancer tumor growth and lymph node metastasis (13–17). However, thus far, genetic evidence of a role for THBS1 in gastric cancer is lacking.
Single-nucleotide polymorphisms (SNPs) have been widely used to search for the association between genetic variations and disease susceptibility. THBS1 SNPs have been associated with a wide range of diseases (18–21), however, the correlation of THBS1 SNPs with individual susceptibility to gastric cancer remains unclear, although recently, our previous study found that THBS1 rs1478604 A>G within the 5′-untranslated region (UTR) of the gene is associated with lymph node metastasis of gastric cancer in a southeast Chinese population (22). To further evaluate a correlation between THBS1 SNPs and the risk of gastric cancer in a southeast Chinese population, a case-control study was conducted to examine the association of rs1478605 and rs3743125 SNPs in the UTRs of THBS1 with the development and progression of gastric cancer. THBS1 rs1478605 is negatively associated with gastric cancer development and lymph node metastasis, while rs3743125 has no significant association with gastric cancer.
Materials and methods
Study population
The study population included 275 patients with gastric carcinoma and 275 cancer-free controls. All the subjects were genetically unrelated ethnic Han Chinese and originated from Fujian (China). Patients were diagnosed with primary incident gastric cancer and were recruited at the Affiliated Hospitals of the Fujian Medical University (Fujian). All the specimens were histopathologically confirmed gastric cancer cases and had detailed clinicopathological data based on post-operative, histopathological examination. Gastric cancer patients were grouped according to the tumor-node-metastasis staging of the American Joint Commission on Cancer (https://cancerstaging.org). Cancer-free control subjects were selected randomly from local residents, who underwent a routine health check with no history of cancer and other known major diseases. There were no differences in age and gender between gastric cancer patients and control participants (Table I). The study was approved by the Institutional Review Board of the Second Affiliated Hospital of Fujian Medical University subsequent to obtaining written, informed consent.
Table I.
Characteristics of the study population.
| Variables | Gastric cancer | Controls | P-valuea |
|---|---|---|---|
| Age, years | |||
| ≤60, n (%) | 130 (47.3) | 112 (40.7) | 0.12 |
| >60, n (%) | 145 (52.7) | 163 (59.3) | |
| Gender | |||
| Male, n (%) | 201 (50.4) | 198 (49.6) | 0.77 |
| Female, n (%) | 74 (49.0) | 77 (51.0) |
P-value was calculated by χ2 test.
Genotyping
For control subjects, venous blood samples (5 ml) were collected from each individual. Genomic DNA was extracted from whole blood cell pellets using the Blood Genomic DNA Extraction kit (Takara, Shiga, Japan). DNA from gastric cancer patients was isolated from paraffin-embedded normal stomach tissue adjacent to the tumor (distance >5 cm) using the proteinase K-phenol/chloroformethanol method. DNA concentration was measured by ultraviolet spectrophotometry (NanoVue Plus®; GE Healthcare, Piscataway, NJ, USA) at 260 nm, and quality was determined using the A260/280 ratio. DNA was stored at −20 ˚ C prior to genotypic analysis.
SNP genotypes were determined using the polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) assay. Primers used for the amplification of rs1478605 [152 base pairs (bp)] were forward, 5′-GCAGGC CAGCTCGGGCGCCG-3′ and reverse, 5′-GGGGGCGGA GAGAGGAGCCCAGAC-3′; and primers used for the amplification of rs3743125 (168 bp) were forward, 5′-GTCAGGGTG GTTTTGTTTGC-3′ and reverse, 5′-GGGGGCGGAGAG AGGAGCCCAGAC-3′ (Invitrogen, Shanghai, China). PCR was performed in a 25-µ1 reaction volume containing template DNA (40 ng), primers (5 pmol/µ1 each) and 2X Taq PCR Master Mix (12.5 µl) (Tiangen, Beijing, China). Amplification was performed using the following cycling conditions: 95˚C for 5 min, and subsequently 35 cycles of 94˚C for 30 sec, 70˚C for 45 sec (for rs1478605) or 55˚C for 45 sec (for rs3743125), 72˚C for 45 sec, and a final extension at 72˚C for 7 min. PCR products were digested overnight at 37˚C with Sac II (2 units) or PvU II (2 units) restriction enzymes for rs1478605 or rs3743125, respectively (Takara) and separated on 4% agarose gels. The T allele of rs1478605 and rs3743125 contained no restriction site, and the C allele of the two SNPs contained one restriction site and produced two fragments (Fig. 1).
Figure 1.
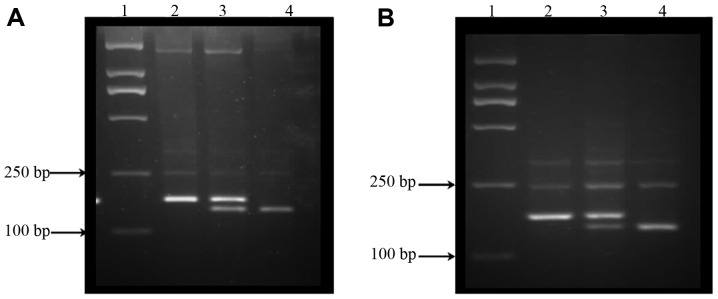
Genotyping patterns of rs1478605 and rs3743125. (A) rs1478605. Lane 1, DNA marker; lane 2, TT genotype [152 base pairs (bp) only]; lane 3, CT genotype (152+132+20 bp); lane 4, CC genotype (132+20 bp). (B) rs3743125: lane 1, DNA marker; lane 2, TT genotype (168 bp only); lane 3, CT genotype (168+145+23 bp); lane 4, CC genotype (145+23 bp).
Statistical analysis
Statistical analysis was performed using SPSS 11.5 software (SPSS, Inc., Chicago, IL, USA). Univariate or multivariate analyses were performed to evaluate the association of genotypic distributions with the development of gastric cancer and clinicopathological features. Univariate analysis was performed using the χ2 test or Fisher's exact test when required. Multivariate analyses were estimated by logistic regression modeling or stratification. Several clinicopathological variables were also dichotomized to avoid the loss of statistical power in logistic regression. All comparisons were two-tailed and P<0.05 was considered to indicate a statistically significant difference. The Haploview 4.2 software (http://www.broad.mit.edu/mpg) was used to measure the pairwise LD values between SNPs. Haplotype frequency was estimated using Phase software version 2.1 (23).
Results
Association of THBS1 rs1478605 and rs3743125 with the development of gastric cancer
To investigate the association between THBS1 rs1478605 and rs3743125 SNPs and the development and progression of gastric cancer, SNP genotyping was performed on DNA obtained from 275 patients with gastric cancer and 275 cancer-free individuals. The allelic and genotypic frequencies of the two groups are listed in Tables II and III. The genotypic distributions of the control group were tested for Hardy-Weinberg equilibrium and showed no significant deviations (P=0.99 for the two SNPs).
Table II.
Association of THBS1 rs1478605 with the development of gastric cancer.
| THBS1 rs1478605 | Controls (n=275) | Gastric cancer (n=275) | Adjusted OR (95% CI)a | P-value | Gastric cancer without lymph node metastasis (n=73) | Adjusted OR (95% CI)a | P-value | Gastric cancer with lymph node P-value metastasis (n=202) | Adjusted OR (95% CI)a | P-value |
|---|---|---|---|---|---|---|---|---|---|---|
| Genotype | ||||||||||
| TT, n (%) | 28 (10.2) | 37 (13.5) | − | − | 9 (12.3) | − | − | 28 (13.9) | − | − |
| CT, n (%) | 120 (43.6) | 148 (53.8) | 0.93 (0.54-1.59) | 0.78 | 29 (39.7) | 0.75 (0.32-1.77) | 0.52 | 119 (58.9) | 0.99 (0.55-1.77) | 0.97 |
| CC, n (%) | 127 (46.2) | 90 (32.7) | 0.52 (0.30-0.92) | 0.02 | 35 (48.0) | 0.86 (0.37-2.00) | 0.73 | 55 (27.2) | 0.42 (0.23-0.78) | 0.006 |
| TT+CT, n (%) | 148 (53.8) | 185 (67.3) | 0.56 (0.39-0.79)b | 0.001 | 38 (52.1) | 1.08 (0.64-1.81)b | 0.78 | 147 (72.8) | 0.43 (0.29-0.63)b | 0.00002 |
| CC+CT, n (%) | 247 (89.8) | 238 (86.5) | 1.40 (0.83-2.36)c | 0.21 | 64 (87.7) | 1.24 (0.55-2.76)c | 0.61 | 174 (86.1) | 1.44 (0.82–2.52)c | 0.21 |
| Allele | ||||||||||
| C, n (%) | 374 (68.0) | 328 (59.6) | 0.71 (0.56-0.92) | 0.008 | 99 (67.8) | 0.99 (0.67-1.47) | 0.97 | 229 (56.7) | 0.62 (0.47-0.80) | 0.0003 |
| T, n (%) | 176 (32.0) | 222 (40.4) | 47 (32.2) | 175 (43.3) | ||||||
Adjusted by age and gender.
Adjusted OR (95% CI) was calculated by logistic regression modeling, comparing CC with the TT+CT genotype.
Adjusted OR (95% CI) was calculated by logistic regression modeling, comparing TT with the CC+CT genotype. THBS1, thrombospondin 1; OR, odds ratio; CI, confidence interval.
Table III.
Association of THBS1 rs3743125 with the development of gastric cancer.
| THBS1 rs3743125 | Gastric cancer (n=275) | Controls (n=275) | Adjusted OR (95% CI)a | P-value |
|---|---|---|---|---|
| Genotype | ||||
| TT, n (%) | 28 (10.2) | 30 (10.9) | − | − |
| CT, n (%) | 133 (48.4) | 123 (44.7) | 1.14 (0.64-2.02) | 0.66 |
| CC, n (%) | 114 (41.5) | 122 (44.4) | 0.98 (0.55-1.75) | 0.95 |
| TT+CT, n (%) | 161 (58.5) | 153 (55.6) | 0.88 (0.63-1.24)b | 0.47 |
| CT+CC, n (%) | 247 (89.8) | 245 (90.1) | 0.94 (0.55-1.63)c | 0.83 |
| Allele | ||||
| C, n (%) | 361 (65.6) | 367 (66.7) | 0.95 (0.74-1.22) | 0.70 |
| T, n (%) | 189 (34.4) | 183 (33.3) | ||
Adjusted by age and gender.
Adjusted OR (95% CI) was calculated by logistic regression modeling, comparing TT+CT with the CC genotype.
Adjusted OR (95% CI) was calculated by logistic regression modeling, comparing CC+CT with the TT genotype. THBS1, thrombospondin 1; OR, odds ratio; CI, confidence interval.
No significant differences in the genotypic distributions and allele frequencies of rs3743125 were observed between gastric cancer patients and control subjects, indicating that rs3743125 is not associated with gastric cancer development (Table III). However, a significant difference in the CC genotype distribution and C allele frequency of rs1478605 between disease and control groups was identified. These results indicate that carriers of the CC genotype exhibit a decreased risk of developing gastric cancer compared to the carriers with the CT and TT genotypes [adjusted odds ratio (OR), 0.56; 95% confidence interval (CI), 0.39-0.79; P=0.001] (Table II). The risk in C allele carriers was lower than that in the T allele carriers (adjusted OR, 0.71; 95% CI, 0.56-0.92; P=0.008) (Table II). Furthermore, comparison of patients with lymph node metastasis and control subjects revealed that carriers of the CC genotype and C allele had a decreased risk of developing this subgroup of gastric cancer (adjusted OR, 0.43; 95% CI, 0.29-0.63; P<0.0001 and adjusted OR, 0.62; 95% CI, 0.47-0.80; P=0.0003, respectively) (Table II). By contrast, there was no significant association between the CC genotype and C allele frequency in gastric cancer patients with and without lymph node metastasis (P=0.78) (Table II).
Association of THBS1 rs1478605 and rs3743125 SNPs with clinicopathological features of gastric cancer
Associations between the genotypic distribution and overall clinicopathological features were analyzed by univariate analysis and are reported in Table IV. No associations between the genotypic distribution of THBS1 rs3743125 and overall patient clinicopathological features were observed. However, a significant association between the genotypic distribution of rs1478605 and the status of gastric cancer lymph node metastasis was observed (P=0.005) (Table V). In comparison with CT and TT genotype carriers, CC genotype carriers exhibited a lower risk of lymph node metastasis (OR, 0.41; 95% CI, 0.23-0.71; P=0.001) (Table V).
Table IV.
Association of the THBS1 polymorphisms with the clinicopathological features of gastric cancer.
| Rs1478605 genotype | Rs3743125 genotype | |||||||
|---|---|---|---|---|---|---|---|---|
| Clinicopathological | TT | CT | CC | TT | CT | CC | ||
| features | n (%) | n (%) | n (%) | P-valuea | n (%) | n (%) | n (%) | P-valuea |
| Gender | ||||||||
| Male (n=201) | 23 (11.4) | 106 (52.7) | 72 (35.8) | 19 (9.5) | 103 (51.2) | 79 (39.3) | ||
| Female (n=74) | 14 (18.9) | 42 (56.8) | 18 (24.3) | 0.1 | 9 (12.2) | 30 (40.5) | 35 (47.3) | 0.29 |
| Age (years) | ||||||||
| ≤60 (n=130) | 16 (12.3) | 69 (53.1) | 45 (34.6) | 11 (8.5) | 64 (49.2) | 55 (42.3) | ||
| >60 (n=145) | 21 (14.5) | 79 (54.5) | 45 (31.0) | 0.79 | 17 (11.7) | 69 (47.6) | 59 (40.7) | 0.67 |
| Differentiation | ||||||||
| Well and moderate (n=100) | 15 (15.0) | 51 (51.0) | 34 (34.0) | 11 (11.0) | 46 (46.0) | 43 (43.0) | ||
| Poor (n=175) | 22 (12.6) | 97 (55.4) | 56 (32.0) | 0.75 | 17 (9.7) | 87 (49.7) | 71 (40.6) | 0.83 |
| TNM | ||||||||
| IA/IB (n=30) | 4 (13.3) | 14 (46.7) | 12 (40.0) | 2 (6.7) | 14 (46.7) | 14 (46.7) | ||
| II (n=65) | 6 (9.2) | 32 (56.7) | 27 (41.5) | 8 (12.3) | 30 (46.2) | 27 (41.5) | ||
| IIIA/IIIB (n=164) | 24 (14.6) | 93 (56.3) | 47 (28.7) | 17 (10.4) | 77 (47.0) | 70 (42.7) | ||
| IV (n=16) | 3 (18.8) | 9 (56.3) | 4 (25.0) | 0.48b | 1 (6.3) | 12 (75.0) | 3 (18.8) | 0.51b |
| Lymph node metastasis | ||||||||
| Without metastasis (n=73) | 9 (12.3) | 29 (39.7) | 35 (47.9) | 7 (9.6) | 33 (45.2) | 33 (45.2) | ||
| With metastasis (n=202) | 28 (13.9) | 119 (58.9) | 55 (27.2) | 0.005 | 21 (10.2) | 100 (48.4) | 81 (41.5) | 0.75 |
| Location | ||||||||
| Cardia (n=76) | 11 (14.5) | 42 (55.3) | 23 (30.3) | 9 (11.8) | 33 (43.4) | 34 (44.7) | ||
| Corpus (n=73) | 7 (9.6) | 39 (53.4) | 27 (37.0) | 10 (13.7) | 41 (56.2) | 22 (30.1) | ||
| Antrum (n=92) | 16 (17.4) | 43 (46.7) | 33 (35.9) | 6 (6.5) | 40 (43.5) | 46 (50.0) | ||
| Pylorus or other (n=34) | 3 (8.8) | 24 (70.6) | 7 (20.6) | 0.28 | 3 (8.8) | 19 (55.9) | 12 (41.5) | 0.18 |
| Invasion | ||||||||
| Within serosa (n=185) | 24 (13.0) | 102 (55.1) | 59 (31.9) | 19 (10.3) | 90 (48.6) | 76 (41.1) | ||
| Serosa and beyond (n=90) | 13 (14.4) | 46 (51.1) | 31 (34.4) | 0.84 | 9 (10.0) | 43 (47.8) | 38 (42.2) | 0.98 |
| Tumor size | ||||||||
| ≤5 cm (n=154) | 16 (10.4) | 81 (52.6) | 57 (37.0) | 14 (9.1) | 73 (47.4) | 67 (43.5) | ||
| >5 cm (n=121) | 21 (17.4) | 67 (55.4) | 33 (27.3) | 0.11 | 14 (11.6) | 60 (49.6) | 47 (41.5) | 0.68 |
P-value was calculated by the χ2 test.
P-value was calculated by the Fisher's exact test. THBS1, thrombospondin 1; TNM, tumor-node-metastasis; OR, odds ratio; CI, confidence interval.
Table V.
Association of THBS1 rs1478605 with gastric cancer lymph node metastasis.
| Genotype | Genotype | Genotype | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lymph node metastasis n (%) | CC n (%) | CT n (%) | TT n (%) | P-valuea | CT+TT n (%) | CC n (%) | Adjusted OR (95% CI) | P-value | CT+CC n (%) | TT n (%) | Adjusted OR (95% CI) | P-value |
| Without metastasis (n=73) | 35 (47.9) | 29 (39.7) | 9 (12.3) | 0.005a | 38 (52.1) | 35 (47.9) | 0.41 (0.23-0.71)b | 0.001 | 64 (87.7) | 9 (12.3) | 1.14 (0.51-2.56)b | 0.74 |
| 0.37 (0.21-0.66)c | 0.001 | 1.13 (0.49-2.56)c | 0.78 | |||||||||
| 0.40 (0.23-0.72)d | 0.002 | 1.04 (0.45-2.39)d | 0.93 | |||||||||
| With metastasis (n=202) | 55 (27.2) | 119 (58.9) | 28 (13.9) | 147 (72.8) | 55 (27.2) | 174 (86.1) | 28 (13.9) | |||||
P-value was calculated by χ2 test.
OR (95% CI) was calculated by χ2 test, comparing CC with TT+CT genotype.
OR (95% CI) was calculated using logistic regression modeling adjusted by age, gender and tumor invasion depth, comparing CC with TT+CT genotype.
OR (95% CI) was calculated using logistic regression modeling adjusted by age, gender and tumor size, comparing TT with CC+CT genotype. THBS1, thrombospondin 1; OR, odds ratio; CI, confidence interval.
To confirm the strength of the association between THBS1 rs1478605 and lymph node metastasis, a multivariate analysis was performed. To identify the potential confounding factors, an analysis of the association between lymph node metastasis and other clinicopathological features was first performed. This analysis revealed that tumor size and tumor invasion depth were significantly associated with lymph node metastasis and were likely to be confounding factors (P=0.002 and P=0.004, respectively) (Table VI). By contrast, the age and gender of patients was included in all the multivariate analyses, as these represented independent variables, and were not associated with lymph node metastasis status (P=0.61 and P=0.49, respectively) (Table VI) or rs1478605 genotypic distribution (P=0.1 and P=0.79, respectively) (Table IV). Following adjustment for tumor invasion depth, patient gender and age, multivariate logistic regression analyses revealed that the homozygous CC genotype was significantly associated with a decreased risk of lymph node metastasis (OR, 0.37; 95% CI, 0.21-0.66; P=0.001) (Table V). Following adjustment for tumor size, patient gender and age, this analysis revealed that the homozygous CC genotype was also associated with a decreased risk of lymph node metastasis (OR, 0.40; 95% CI, 0.23-0.72; P=0.002) (Table V).
Table VI.
Associations of the clinicopathological features with gastric cancer lymph node metastasis.
| Lymph node metastasis | All cases, n (%) | Cases with no metastasis, n (%) | Cases with metastasis, n (%) | P-valuea |
|---|---|---|---|---|
| Gender | ||||
| Male | 201 (73.1) | 55 (75.3) | 146 (72.3) | |
| Female | 74 (26.9) | 18 (24.7) | 56 (27.7) | 0.61 |
| Age (year) | ||||
| 1 (≤60) | 130 (47.3) | 32 (43.8) | 98 (48.5) | |
| 2 (>60) | 145 (52.7) | 41 (56.2) | 104 (51.5) | 0.49 |
| Differentiation | ||||
| Well and moderate | 100 (36.4) | 32 (43.8) | 68 (33.7) | |
| Poor | 175 (63.6) | 41 (56.2) | 134 (66.3) | 0.12 |
| Location | ||||
| Cardia | 76 (27.6) | 19 (26.0) | 57 (28.2) | |
| Corpus | 73 (26.5) | 24 (32.9) | 49 (24.3) | |
| Antrum | 92 (33.5) | 23 (31.5) | 69 (34.2) | |
| Pylorus or other | 34 (12.4) | 7 (9.6) | 27 (13.4) | 0.51 |
| Invasion | ||||
| Within serosa | 185 (67.3) | 59 (80.8) | 126 (62.4) | |
| Serosa and beyond | 90 (32.7) | 14 (19.2) | 76 (37.6) | 0.004 |
| Tumor size | ||||
| ≤5 cm | 154 (56.0) | 52 (71.2) | 102 (50.5) | |
| >5 cm | 121 (44.0) | 21 (28.8) | 100 (49.5) | 0.002 |
P-value was calculated by χ2 test.
To further validate the results from this logistic regression modeling analyses and to investigate the interaction between factors, stratification was applied to analyze the association of rs1478605 with lymph node metastasis, using other clinicopathological features as stratification factors. The majority of these analyses did not yield significant results, including those using age, differentiation status, tumor location, size and invasion depth as stratification factors (data not shown), as each OR value between strata was not significantly different, indicating that the associations were independent of these factors. However, when patient gender was used as the stratification factor, a clear association between rs1478605 and lymph node metastasis was observed in the two strata. There was a synergistic effect of the rs1478605 SNP and male gender on gastric cancer lymph node metastasis without any adjustments (OR, 0.27; 95% CI, 0.14-0.52; P<0.001) (Table VII). By contrast, the effect of the rs1478605 SNP and female gender on lymph node metastasis was not significant (OR, 1.83; 95% CI, 0.46-7.22; P=0.38) (Table VII). A significant difference in the effect of the rs1478605 SNP between the two strata was observed (P=0.01) (Table VII). These results indicate that the CC genotype is associated with a reduced risk of lymph node metastasis in male patients.
Table VII.
Stratified analyses of the associations of rs1478605 with lymph node metastasis.
| Gender | Lymph node metastasis | Rs1478605 CT+TT, n (%) | Genotype CC, n (%) | OR (95% CI)a | P-value | P-value between stratab |
|---|---|---|---|---|---|---|
| Male | Without metastasis (n=55) | 23 (41.8) | 32 (58.2) | 0.27 (0.14-0.52) | 0.00005 | 0.01 |
| With metastasis (n=146) | 106 (72.6) | 40 (27.4) | ||||
| Female | Without metastasis (n=18) | 15 (83.3) | 3 (16.7) | 1.83 (0.46-7.22) | 0.38 | |
| With metastasis (n=56) | 41 (73.2) | 15 (26.8) |
OR (95% CI) was calculated by χ2 test, using gender as the stratification factor.
P-value was calculated by Mantel-Haenszel's test. OR, odds ratio; CI, confidence interval.
Association of THBS1 haplotypes with the development and progression of gastric cancer
THBS1 rs1478605 and rs3743125 exist in a single block of disequilibrium, with r2 and D′ values of 0.91 and 1, respectively. The four major haplotypes and their frequencies identified in gastric cancer patients and control subjects are shown in Table VIII. A significant difference in the THBS1 CT haplotype distribution was identified between patient and control groups (OR, 0.56; 95% CI, 0.33-0.93; P=0.02) (Table VIII), indicating that CT haplotype carriers had a lower risk of developing gastric cancer, compared to CC+TT+TC haplotype carriers. However, no association between THBS1 haplotypes and the overall clinicopathological features of gastric cancer were observed (data not shown).
Table VIII.
Correlation between the THBS1 haplotypes and the development of gastric cancer.
| Haplotypes | Gastric cancer, n (%) | Controls, n (%) | P-value | OR (95% CI)a |
|---|---|---|---|---|
| CT | 27 (10) | 45 (16) | 0.02 | 0.56 (0.33-0.93) |
| CC | 137 (50) | 141 (51) | ||
| TT | 64 (23) | 49 (18) | ||
| TC | 47 (17) | 40 (15) |
OR (95% CI) was calculated by χ2 test, comparing CT haplotype with the CC+TT+TC haplotypes. THBS1, thrombospondin 1; OR, odds ratio; CI, confidence interval.
Discussion
The present study demonstrated that the THBS1 rs1478605 CC genotype is associated with a decreased risk of developing gastric cancer, particularly gastric cancer with lymph node metastasis. Among the patients with gastric cancer, the rs1478605 CC genotype was negatively associated with lymph node metastasis. The THBS1 CT haplotype is associated with a reduced risk of gastric cancer. However, no associations between the THBS1 rs3743125 genotypic distribution and the development or clinicopathological features of gastric cancer.
In the present study, careful statistical analyses were performed to avoid spurious results caused by artificial bias in the study design and by confounding factors. A multitude of univariate or multivariate analyses were performed to detect inconsistencies reflecting the presence of biases. The similar results derived from these analyses confirmed and reinforced our findings.
The study focused on THBS1 rs1478605 and rs37431125 SNPs, which are located in gene UTRs. UTRs are known to play crucial roles in the post-transcriptional regulation of gene expression (22, 24–25), which is important for normal cell function, and dysfunctions have been linked to the pathophysiology of numerous diseases (26–30). In recent years, gene polymorphisms in UTRs have also been extensively studied and reported to be associated with cancer susceptibility (31–34). However, there has been relatively little study associated with the correlation of SNPs in THBS1 UTRs with individual susceptibility to gastric cancer until recently. The present data demonstrate that THBS1 rs3743125 has no association with the development and progression of gastric cancer, which was similar to our previous study (22). Notably, in the previous study (22), the data showed that the AG and GG genotypes of THBS1 rs1478604 A>G, the other SNP loci located in 5′-UTRs, was positively associated with lymph node metastasis in gastric cancer. However, our data suggested that the rs1478605 CC genotype was negatively associated with lymph node metastasis. To identify the synergistic effect of the two SNPs, a haplotype-based association study was necessary. However, the data of rs1478604 was unavailable in this study, which requires to be completed in future studies. Despite the limitation, the present study adds to the evidence that SNPs in THBS1 UTRs may affect the development and progression of gastric cancer.
As non-coding polymorphisms, the SNPs of THBS1 do not lead to the alteration of the THBS1 protein. However, they may affect the susceptibility to gastric cancer through the following mechanisms: i) The polymorphism locus may change the regulatory sequences of the UTRs, which influence expression of THBS1 at the level of translation; and ii) other nearby polymorphisms in linkage disequilibrium with the SNPs may have a functional role. Clearly, further studies are required to confirm these hypotheses.
THBS1 has been reported to exert a pro-angiogenic effect in gastric cancer (13–17), which plays an important role in cancer metastasis. Tumor metastasis is well established as a critical event affecting patient prognosis. The present study suggests that the rs1478605 CC genotype is negatively associated with lymph node metastasis, indicating that gastric cancer patients with the THBS1 rs1478605 CC genotype may have an improved prognosis than patients with CT and TT genotypes.
In conclusion, the present study demonstrates that THBS1 rs1478605 may be a protective factor in gastric cancer. The study had 89 and 97% power to detect an effect with an OR of 0.56 in the case and control groups under a dominant genetic model and an OR of 0.27 in the lymph node metastasis and non-metastasis groups in male gastric cancer patients. Further studies using larger patient cohorts are required to confirm our findings, and mechanistic studies are required to improve the understanding of the complex mechanisms involved in the development and progression of gastric cancer. Despite several limitations in the present study, our data provide additional information that is necessary for genetic risk assessments, and confirm the important role of THBS1 in the development and progression of gastric cancer.
Acknowledgements
The present study was supported by the National Natural Science Foundation of China (grant no. 81101893 to S.C.). The authors are grateful to all the patients involved in the study and for the collaboration with participating hospitals and their staff.
References
- 1.Malfertheiner P, Bornschen J, Selgrad M. Role of Helicobacter pylori infection in gastric cancer pathogenesis: a chance for prevention. J Dig Dis. 2010;11:2–11. doi: 10.1111/j.1751-2980.2009.00408.x. [DOI] [PubMed] [Google Scholar]
- 2.Lee DS, Yang HK, Kim JW, et al. Identifying the risk factors through the development of a predictive model for gastric cancer in South Korea. Cancer Nurs. 2009;32:135–142. doi: 10.1097/NCC.0b013e3181982c2e. [DOI] [PubMed] [Google Scholar]
- 3.Shen X, Zhang J, Yan Y, et al. Analysis and estimates of the attributable risk for environmental and genetic risk factors in gastric cancer in a Chinese population. J Toxicol Environ Health A. 2010;72:759–766. doi: 10.1080/15287390902841599. [DOI] [PubMed] [Google Scholar]
- 4.Yamashita K, Sakuramoto S, Watanabe M. Genomic and epigenetic profiles of gastric cancer: potential diagnostic and therapeutic applications. Surg Today. 2011;41:24–38. doi: 10.1007/s00595-010-4370-5. [DOI] [PubMed] [Google Scholar]
- 5.Hudler P. Genetic aspects of gastric cancer instability. Scientific World Journal. 2012;2012:761909. doi: 10.1100/2012/761909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.González CA, Sala N, Rokkas T. Gastric cancer: epidemiologic aspects. Helicobacter. 2013;18(Suppl 1):34–38. doi: 10.1111/hel.12082. [DOI] [PubMed] [Google Scholar]
- 7.Adams JC, Lawler J. The thrombospondins. Int J Biochem Cell Biol. 2004;36:961–968. doi: 10.1016/j.biocel.2004.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kazerounian S, Yee KO, Lawler J. Thrombospondins in cancer. Cell Mol Life Sci. 2008;65:700–712. doi: 10.1007/s00018-007-7486-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yee KO, Connolly CM, Duquette M, et al. The effect of thrombospondin-1 on breast cancer metastasis. Breast Cancer Res Treat. 2009;114:85–96. doi: 10.1007/s10549-008-9992-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou ZQ, Cao WH, Xie JJ, et al. Expression and prognostic significance of THBS 1, Cyr61 and CTGF in esophageal squamous cell carcinoma. BMC Cancer. 2009;9:291. doi: 10.1186/1471-2407-9-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Streit M, Velasco P, Brown LF, et al. Overexpression of thrombospondin-1 decreases angiogenesis and inhibits the growth of human cutaneous squamous cell carcinomas. Am J Pathol. 1999;155:441–452. doi: 10.1016/S0002-9440(10)65140-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miyanaga K, Kato Y, Nakamura T, et al. Expression and role of thrombospondin-1 in colorectal cancer. Anticancer Res. 2002;22:3941–3948. [PubMed] [Google Scholar]
- 13.Kiyono K, Suzuki HI, Morishita Y, et al. c-Ski overexpression promotes tumor growth and angiogenesis through inhibition of transforming growth factor-beta signaling in diffuse-type gastric carcinoma. Cancer Sci. 2009;100:1809–1816. doi: 10.1111/j.1349-7006.2009.01248.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang J, Ito R, Oue N, et al. Expression of thrombospondin-1 is correlated with microvessel density in gastric carcinoma. Virchows Arch. 2003;442:563–568. doi: 10.1007/s00428-003-0810-6. [DOI] [PubMed] [Google Scholar]
- 15.Miyamoto N, Yamamoto H, Taniguchi H, et al. Differential expression of angiogenesis-related genes in human gastric cancers with and those without high-frequency microsatellite instability. Cancer Lett. 2007;254:42–53. doi: 10.1016/j.canlet.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 16.Nakao T, Kurita N, Komatsu M, et al. Expression of thrombospondin-1 and Ski are prognostic factors in advanced gastric cancer. Int J Clin Oncol. 2011;16:145–152. doi: 10.1007/s10147-010-0147-5. [DOI] [PubMed] [Google Scholar]
- 17.Lin XD, Chen SQ, Qi YL, et al. Overexpression of thrombospondin-1 in stromal myofibroblasts is associated with tumor growth and nodal metastasis in gastric carcinoma. J Surg Oncol. 2012;106:94–100. doi: 10.1002/jso.23037. [DOI] [PubMed] [Google Scholar]
- 18.Sfar S, Saad H, Mosbah F, Chouchane L. Combined effects of the angiogenic genes polymorphisms on prostate cancer susceptibility and aggressiveness. Mol Biol Rep. 2009;36:37–45. doi: 10.1007/s11033-007-9149-4. [DOI] [PubMed] [Google Scholar]
- 19.Zwicker JI, Peyvandi F, Palla R, et al. The thrombospondin-1 N700S polymorphism is associated with early myocardial infarction without altering von Willebrand factor multimer size. Blood. 2006;108:1280–1283. doi: 10.1182/blood-2006-04-015701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ashokkumar M, Anbarasan C, Saibabu R, et al. An association study of thrombospondin 1 and 2 SNPs with coronary artery disease and myocardial infarction among South Indians. Thromb Res. 2011;128:49–53. doi: 10.1016/j.thromres.2011.05.026. [DOI] [PubMed] [Google Scholar]
- 21.Koch W, Hoppmann P, de Waha A, et al. Polymorphisms in thrombospondin genes and myocardial infarction: a case-control study and a meta-analysis of available evidence. Hum Mol Genet. 2008;17:1120–1126. doi: 10.1093/hmg/ddn001. [DOI] [PubMed] [Google Scholar]
- 22.Lin XD, Chen SQ, Qi YL, et al. Polymorphism of THBS1 rs1478604 A>G in 5-untranslated region is associated with lymph node metastasis of gastric cancer in a Southeast Chinese population. DNA Cell Biol. 2012;31:511–519. doi: 10.1089/dna.2011.1344. [DOI] [PubMed] [Google Scholar]
- 23.Scheet P, Stephens M. A fast and flexible statistical model for large-scale population genotype data: applications to inferring missing genotypes and haplotypic phase. Am J Hum Genet. 2006;78:629–644. doi: 10.1086/502802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van der Velden AW, Thomas AA. The role of the 5-untranslated region of an mRNA in translation regulation during development. Int J Biochem Cell Biol. 1999;31:87–106. doi: 10.1016/S1357-2725(98)00134-4. [DOI] [PubMed] [Google Scholar]
- 25.Jansen RP. mRNA localization: message on the move. Nat Rev Mol Cell Biol. 2001;2:247–256. doi: 10.1038/35067016. [DOI] [PubMed] [Google Scholar]
- 26.Bashirullah A, Cooperstock RL, Lipshitz HD. Spatial and temporal control of RNA stability. Proc Natl Acad Sci USA. 2001;98:7025–7028. doi: 10.1073/pnas.111145698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mignone F, Gissi C, Liuni S, Pesole G. Untranslated regions of mRNAs. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-3-reviews0004. REVIEWS0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cazzola M, Skoda RC. Translation pathophysiology: a novel molecular mechanism of human disease. Blood. 2000;95:3280–3288. [PubMed] [Google Scholar]
- 29.Mihailovich M, Thermann R, Grohovaz F, et al. Complex translational regulation of BACE1 involves upstream AUGs and stimulatory elements within the 5′ untranslated region. Nucleic Acids Res. 2007;35:2975–2985. doi: 10.1093/nar/gkm191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Welch EM, Barton ER, Zhuo J, et al. PTC124 targets genetic disorders caused by nonsense mutations. Nature. 2007;447:87–91. doi: 10.1038/nature05756. [DOI] [PubMed] [Google Scholar]
- 31.Chen JM, Férec C, Cooper DN. A systematic analysis of disease-associated variants in the 3′ regulatory regions of human protein-coding genes II: the importance of mRNA secondary structure in assessing the functionality of 3′UTR variants. Hum. Genet. 2006;120:301–333. doi: 10.1007/s00439-006-0180-7. [DOI] [PubMed] [Google Scholar]
- 32.Tian X, Tian Y, Ma P, et al. Association between MDM2 SNP 309 T>G and risk of gastric cancer: a meta-analysis. Asian Pac J Cancer Prev. 2013;14:1925–1929. doi: 10.7314/APJCP.2013.14.3.1925. [DOI] [PubMed] [Google Scholar]
- 33.Zhuang W, Wu XT, Zhou Y, et al. Polymorphisms of thymidylate synthase in the 5′- and 3′-untranslated regions and gastric cancer. Dig Dis Sci. 2009;54:1379–1385. doi: 10.1007/s10620-008-0511-8. [DOI] [PubMed] [Google Scholar]
- 34.Hamai Y, Matsumura S, Matsusaki K, et al. A single nucleotide polymorphism in the 5′ untranslated region of the EGF gene is associated with occurrence and malignant progression of gastric cancer. Pathobiology. 2005;72:133–138. doi: 10.1159/000084116. [DOI] [PubMed] [Google Scholar]


