Abstract
Degradation rates of extracellular DNA determined in marine sediments were much higher than those in the water column. However, due to the high sediment DNA content, turnover times were much shorter in seawater. Results reported here provide new insights into the role of extracellular DNA in P cycling in marine ecosystems.
In all aquatic ecosystems, extracellular DNA concentrations in the sediment are 3 to 4 orders of magnitude higher than those in the water column (4, 7, 8, 9). Extracellular DNA accumulation in sediments could depend on a complex array of biotic factors (e.g., bacteria-mediated degradation rates) and abiotic factors (e.g., DNA binding to mineral and refractory organic particles) which are responsible for a half-life of sediment extracellular DNA longer than that in the water column (2, 10, 14).
Despite the potential importance of extracellular DNA in lateral gene transfer and biogeochemical cycles (6, 10), information dealing with degradation and turnover rates of extracellular DNA in marine sediments is practically nonexistent (11).
In this study, we investigated degradation and turnover rates of extracellular DNA in coastal sediments of the Mediterranean Sea in order to provide new insights into P cycling through extracellular DNA diagenesis.
Study area and sampling.
Sediment samples were collected at three coastal stations located in the Adriatic Sea (hereinafter identified as Goro Lagoon, Ancona Harbor, and outside Ancona Harbor). Sediment samples were collected for the analysis of DNase activity and extracellular DNA content as well as the determination of bacterial abundance, biomass, and carbon production. Moreover, surface water samples were collected for the analyses of DNase activity and dissolved extracellular DNA.
Determination of DNase activity.
DNase activity was determined fluorometrically by using a fluorescent DNA analog, poly deoxyribo-1-N6ethenoadenylic acid [poly(dɛA)] (16). Poly(dɛA) substrate was prepared by a chemical modification of poly(dA) (fragment length, 50 bases) with chloroacetaldehyde according to the procedure of Cazenave et al. (3). Kinetic parameters of extracellular DNA hydrolysis were determined by incubating sediment and seawater samples with increasing concentrations of poly(dɛA) that had previously been diluted with prefiltered and autoclaved seawater. Autoclaved sediment and seawater samples were utilized as blanks. All samples were incubated in the dark at the in situ temperature and subsequently analyzed fluorometrically. The fluorescence of the samples was converted into the amount of released mononucleotides by using calibration curves obtained from standard solutions of 1,N6-ethenoadenine deoxyribose-5′-monophosphate. The maximum velocity and half-saturation constant were calculated by using Lineweaver-Burke plots of the reaction velocity versus substrate concentrations. In order to test the specificity of poly(dɛA) for the detection of extracellular nuclease activity, sediment samples treated with commercial nucleases were mixed before incubation with the fluorescent DNA analog and compared with samples without enzyme addition.
Extracellular DNA in sediments.
Extracellular-DNA analyses were carried out according to the nuclease-based procedure described by Dell'Anno et al. (9). This procedure allows quantification of the bioavailable (i.e., hydrolyzable by nucleases) fraction of the extracellular DNA pool.
Extracellular DNA in seawater.
In order to quantify the bioavailable fraction of the dissolved extracellular DNA pool in seawater samples, we utilized the nuclease-based procedure described by Siuda and Chrost (15).
Bacterial parameters.
Benthic bacterial abundance and biomass were determined by epifluorescence microscopy according to Danovaro et al. (5). Bacterial production was measured by [3H]leucine incorporation according to the procedure described by van Duyl and Kop (17) for sediments.
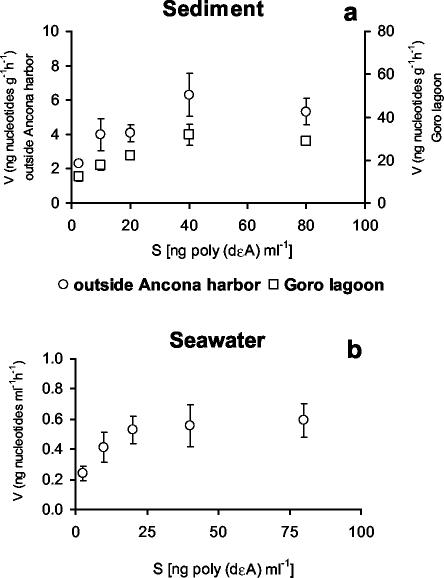
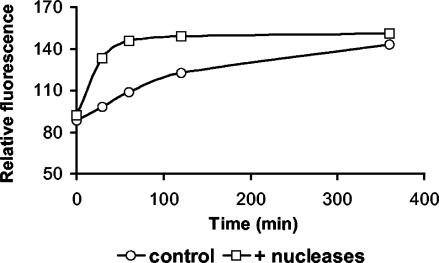
Kinetic experiments revealed that the half-saturation constant was low for both seawater and sediment samples (Fig. 1a and b). DNase activity increased linearly with time in both sediment and seawater samples (Fig. 2). Sediment samples incubated with commercial nucleases reached a plateau of fluorescence after 60 min (Fig. 3). Results obtained by fluorometric assay with the fluorescent DNA analog and by high-pressure liquid chromatography analysis with high-molecular-weight DNA did not show significant differences (data not shown).
FIG. 1.
Velocity (V) of poly(dɛA) degradation as a function of substrate concentration in the top 1 cm of sediments collected outside Ancona Harbor and at Goro Lagoon (a) and in surface seawater samples collected outside Ancona Harbor (b). Standard deviations are reported.
FIG. 2.
Time course analysis of poly(dɛA) degradation in sediment and seawater samples. Data, expressed in relative fluorescence units, refer to time course experiments carried out with the top 1 cm of sediments collected in Ancona Harbor and on surface seawater samples collected outside Ancona Harbor.
FIG. 3.
Time course analysis of poly(dɛA) degradation in treated (i.e., with added nucleases) and control (i.e., non-enzymatically treated) sediment samples. Data, expressed in relative fluorescence units, refer to time course experiments carried out with the top 1 cm of sediments collected outside Ancona Harbor.
DNase activities and extracellular DNA concentrations in sediments were characterized by clear changes among the investigated sites with the highest values at Goro Lagoon and the lowest values outside Ancona Harbor (Table 1). Turnover times for extracellular DNA (calculated as the ratio between extracellular DNA concentrations and degradation rates [12, 13]) were much longer in sediments than in the water column (Table 1).
TABLE 1.
DNase activity, extracellular DNA concentrations (enzymatically hydrolyzed), and extracellular DNA turnover times for sediments collected at the three sampling sites and in seawater samples collected outside Ancona Harbora
| Sample | Sampling site | DNase activity (ng of nucleotides g−1 h−1)b | Extracellular DNA (μg g−1)b | Extracellular DNA turnover time (days) | Bacterial abundance (108 cells g−1)b | Bacterial biomass (μg of C g−1)b | Bacterial C production (μg of C g−1 h−1)b |
|---|---|---|---|---|---|---|---|
| Sediment | Outside Ancona Harbor | 3.9 ± 1.5 | 8.6 ± 1.5 | 93 | 3.1 ± 0.13 | 25.6 ± 3.4 | 0.15 ± 0.01 |
| Ancona Harbor | 20.5 ± 6.9 | 22.3 ± 4.2 | 45 | 30.2 ± 5.3 | 61.1 ± 9.8 | 0.18 ± 0.07 | |
| Goro Lagoon | 50.7 ± 12.3 | 35.0 ± 3.9 | 29 | 47.5 ± 4.6 | 84.2 ± 5.2 | 0.30 ± 0.1 | |
| Seawater | Outside Ancona Harbor | 0.52 ± 0.07c | 5.1 ± 0.6d | 0.41 | ND | ND | ND |
For sediments collected at the three sampling sites, bacterial abundance, biomass, and carbon production are also reported. ND, not determined.
Mean ± standard error.
Values are expressed in nanograms of nucleotides per milliliter per hour.
Values are expressed in micrograms per liter.
Degradation and turnover rates for extracellular DNA.
Information on degradation and turnover rates for extracellular DNA in marine environments is still limited and almost completely restricted to the plankton domain (12, 13).
In this study, we found that degradation rates for extracellular DNA in sediments were 7 to 100 times higher than those for extracellular DNA in the water column. However, due to the high extracellular DNA content in sediments, turnover times were much shorter in seawater (29 to 93 days in sediments versus about 10 h in seawater). Calculation of turnover times could be biased by quantitative estimates of extracellular DNA pools, which do not reflect actual bioavailability (9, 10). This was not the case in this study, since we quantified only the bioavailable fraction of the extracellular DNA. Therefore, long turnover times for extracellular DNA in marine sediments are likely the result of the unbalanced ratio between its supply-production and degradation rates.
Extracellular DNA concentrations in sediments were, on average, 4.3 times higher than concentrations of DNA associated with total bacterial cells (assuming a conversion factor of 3.3 fg of DNA cell−1 [7]) (Table 1), indicating that the majority of the sedimentary DNA pool is not accounted for by living biomass.
Previous studies have hypothesized that extracellular DNA in marine sediments might represent an important source of P for bacterial growth and turnover (4, 7, 9). In order to explore the potential ecological implication of extracellular DNA in benthic biogeochemical processes, we estimated the daily bacterial P requirement on the basis of bacterial C production (assuming a bacterial C/N/P ratio of 40:10:1 [18]) and the daily P supply derived from DNA degradation rates (assuming for DNA an average P content of 10% [1]). From these estimates, we calculated that extracellular DNA might potentially supply, on average for the three sampling sites, 41% of the daily bacterial P requirement.
Although further studies are needed to clarify the ecological role of extracellular DNA, our results suggest that extracellular DNA degradation may play an important role in P cycling in marine ecosystems.
Acknowledgments
This work was carried out in the frame of the program ADIOS (Atmospheric Deposition and Impact of pollutants, key elements and nutrients on the Open Mediterranean Sea), financially supported by EC funding under contract no. EVK3-CT-2000-00035.
We thank E. Manini and G. M. Luna for their help during sampling activities and S. Bompadre for his precious assistance during high-pressure liquid chromatography analyses.
REFERENCES
- 1.Blackburn, N., U. L. Zweife, and A. Hagstrom. 1996. Cycling of marine dissolved organic matter. II. A model analysis. Aquat. Microb. Ecol. 11:79-90. [Google Scholar]
- 2.Blum, S. A. E., M. G. Lorenz, and W. Wackernagel. 1997. Mechanism of retarded degradation and prokaryotic origin of DNases in non-sterile soils. Syst. Appl. Microbiol. 20:513-521. [Google Scholar]
- 3.Cazenave, C., J.-J. Toulmè, and C. Helene. 1983. Binding of recA protein to single-stranded nucleic acids: spectroscopic studies using fluorescent polynucleotides. EMBO J. 2:2247-2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Danovaro, R., A. Dell'Anno, A. Pusceddu, and M. Fabiano. 1999. Nucleic acid concentrations (DNA, RNA) in the continental and deep-sea sediments of the eastern Mediterranean: relationships with seasonal varying organic inputs and bacterial dynamics. Deep-Sea Res. Part I 46:1077-1094. [Google Scholar]
- 5.Danovaro, R., M. Manini, and A. Dell'Anno. 2002. Higher abundance of bacteria than of viruses in deep Mediterranean sediments. Appl. Environ. Microbiol. 68:1468-1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davison, J. 1999. Genetic exchange between bacteria in the environment. Plasmid 42:73-91. [DOI] [PubMed] [Google Scholar]
- 7.Dell'Anno, A., M. Fabiano, G. C. A. Duineveld, A. Kok, and R. Danovaro. 1998. Nucleic acid (DNA, RNA) quantification and RNA/DNA ratio determination in marine sediments: comparison of spectrophotometric, fluorometric, and high-performance liquid chromatography methods and estimation of detrital DNA. Appl. Environ. Microbiol. 64:3238-3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dell'Anno, A., M. L. Mei, and R. Danovaro. 1999. Pelagic-benthic coupling of nucleic acids in an abyssal location of the northeastern Atlantic Ocean. Appl. Environ. Microbiol. 65:4451-4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dell'Anno, A., S. Bompadre, and R. Danovaro. 2002. Quantification, base composition and fate of extracellular DNA in marine sediments. Limnol. Oceanogr. 47:899-905. [Google Scholar]
- 10.Lorenz, M. G., and W. Wackernagel. 1994. Bacterial gene transfer by natural transformation in the environment. Microbiol. Rev. 58:563-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Novitsky, J. A. 1986. Degradation of dead microbial biomass in a marine sediment. Appl. Environ. Microbiol. 52:504-509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paul, J. H., W. H. Jeffrey, and M. F. DeFlaun. 1987. Dynamics of extracellular DNA in the marine environment. Appl. Environ. Microbiol. 53:170-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paul, J. H., A. W. David, M. F. DeFlaun, and L. H. Cazares. 1989. Turnover of extracellular DNA in eutrophic and oligotrophic freshwater environments of southwest Florida. Appl. Environ. Microbiol. 55:1823-1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Romanowski, G., M. G. Lorenz, and W. Wackernagel. 1991. Adsorption of plasmid DNA to mineral surfaces and protection against DNase I. Appl. Environ. Microbiol. 57:1057-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Siuda, W., and R. J. Chrost. 2000. Concentration and susceptibility of dissolved DNA for enzyme degradation in lake water: some methodological remarks. Aquat. Microb. Ecol. 21:195-201. [Google Scholar]
- 16.Takahashi, M., and C. Ling. 1991. Use of a fluorescent DNA analog for fluorometric detection of DNase activity. Anal. Biochem. 198:246-249. [DOI] [PubMed] [Google Scholar]
- 17.van Duyl, F. C., and A. J. Kop. 1994. Bacterial variation in North Sea sediments: clues to seasonal and spatial variations. Mar. Biol. 120:323-337. [Google Scholar]
- 18.Whitman, W. B., D. C. Coleman, and W. J. Wiebe. 1998. Prokaryotes: the unseen majority. Proc. Natl. Acad. Sci. USA 95:6578-6583. [DOI] [PMC free article] [PubMed] [Google Scholar]