Abstract
The effect of tylosin on erythromycin-resistant enterococci was examined on three farms; farm A used tylosin for growth promotion, farm B used tylosin for treatment of disease, and farm C did not use tylosin for either growth promotion or disease treatment. A total of 1,187 enterococci were isolated from gestation, farrowing, suckling, nursery, and finishing swine from the farms. From a subset of those isolates (n = 662), 59% (124 out of 208), 28% (80 out of 281), and 2% (4 out of 170) were resistant to erythromycin (MIC ≥ 8 μg/ml) from farms A, B, and C, respectively. PCR analysis and Southern blotting revealed that 95% (65 out of 68) of isolates chosen from all three farms for further study were positive for ermB, but all were negative for ermA and ermC. By using Southern blotting, ermB was localized to the chromosome in 56 of the isolates while 9 isolates from farms A and B contained ermB on two similar-sized plasmid bands (12 to 16 kb). Pulsed-field gel electrophoresis revealed that the isolates were genetically diverse and represented a heterogeneous population of enterococci. This study suggests that although there was resistance to a greater number of enterococcal isolates on a farm where tylosin was used as a growth promotant, resistant enterococci also existed on a farm where no antimicrobial agents were used.
Emerging problems in antimicrobial resistance include the possibility of transfer of resistant genes from bacterial isolates recovered from animals to bacterial isolates harbored by humans; the proposed increase in resistance is thought to be a result of antimicrobial use in animals (14). Enterococci have been recognized not only as one of the primary causes of nosocomial infections but also as a reservoir of antimicrobial resistance genes (10, 13). Although macrolides are not used to treat enterococcal infections, they are used to treat other bacterial diseases in humans and serve as second-line treatments in cases of allergic reactions to other antimicrobials (11). In swine, the macrolide tylosin is used for a number of purposes. Tylosin phosphate or tylosin injections are administered to treat bacterial infections, including swine arthritis, ileitis, erysipelas, and, most important, swine dysentery (16). Although discontinued as a growth promotant by the European Union in 1999, tylosin is used in the United States in medicated feed to improve feed efficiency and to increase weight gain in swine (3).
Resistance to macrolides in human clinical enterococci and enterococci from animal sources in Europe has been well documented (2, 3, 11). Acquired resistance to the macrolides can be due to alteration of the antimicrobial, pumping of the antimicrobial from the cell, or modification of the target. Of these mechanisms, the most widespread appears to be target modification mediated by erythromycin resistance methylase (erm) genes, primarily ermB (11). The ermB gene confers cross-resistance to macrolide, lincosamide, and streptogramin type B antimicrobials, the MLSB phenotype (17). Resistance among enterococci to macrolides in swine and cross-resistance to erythromycin are thought to be due to tylosin use in this group of animals.
While European studies have investigated the effects of growth promoters such as tylosin on resistance in swine, few such studies have been conducted in the United States (1, 5, 6). Even fewer of those reports compared enterococci recovered from animals where the animals had not been fed or administered growth promotant to those that were fed antimicrobials for growth promotion, disease prevention, or disease treatment. In this study, erythromycin-resistant enterococci isolated from swine from a farm where tylosin was not used were compared to resistant enterococci isolated from swine from two farms where tylosin was used for either growth promotion or prevention of disease. Enterococci were collected from five stages of swine production, including gestation, farrowing, suckling, nursery, and finishing in order to determine enterococci resistance for animals prior to, during, and subsequent to tylosin use.
(This study was presented in part at the 16th Congress of the International Pig Veterinary Society, Ames, Iowa, 2 to 5 June 2002.)
MATERIALS AND METHODS
Study design.
Enterococci used in this study represented a subset of enterococcal isolates collected from an on-farm epidemiologic investigation conducted during 1999 to 2000 (8). Enterococci were isolated from swine fecal samples collected on farm from three geographically separate locations within the United States. Each farm was visited a total of five times, with 150 fecal samples collected per visit for farms A and B and 100 fecal samples per visit for farm C. Thirty (farms A and B) or 20 (farm C) fecal samples were collected from each stage of production, including gestation, farrowing, suckling, nursery, and finishing. On farm A, finisher swine were fed diets of Tylan 10 (tylosin phosphate) at 10 g/ton, and on farm B Tylan 10 was used for 5 days or less as pigs entered the nursery to reduce the incidence of scours; no tylosin was used on farm C. Use of streptogramin or lincosamide antimicrobials on the farms was not reported.
Bacterial isolation.
One gram of fecal sample was diluted 1:10 in phosphate-buffered saline (pH 7.4) and vortexed, and 100 μl of solution was inoculated onto BBL Enterococcosel agar (Becton Dickinson, Sparks, Md.) and incubated for 24 h at 37°C. Presumptive enterococcal isolates were subcultured onto Trypticase soy agar containing 5% defibrinated sheep blood. Enterococcal species identification was performed twice for each isolate with the BBL Crystal kit (Becton Dickinson) according to the manufacturer's instructions.
Antimicrobial susceptibility testing.
MICs for enterococci were determined by broth microdilution using the Sensititre automated antimicrobial susceptibility system (Trek Diagnostic Systems Limited, Westlake, Ohio) according to the manufacturer's directions. A customized 96-well panel of antimicrobials for the National Antimicrobial Resistance Monitoring System program was used. Results were interpreted according to NCCLS guidelines (15). Erythromycin resistance was defined as MIC ≥ 8 μg/ml. Enterococcus faecalis ATCC 29212 and ATCC 51299 were the qualitative control strains for determination of erythromycin MIC.
PCR.
Template for PCR was prepared by suspending a single bacterial colony in 100 μl of sterile deionized water. Five microliters of template was used in amplification reactions with primers for ermA, ermB, or ermC as previously described (19). Positive controls for PCR were Staphylococcus aureus RN1389 (ermA), Streptococcus pyogenes AC1 (ermB), and S. aureus RN4220 (ermC). Probes for Southern hybridization were generated by replacing standard deoxynucleoside triphosphates with digoxigenin (DIG)-labeled deoxynucleoside triphosphates (Roche, Indianapolis, Ind.) in the amplification reaction mixture according to the manufacturer's instructions. DNA molecular size marker XVII (500 bp; Roche) was used as the standard.
Plasmid isolation.
Plasmids were extracted with the QIAprep Spin Miniprep kit (QIAGEN, Valencia, Calif.) according to the manufacturer's instructions with the following modifications. Pelleted bacterial cells were preincubated in buffer P1 containing 20 mg of lysozyme (Sigma, St. Louis, Mo.)/ml for 15 min at 37°C. One hundred microliters of proteinase K (5 mg/ml; Roche) was then added, and the mixture was incubated an additional 15 min at 50°C. Subsequent steps were conducted per the manufacturer's directions. Plasmids (20 μl) were separated on a 0.8% 1× Tris-acetate-EDTA agarose gel for 2 h at 80 V and were visualized by ethidium bromide staining. A supercoiled DNA ladder (Invitrogen, Carlsbad, Calif.) and DIG-labeled HindIII-cleaved lambda DNA (Roche) were used as molecular weight markers.
PFGE, Southern hybridization, and cluster analysis.
Pulsed-field gel electrophoresis (PFGE) using SmaI-digested DNA was performed as previously described (21). Megabase DNA from PFGE and plasmid extractions were transferred to nylon as previously described and were probed with DIG-labeled PCR products for ermA, ermB, and ermC (26). Saccharomyces cerevisiae chromosomes (BioWhittaker, Rockland, Maine) were used as molecular standards for PFGE. Cluster analysis was determined with Bionumerics software (Applied Maths, Sint-Martens-Latem, Belgium) with Dice coefficient and the unweighted pair group method. Optimization settings for dendrograms were 4% with a band tolerance of 2 to 5%.
RESULTS
Bacterial isolates and antimicrobial susceptibility.
A total of 1,187 enterococcal isolates were recovered during the course of the study. Four hundred twenty-three (56.4%), 534 (71.2%), and 230 (46.0%) enterococci were isolated from fecal samples from farms A, B, and C, respectively. Because of the large number of enterococci in the study, samples positive for enterococci were chosen from the first two collection periods for each farm (n = 211, 281, and 170 for farms A, B, and C, respectively) (Table 1). These isolates were representative of the overall population of enterococci for farm and source. From those samples, eight species of enterococci (E. avium, E. casseliflavus, E. durans, E. gallinarum, E. hirae, E. faecalis, E. faecium, and E. solitarius) were isolated, with E. durans (n = 260), E. faecalis (n = 136), and E. faecium (n = 101) identified most frequently. Two hundred eight isolates (31.4%) were resistant to erythromycin (MIC ≥ 8 μg/ml). Approximately 59% (124 out of 208) of the isolates from farm A, which used tylosin for growth promotion, were resistant to erythromycin, while 28.5% (80 out of 208) of the isolates from farm B, which used tylosin for disease prevention, were found to be resistant to erythromycin (Table 2). On farm C, where no tylosin was used, 2.4% (4 out of 208) of the isolates were erythromycin resistant. Although second in total number isolated, more E. faecalis isolates (85 out of 136; 63%) were resistant to erythromycin than any other species when data for all three farms were combined. E. durans (n = 67), however, was the predominant species detected on farm A (Table 2).
TABLE 1.
Species distribution of enterococci isolated from swine
| Farm (n) | No. of isolates (%)
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| E. avium | E. casseliflavus | E. durans | E. gallinarum | E. hirae | E. faecalis | E. faecium | E. solitarius | Enterococcus species | |
| A (211) | 5 (2.4) | 8 (3.8) | 91 (43.1) | 0 (0) | 15 (7.1) | 59 (28.0) | 29 (13.7) | 1 (0.5) | 3 (1.4) |
| B (281) | 5 (1.8) | 3 (1.1) | 126 (44.8) | 1 (0.4) | 21 (7.5) | 70 (24.9) | 37 (13.2) | 0 (0) | 18 (6.4) |
| C (170) | 0 (0) | 5 (2.9) | 43 (25.3) | 3 (1.8) | 14 (8.2) | 7 (4.1) | 35 (20.6) | 0 (0) | 63 (37.1) |
TABLE 2.
Species distribution of erythromycin-resistant enterococci isolated from swine
| Farm (n) | No. of erythromycin-resistant isolates (%)a
|
||||||
|---|---|---|---|---|---|---|---|
| E. avium | E. casseliflavus | E. durans | E. hirae | E. faecalis | E. faecium | Enterococcus species | |
| A (124) | 3 (2.4) | 4 (3.2) | 67 (54.0) | 7 (5.6) | 32 (25.8) | 9 (7.3) | 2 (1.6) |
| B (80) | 0 (0) | 1 (1.3) | 7 (8.8) | 1 (1.3) | 52 (65.0) | 13 (16.3) | 6 (7.5) |
| C (4) | 0 (0) | 1 (25.0) | 0 (0) | 0 (0) | 1 (25.0) | 2 (50.0) | 0 (0) |
Percent erythromycin-resistant isolates by species resistance was calculated by dividing the number of erythromycin-resistant isolates per species by the total number of erythromycin isolates per farm.
Sixty-five erythromycin-resistant isolates were recovered from all three farms during the same month. These isolates and the remaining three resistant isolates from farm C (C2, C12, and C59) were selected for further analysis (Table 3). The additional three erythromycin-resistant isolates from farm C were included because of the low number of erythromycin-resistant isolates recovered there and for comparison to other isolates in the analysis. Only four species of enterococci (E. casseliflavus, E. durans, E. faecalis, and E. faecium) were recovered from these samples; E. faecalis and E. durans predominated. Farm A contained almost equal numbers of resistant isolates from farrowing and nursery swine, with lower numbers of isolates from gestation, suckling, and finishing swine (Table 3). Likewise, on farm B, where tylosin was only used in the nursery, the highest number of resistant isolates was from suckling swine. From the 68 selected erythromycin-resistant isolates, the majority were isolated from farrowing pigs (n = 22), including three resistant isolates from farm C (Table 3). In addition, regardless of farm, erythromycin resistance appeared to be low in the gestational sows and then increased in farrowing sows as well as suckling and nursery piglets, and then erythromycin resistance decreased in swine in the finisher stage.
TABLE 3.
Erythromycin-resistant enterococci by swine production stage
| Species isolated (n) | No. of erythromycin-resistant isolates (%) by stage and farm
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gestation
|
Farrowing
|
Suckling
|
Nursery
|
Finishing
|
|||||||||||
| A | B | C | A | B | C | A | B | C | A | B | C | A | B | C | |
| E. casseliflavus (2) | 1 (50.0) | 1 (50.0) | 0 (0) | 0 (0) | 0 (0) | ||||||||||
| E. durans (25) | 1 (4.0) | 1 (4.0) | 2 (8.0) | 1 (4.0) | 0 (0) | 13 (52.0) | 0 (0) | 7 (28.0) | 0 (0) | ||||||
| E. faecalis (30) | 3 (10.0) | 1 (3.3) | 10 (33.3) | 3 (10.0) | 1 (3.3) | 2 (6.7) | 8 (26.7) | 0 (0) | 0 (0) | 2 (6.7) | 0 (0) | ||||
| E. faecium (11) | 1 (9.1) | 1 (9.1) | 4 (36.4) | 1 (9.1) | 1 (9.1) | 2 (18.2) | 0 (0) | 1 (11.1) | 0 (0) | 0 (0) | |||||
Percent erythromycin-resistant isolates by species resistance was calculated by dividing the number or erythromycin resistant-isolates per species by the total number of erythromycin-resistant isolates per farm.
Detection of ermB by PCR and Southern analysis.
All 68 erythromycin-resistant isolates were tested by PCR for the presence of ermA, ermB, and ermC. With the exception of three isolates from farm C, a 639-bp product was detected in all isolates by using primers to ermB. No products were detected in any of the isolates when primers directed against ermA or ermC were used (data not shown). In order to ensure that sequence variations in ermA or ermC were not the cause of negative PCR results, probes to those genes were used in Southern hybridizations using total genomic DNA from PFGE. No hybridization products were detected with either ermA or ermC, indicating that the strains did not contain those two genes.
In order to determine if ermB was located on the chromosome or plasmids in the isolates, total genomic DNA and plasmids from the strains were probed with the 639-bp labeled portion of ermB by using Southern blotting. The ermB probe hybridized to SmaI fragments ranging in size from 90 to 225 kb in 56 of the isolates; 12 isolates (3 isolates were ermB negative by PCR from farm C) of the isolates were negative when probed with ermB. When plasmids from all strains were probed with ermB, ermB hybridized only to plasmid DNA from the nine previously negative isolates from farms A and B, indicating that the gene was located on a plasmid in those strains. Plasmid DNA from the three ermB-negative strains from farm C was not obtained. Multiple plasmids of various sizes were isolated from the remaining strains (data not shown). The ermB fragment hybridized to two plasmids from each of seven isolates from farm A and two isolates from farm B (data not shown). Both plasmids from all isolates were similar in size, the smaller plasmid ranging in size from 12 to 14 kb, while the larger plasmid was between 14 and 16 kb. Neither erythromycin-susceptible isolates nor isolates with ermB located on the chromosome contained ermB-positive plasmids.
Genetic relatedness of erythromycin-resistant isolates.
Erythromycin-resistant isolates were subjected to PFGE and were examined on the basis of farm, species, and stage of production in order to determine the genetic relationship between the isolates (Fig. 1). Date of isolation was not considered a factor because the majority of isolates were recovered from fecal samples during the same month.
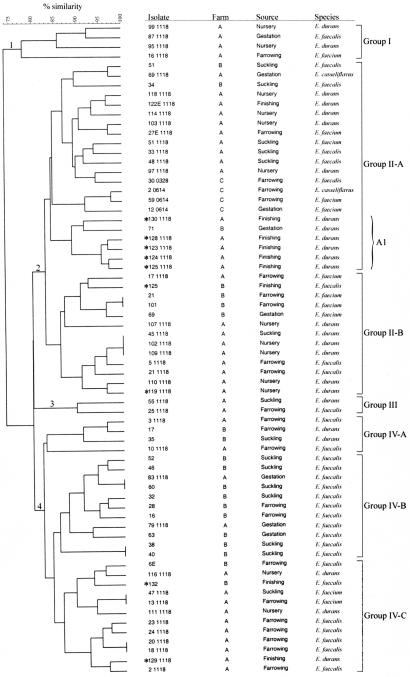
FIG. 1.
Phylogenetic tree of erythromycin-resistant enterococci from swine. DNA for PFGE was digested with SmaI. Farm, species, and source for each isolate are shown. One cluster of >75% similarity and six clusters of ≥80% similarity were identified. Levels of similarity were determined by using Dice coefficient and the unweighted pair group method. Asterisks indicate isolates that contain ermB-positive plasmids.
With the exception of Group I, which had 77% similarity to other clusters, grouping of the isolates based upon at least 80% similarity resulted in three distinct branches (groups II, III, and IV) (Fig. 1). Groups II and IV were further subdivided into clusters of Group II-A and -B and Group IV-A, -B, and -C. Groups I and III were very small, including only four and two isolates, respectively. The largest cluster was Group II-A, consisting of 22 (32%) of the isolates. A minicluster (A1) within Group II-A contained five of the nine isolates in which ermB was located on a plasmid instead of the chromosome. The six isolates in the minicluster were 90% similar to each other, and five shared several characteristics, including originating from farm A, being identified as E. durans, and being isolated from finishing swine. Even though the only other isolate in the minicluster (isolate 71) shared a similar PFGE pattern with isolate 130-1118 and was also identified as E. durans, isolate 71 was from farm B, contained ermB on the chromosome, and was from a gestation stage swine. The remaining four isolates with plasmid-localized ermB were in Group II-B and Group IV-B (Fig. 1). All four isolates from farm C clustered in Group II-A; three clustered on the same minor branch. Although isolate 30-0328 was also in Group II-A, it appeared to be more closely related to isolates from farm A than from farm C. Interestingly, this was the only ermB-positive isolate from farm C.
Weak associations between genetic type and farm or species were observed, but no definite association between genetic type and farm, species, or source could be discerned. For example, a majority of isolates in Group IV-B were from farm B and all were E. faecalis, but isolate 60 shared an identical PFGE pattern with isolate 83-1118, which was from farm A (Fig. 1). Isolates from farm A clustered primarily in groups II-A and IV-C, while isolates from farm B clustered mainly in Group IV-B. Regarding species, E. faecalis was the only species identified in Group IV-B and E. durans was the predominant species in Group II-A. Stage of production of the isolate appeared to have the weakest association with genotype, as an almost even distribution from the stages was seen throughout the seven groups (Fig. 1).
DISCUSSION
Few studies have been conducted that examine the effects of the use of antimicrobials on bacterial resistance in the natural farm setting. In this study, the effect of tylosin use and erythromycin resistance of enterococci from swine on farms was investigated. As susceptibility breakpoints have not yet been established for growth-promoting antimicrobials, other means of providing meaningful data when studying these drugs must be utilized. Two such ways include indication of a breakpoint for these antimicrobials and susceptibility testing of an antimicrobial in the same class for which a breakpoint has been established. Because cross-resistance between tylosin and erythromycin exists as well as a susceptibility breakpoint for erythromycin, the breakpoint for erythromycin (≥8 μg/ml) was used to determine resistance in this study (17).
From the fecal samples tested, eight enterococcal species were identified. However, only five species were previously identified in other studies involving swine fecal samples (E. cecorum, E. faecalis, E. faecium, E. hirae, and E. malodoratus) (7, 20). The predominant enterococcal species from this study were E. durans (39.2%), E. faecalis (20.5%), and E. faecium (15.3%), whereas in other studies the predominant species were E. faecium (50%), E. cecorum (28%), and E. hirae (11%) (7, 20). The differences in species could be due to several factors, including method of identifying species, the stage of production from which the sample was taken, and the geographical location of the farm. Because samples for this investigation were taken from swine during different stages of production, they should be representative of species that may be found in the overall population of swine and not only from species found in a geographically limited sample of animals.
Conflicting reports on the consequences of tylosin use in swine exist (1, 2, 6). Some reports indicated that resistant enterococci were selected for by tylosin use, while other reports concluded that there was no difference in resistant enterococci from animals for which tylosin was used versus those for which it was not used. In this study, resistance was more frequently observed on farm A, where swine were administered feed containing tylosin, than on farm B, where swine were injected with tylosin for a short period of time. It is possible that susceptible enterococci in nursery pigs on farm B could have been eliminated by the levels of tylosin used. Higher levels of resistance in gestation, farrowing, and suckling pigs than in the nursery pigs support this observation. Because low, nonlethal levels of some macrolides can cause expression of MLSB genes via induction, it is also possible that pigs on farm A were exposed to low levels of tylosin, resulting in induction of ermB and insufficient elimination of resistant enterococci (22, 24). This phenomenon has been described in many studies of the upstream region of erm genes.
Although no antimicrobials were used on farm C, four isolates were resistant to erythromycin. Persistence of antimicrobial resistance genes in the absence of usage of a specific antimicrobial has been attributed to genetic linkage of resistance genes. For example, linkage of genes conferring resistance to macrolides, streptogramins, and glycopeptides has been previously described (9, 12, 25). A decrease in resistance to one antimicrobial could not be effectively achieved without a decrease in the use of another because of the linkage of resistance genes. To our knowledge, no other antimicrobial was used on farm C that could account for erythromycin resistance.
Erythromycin resistance in European countries has ranged from 14 to 82% for E. faecium and 86 to 94% for E. faecalis (2-4). In Denmark, a decrease in resistance to 46.7% for E. faecium and 28.1% for E. faecalis from approximately 90% was observed after tylosin usage was reduced. Although sample size may account for differences in the studies, in this study resistance was remarkably lower for both E. faecium (3.6%) and E. faecalis (12.7%). When only resistant isolates were calculated, resistance in E. faecium (11.5%) and E. faecalis (40.9%) still remained lower than that reported for European isolates.
Not unexpected was the detection of ermB in 96% (65 out of 68) of the resistant isolates. This gene is one of the most common macrolide resistance genes found in both animal and human isolates resistant to erythromycin (11, 17). It not only mediates resistance to erythromycin but also confers resistance to more active macrolides used in human medicine (23). While often located on plasmids, the majority of ermB sequences detected in isolates in this study were on the chromosome. The ermB gene was found on plasmids in only nine of the isolates examined, suggesting that dissemination of the gene via plasmids was not likely. This does not discount the presence of transposable elements, such as Tn917, that could disseminate ermB among the isolates, but it suggests the probability is low (18). Although the plasmids have not been fully characterized, the presence of similar-sized plasmids from two separate farms does suggest that the plasmids may be the same but have yet to be widely distributed to other enterococcal isolates. In addition, although two plasmid bands were present, further work must be performed to determine if there are two distinct plasmids or different forms of the same element. The three isolates from farm C will be studied further in order to elucidate the mechanism of erythromycin resistance in those strains.
PFGE analysis did not reveal definitive factors influencing genetic relatedness. Isolates from all farms appeared to represent a heterogeneous population without regard to farm, species, or stage of pig maturation. Farms used for this study were geographically separated, thereby decreasing the chance of cross-contamination between farms. This does not exclude the possibility that farms may obtain pigs from the same source. It is more probable that swine contain diverse populations of enterococci, as several species can easily be recovered from swine in the same stage of maturation. Population changes may occur, and some species of enterococci may be better able to thrive in the swine population than others. These are areas that need to be further investigated in order to understand the population dynamics of enterococci in this group of animals.
REFERENCES
- 1.Aarestrup, F. M., and B. Carstensen. 1998. Effect of tylosin used as a growth promoter on the occurrence of macrolide-resistant enterococci and staphylococci in pigs. Microb. Drug Resist. 4:307-312. [DOI] [PubMed] [Google Scholar]
- 2.Aarestrup, F. M., H. Kruse, E. Tast, A. M. Hammerum, and L. B. Jensen. 2000. Associations between the use of antimicrobial agents for growth promotion and the occurrence of resistance among Enterococcus faecium from broilers and pigs in Denmark, Finland, and Norway. Microb. Drug Resist. 6:63-70. [DOI] [PubMed] [Google Scholar]
- 3.Aarestrup, F. M., A. M. Seyfarth, H. D. Emborg, K. Pedersen, R. S. Hendriksen, and F. Bager. 2001. Effect of abolishment of the use of antimicrobial agents for growth promotion on occurrence of antimicrobial resistance in fecal enterococci from food animals in Denmark. Antimicrob. Agents Chemother. 45:2054-2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Butaye, P., L. A. Devriese, and F. Haesebrouck. 2001. Differences in antibiotic resistance patterns of Enterococcus faecalis and Enterococcus faecium strains isolated from farm and pet animals. Antimicrob. Agents Chemother. 45:1374-1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Christie, P. J., J. N. Davidson, R. P. Novick, and G. M. Dunny. 1983. Effects of tylosin feeding on the antibiotic resistance of selected gram-positive bacteria in pigs. Am. J. Vet. Res. 44:126-128. [PubMed] [Google Scholar]
- 6.Davies, R., and T. A. Roberts. 1999. Antimicrobial susceptibility of enterococci recovered from commercial swine carcasses: effect of feed additives. Lett. Appl. Microbiol. 29:327-333. [DOI] [PubMed] [Google Scholar]
- 7.Devriese, L. A., J. Hommez, B. Pot, and F. Haesebrouck. 1994. Identification and composition of the streptococcal and enterococcal flora of tonsils, intestines and faeces of pigs. J. Appl. Bacteriol. 77:31-36. [DOI] [PubMed] [Google Scholar]
- 8.Fedorka-Cray, P. J., M. D. Englen, J. T. Gray, C. R. Hudson, and M. L. Headrick. 2002. Programs for monitoring antimicrobial resistance. Animal Biotechnol. 13:43-55. [DOI] [PubMed] [Google Scholar]
- 9.Hammerum, A. M., S. E. Flannagan, D. B. Clewell, and L. B. Jensen. 2001. Indication of transposition of a mobile DNA element containing the vat(D) and erm(B) genes in Enterococcus faecium. Antimicrob. Agents Chemother. 45:3223-3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huycke, M. M., D. F. Sahm, and M. S. Gilmore. 1998. Multiple-drug resistant enterococci: the nature of the problem and an agenda for the future. Emerg. Infect. Dis. 4:239-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jensen, L. B., N. Frimodt-Moller, and F. M. Aarestrup. 1999. Presence of erm gene classes in gram-positive bacteria of animal and human origin in Denmark. FEMS Microbiol. Lett. 170:151-158. [DOI] [PubMed] [Google Scholar]
- 12.Jensen, L. B., A. M. Hammerum, and F. M. Aarestrup. 2000. Linkage of vat(E) and erm(B) in streptogramin-resistant Enterococcus faecium isolates from Europe. Antimicrob. Agents Chemother. 44:2231-2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Murray, B. E. 1990. The life and times of the Enterococcus. Clin. Microbiol. Rev. 3:46-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murray, B. E. 1998. Diversity among multidrug-resistant enterococci. Emerg. Infect. Dis. 4:37-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.National Committee for Clinical Laboratory Standards. 2001. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 4th ed. Approved standard M7-A4. National Committee for Clinical Laboratory Standards, Villanova, Pa.
- 16.Prescott, J. F. 2000. Lincosamides, macrolides, and pleuromutilins, p. 229-262. In J. F. Prescott, J. D. Baggott, and R. D. Walker (ed.), Antimicrobial therapy in veterinary medicine. Iowa State University Press, Ames, Iowa.
- 17.Roberts, M. C., J. Sutcliffe, P. Courvalin, L. B. Jensen, J. Rood, and H. Seppala. 1999. Nomenclature for macrolide and macrolide-lincosamide-streptogramin B resistance determinants. Antimicrob. Agents Chemother. 43:2823-2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rollins, L. D., L. N. Lee, and D. J. LeBlanc. 1985. Evidence for a disseminated erythromycin resistance determinant mediated by Tn917-like sequences among group D streptococci isolated from pigs, chickens, and humans. Antimicrob. Agents Chemother. 27:439-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sutcliffe, J., T. Grebe, A. Tait-Kamradt, and L. Wondrack. 1996. Detection of erythromycin-resistant determinants by PCR. Antimicrob. Agents Chemother. 40:2562-2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thal, L. A., J. W. Chow, R. Mahayni, H. Bonilla, M. B. Perri, S. A. Donabedian, J. Silverman, S. Taber, and M. J. Zervos. 1995. Characterization of antimicrobial resistance in enterococci of animal origin. Antimicrob. Agents Chemother. 39:2112-2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Turabelidze, D., M. Kotetishvili, A. Kreger, J. G. Morris, Jr., and A. Sulakvelidze. 2000. Improved pulsed-field gel electrophoresis for typing vancomycin-resistant enterococci. J. Clin. Microbiol. 38:4242-4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weisblum, B. 1984. Inducible erythromycin resistance in bacteria. Br. Med. Bull. 40:47-53. [DOI] [PubMed] [Google Scholar]
- 23.Weisblum, B. 1995. Erythromycin resistance by ribosome modification. Antimicrob. Agents Chemother. 39:577-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weisblum, B., C. Siddhikol, C. J. Lai, and V. Demohn. 1971. Erythromycin-inducible resistance in Staphylococcus aureus: requirements for induction. J. Bacteriol. 106:835-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Werner, G., B. Hildebrandt, I. Klare, and W. Witte. 2000. Linkage of determinants for streptogramin A, macrolide-lincosamide-streptogramin B, and chloramphenicol resistance on a conjugative plasmid in Enterococcus faecium and dissemination of this cluster among streptogramin-resistant enterococci. Int. J. Med. Microbiol. 290:543-548. [DOI] [PubMed] [Google Scholar]
- 26.White, D. G., C. Hudson, J. J. Maurer, S. Ayers, S. Zhao, M. D. Lee, L. Bolton, T. Foley, and J. Sherwood. 2000. Characterization of chloramphenicol and florfenicol resistance in Escherichia coli associated with bovine diarrhea. J. Clin. Microbiol. 38:4593-4598. [DOI] [PMC free article] [PubMed] [Google Scholar]