Abstract
The Nuclear factor I (NFI) family of transcription factors regulates proliferation and differentiation throughout the developing central nervous system. In the developing telencephalon of humans and mice, reduced Nfi expression is associated with agenesis of the corpus callosum and other neurodevelopmental defects. Currently, little is known about how Nfi expression is regulated during early telencephalic development. PAX6, a transcription factor important for telencephalic development, has been proposed as an upstream regulator of Nfi expression in the neocortex. Here we demonstrate that, in the developing neocortex of mice, NFIA and NFIB are endogenously expressed in gradients with high caudo-medial to low rostro-lateral expression and are most highly expressed in the cortical plate. We found that this expression pattern deviates from that of PAX6, suggesting that PAX6 does not drive Nfi expression. This is supported by in vitro reporter assays showing that PAX6 over-expression does not regulate Nfi promoter activity. Similarly, we also found that in the Pax6 Small Eye mutant, no changes in Nfi mRNA or protein expression are observed in the neocortical ventricular zone where PAX6 and the NFIs are expressed. Together these data demonstrate that in mice, PAX6 is not a transcriptional activator of Nfi expression during neocortical development.
Transcriptional regulatory networks are fundamental to the development of distinct cellular populations throughout embryogenesis. These networks consist of interconnected and independent transcriptional regulatory cascades that are conventionally inferred from gene expression and phenotypic analyses of loss-of-function animal models. Identifying these regulatory pathways during neurodevelopment may implicate novel genes in neurodevelopmental disorders, or determine if similar disorders are the result of interconnected or distinct causes. In the developing telencephalon, the Nuclear factor I (NFI) family of transcription factors forms part of such an undefined network, with the majority of its regulatory components currently unknown. In mice the NFI transcription factors are expressed from embryonic day (E)12 within the dorsal telencephalon1, and function to regulate the proliferation and differentiation of neural progenitor cells2,3,4,5,6,7,8. Knockout mice for Nfia, Nfib or Nfix display similar forebrain phenotypes2,3,4,5,6,7,8. These mice are characterized by delayed glial differentiation, consequently resulting in malformations of the corpus callosum and hippocampus2,3,4,5,6,7,8. In humans, haploinsufficiency for NFI has been similarly associated with agenesis of the corpus callosum and other neurodevelopmental disorders9,10,11,12,13,14,15,16. Thus, precise regulation of Nfi expression is required for normal forebrain development in both humans and mice.
The transcription factor PAX6 has been implicated as a potential transcriptional activator of Nfia and Nfib expression in the developing neocortex17. PAX6 expression precedes both NFIA and NFIB, and is expressed within the proliferative ventricular zone (VZ) of the dorsal telencephalon throughout embryonic development1,18. The function of PAX6 has been examined in a number of different Pax6 loss-of-function mouse models, most notably the Pax6 Small Eye (Pax6Sey) mutant strain19,20,21. This model expresses a truncated PAX6 protein that lacks the homeobox DNA-binding domain due to a mutation that introduces a premature stop codon20. Gene expression array analyses comparing Pax6Sey/Sey mutant and wildtype neocortices have identified potential downstream candidates regulated by PAX6 during prenatal development. In the Pax6Sey/Sey neocortex, expression of both Nfia and Nfib are reduced at E12, with Nfia expression similarly reduced at E1517. As Pax6 overexpression in cultured E13 cortical cells correspondingly increased Nfia expression, Nfia appears to be a bona fide transcriptional target of PAX617. In this paper, we investigated the hypothesis that PAX6 functions as a transcriptional activator of Nfia and Nfib (hereafter referred to as Nfi) within the developing neocortex.
Results
NFI expression opposes the gradient of PAX6 expression during neocortical development
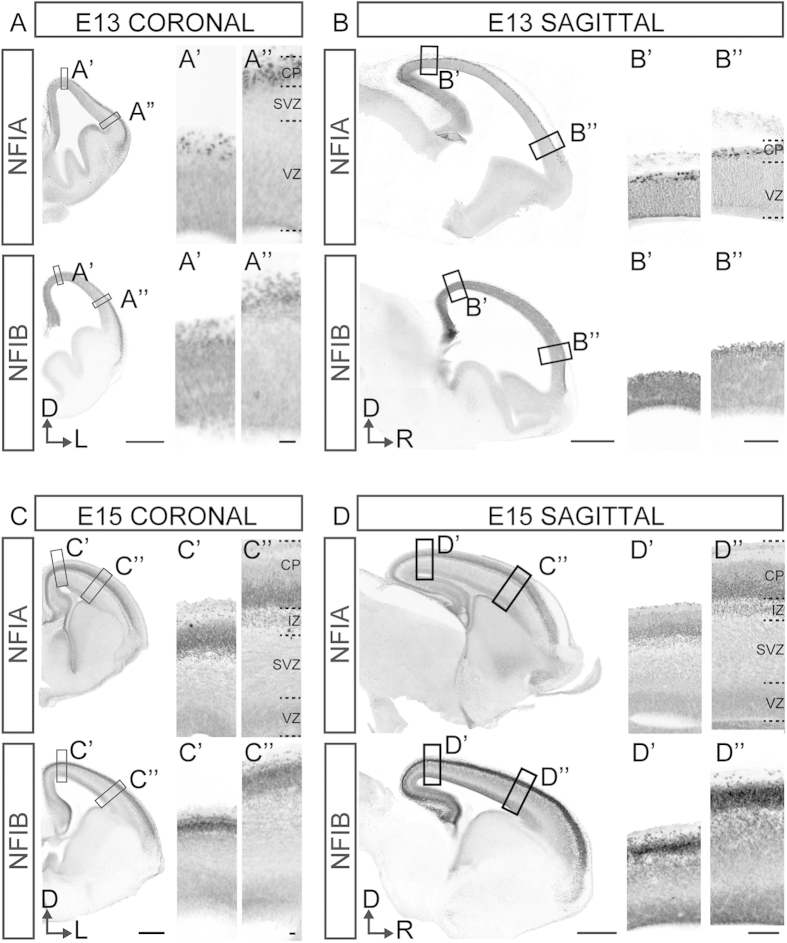
It has previously been shown that PAX6 expression in the developing neocortex is limited to the VZ, where it is expressed in a high rostro-lateral to low caudo-medial gradient22. We reasoned that if PAX6 transcriptionally activates Nfia or Nfib expression, the expression patterns of these transcription factors would resemble each other. To test this, we analyzed NFI protein expression across the neocortex of wildtype C57Bl/6J mice at E13 and E15. In contrast to PAX6 expression, we observed that in coronal sections at E13, both NFIA and NFIB are expressed in high medial to low lateral gradients (Fig. 1A). Similarly, analyses of sagittal sections at this age show that both NFIA and NFIB are expressed higher caudally and lower rostrally (Fig. 1B). At E15, the graded expression of NFIA and NFIB is sustained in the medial-to-lateral axis (Fig. 1C), but not in the rostral-to-caudal axis, where NFI protein expression was now more evenly distributed (Fig. 1D). The graded expression of Nfia and Nfib in the sagittal plane was confirmed at the mRNA level at E11.5 and E13.5 by examining in situ hybridization data available from the Allen Brain Atlas (Supplementary Figure 1)23.
Figure 1. NFIA and NFIB are expressed in a high caudo-medial to low rostro-lateral gradient in the developing neocortex.
Coronal and sagittal sections of E13 (A and B) and E15 (C and D) C57Bl/6J wildtype brains were analyzed for NFI protein expression in the medial-to-lateral and rostral-to-caudal axes respectively. NFIA and NFIB were expressed in a high medial to low lateral gradient at both ages (A and C) and in a high caudal to low rostral gradient at E13, but not at E15 (B and D). The developing cortical plate (CP) showed highest expression across the cortical laminae, with intermediate expression in the ventricular zone (VZ). For coronal sections, A’ and C’ represent medial insets and A” and C” represent lateral insets. For sagittal sections, B’ and D’ represent caudal insets and B” and D” represent rostral insets. Scale bars represent 500 μm in low-powered magnifications and 100 μm (A and C) or 20 μm (B and D) in high-powered magnification panels. SVZ = subventricular zone; IZ = intermediate zone; D = dorsal; L = lateral; R = rostral.
Similarly, we note that the NFI transcription factors were more broadly expressed across the cortical laminae as compared to PAX6 expression. In line with our previous characterization of NFI expression in the developing forebrain1, we observed that both NFIA and NFIB are most highly expressed within the cortical plate of the neocortex (Fig. 1 and S1), where PAX6 is not expressed18. Thus, in mice, the graded expression pattern of NFIA and NFIB across the wildtype neocortex does not resemble that of PAX6 expression.
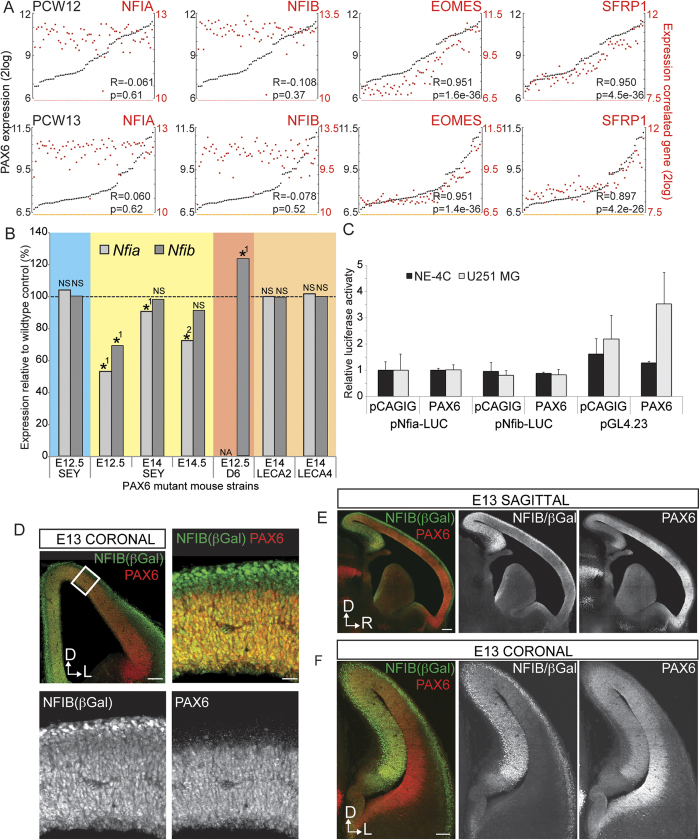
To determine whether NFI expression correlates with PAX6 expression during human neocortical development, we analyzed publically available mRNA expression data obtained from neocortical regions at 12 and 13 weeks post-conception24 (Fig. 2A), which correspond to our E13 and E15 analyses in mice25. At both ages, NFIA and NFIB expression levels in the neocortical areas showed no significant correlation with PAX6 expression levels in the neocortex (Fig. 2A), whereas SFRP1 and EOMES showed significant correlation as reported previously in mice17,26,27. Thus, endogenous expression of NFIA and NFIB do not appear to be positively correlated with PAX6 expression in the developing neocortex of both mice and humans.
Figure 2. NFI and PAX6 expression are not correlated in the developing neocortex and PAX6 does not activate Nfi promoter activity in vitro.
(A). Correlation of PAX6 mRNA expression (black) with NFIA, NFIB, EOMES or SFRP1 mRNA expression (red) at 12 (upper panels) and 13 (lower panels) weeks post-conception in the human neocortex24. (B). Relative expression of Nfia and Nfib in the Pax6 loss-of-function mouse models Pax6Sey (SEY), Pax6Leca2 (LECA2) and Pax6Leca4 (LECA4) and the transgenic PAX6 over-expression model Dach1-PAX6 (D6-PAX6) from published expression datasets27,29,30,34, normalized to wildtype control (100%; dashed line) for comparison between datasets. All datasets represent rostral cortex/neocortex, with the exception of the first set (blue) that was generated from the PAX6-positive fraction only34. Statistical significance was determined using one-way ANOVA for genotypes with n = 3 or more. NA = raw data not available from original dataset, *1 = reported significant in original dataset, *2 = p < 0.05 by one-way ANOVA, NS = non-significant (one-way ANOVA). (C). Relative luciferase activity of NE-4C and U251 MG cells transfected with the pGL4.23 luciferase construct driven by the mouse Nfia or Nfib promoter (pNfia-LUC and pNfib-LUC respectively) or its endogenous minimal promoter. PAX6 and pCAGIG denote cells co-transfected with the PAX6-over-expression or empty control vectors. Luciferase activity was determined 48 hours after transfection and normalized to Renilla luciferase that was co-trasfected as an internal control. pNfia-LUC with pCAGIG was scaled to ‘1’ and all other conditions were then normalized to this to determine relative luciferase activity. No statistical significance was observed between pCAGIG and PAX6 for all experimental conditions (n = 3, figure depicts a representative sample with three technical replicates). (D-F). Representative immunofluorescence images of coronal (D and F) and sagittal (E) sections of E13 Nfib heterozygous brains stained for PAX6 and the β-galactoside reporter knocked into the Nfib locus. The boxed region in the low-magnification panel in (D) represents subsequent panels with high-powered magnification showing PAX6 and β-galactoside co-expression. Scale bars represent 100 μm (low-powered) and 20 μm (high-powered magnification) in D, 200 μm (E) or 100 μm (F). D = dorsal; R = rostral; L = lateral.
Differential expression of Nfia and Nfib in Pax6 mouse models
We next investigated the reproducibility of reduced Nfi expression previously reported in microarray analyses of the Pax6Sey/Sey mutant17. Using validated independent primer sets for quantitative PCR28, we analyzed Nfi mRNA levels in microdissected dorsolateral cortex/neocortex of E13 Pax6Sey/Sey and wildtype embryos. We observed reduced Nfia and Nfib expression in the Pax6Sey/Sey neocortex (-3.2 and -4.3 fold, respectively when compared within litters). These reductions did not achieve statistical significance when litters were pooled due to the large variation between litters (p = 0.32 and p = 0.22, respectively; one-way ANOVA). To determine if the down-regulation of Nfi expression observed in the Pax6Sey/Sey neocortex is reproducible in microarray analyses, we analyzed independent Pax6Sey/Sey expression datasets of E12.5 neocortex27, E14 rostral cortex29 and E14.5 neocortex30 (Fig. 2B; yellow). In line with the observations of Holm et al.17, Nfia expression was significantly reduced at all ages. In contrast, Nfib expression was significantly reduced only in the E12.5 dataset.
As the reduction of Nfi expression may be specific to the Pax6Sey mouse model and its phenotype, we also analyzed available expression data from other Pax6 mutants, namely the D6-PAX627, Pax6Leca2 and Pax6Leca4 31,32 mouse mutants. A significant increase in Nfib expression was observed in E12.5 neocortex for the D6-PAX6 mouse model that over-expresses wildtype PAX6 (Fig. 2B; red, Sansom and colleagues also reported increased Nfia expression but no raw data is available to validate this)27. In the Pax6Leca2 and Pax6Leca4 mouse models, no reduction in Nfia or Nfib expression was detected in E14 rostral cortex in contrast to reduced Nfia expression in the Pax6Sey rostral cortex collected and analyzed within the same experiment (Fig. 2B; orange)29. These mice differ from the Pax6Sey mice as they both carry single missense mutations that result in the expression of functional PAX6 protein with reduced DNA binding capability31,32. Thus the mis-regulation of Nfi expression is not consistently observed in other Pax6 mouse models.
Mutation of PAX6 does not alter radial glial expression of NFI
Our analyses thus far suggest that PAX6 is unlikely to function as a direct transcriptional activator of Nfi expression in the developing neocortex. To further test this hypothesis, we used an in vitro reporter assay to determine if PAX6 over-expression regulates Nfi promoter activity. We transfected mouse Nfia or Nfib promoter-driven luciferase constructs (pNfia-LUC or pNfib-LUC), or a control reporter plasmid into the NE-4C (mouse neuroepithelial cell line) and U251 MG (human glioma cell line) cell lines. No change in Nfi promoter activities were observed when these cells were co-transfected with a PAX6 overexpression construct (pCAGIG-PAX6) as compared to an empty control vector (Fig. 2C).
Nevertheless, if PAX6 autonomously regulates Nfi expression in vivo, this regulation would likely be limited to the VZ where PAX6 and the NFIs are expressed22. To test this, we first determined whether PAX6 and NFIB are co-expressed within the same cells in the neocortical VZ. Using a knock-in reporter gene for NFIB substituted into the deleted exon of the Nfib knockout mice3,7,33, we observed co-expression of the β-galactosidase reporter gene and PAX6 in the radial glial cells of E13 heterozygous Nfib knockout mice, as has been reported at E18.5 (Fig. 2D)3,7,33. In keeping with our analyses of endogenous NFI expression in wildtype mice (Fig. 1), PAX6 expression across the neocortical VZ negatively correlated with β-galactosidase expression in Nfib heterozygous mice when observed in both the sagittal (Fig. 2E) and coronal (Fig. 2F) planes.
To investigate whether Nfi expression is specifically mis-regulated in the radial glial cells of Pax6Sey mice, we analyzed Nfi expression using previously published data obtained from flow cytometry-sorted populations of PAX6-positive cells. Price and colleagues generated this data by crossing the Pax6Sey mice to the DTy54 reporter mouse line with GFP expression in PAX6-positive cells34. Analyses of Nfia and Nfib expression in GFP-positive cells obtained from E12.5 Pax6Sey/Sey mutant and wildtype neocortices revealed no difference in Nfi expression (Fig. 2B; blue). Thus, the reduction of Nfi expression observed in microarray analyses of Pax6Sey/Sey mutants cannot be justified by the mis-regulation of Nfi expression in radial glial cells.
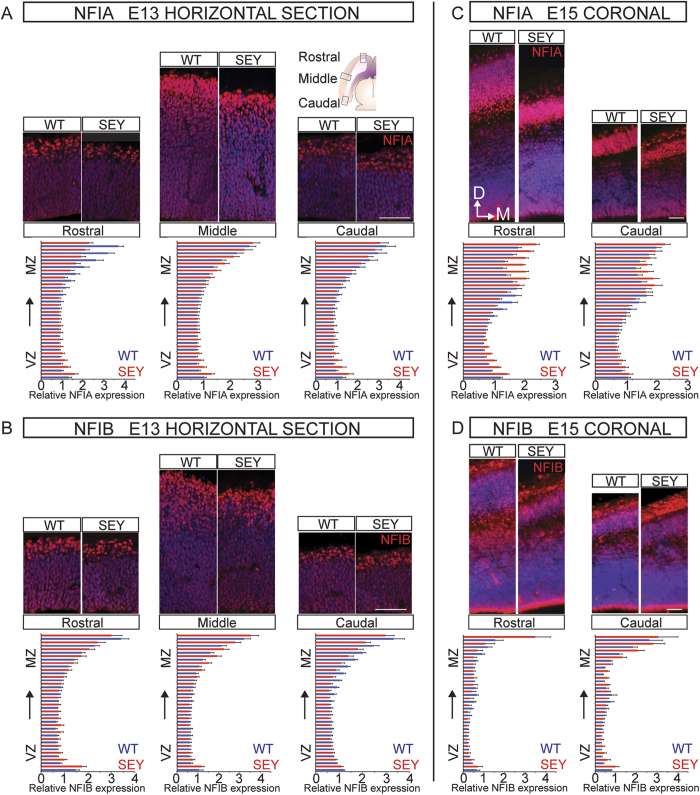
To validate this at the protein level, we quantified NFIA and NFIB protein expression in horizontal sections of E13 Pax6Sey/Sey mutant and wildtype brains (Fig. 3A,B). For each section analyzed, the neocortex from the ventricular to pial surfaces were divided into 20 equally spaced bins, with strict precautions taken to account for variability between sections (see Methods for a full description). These analyses were independently repeated at three different regions along the rostral to caudal axis to account for the graded expression observed in this axis. In all three analyses, we observed no significant reduction in NFI expression in the bins that represent the VZ where PAX6 is expressed. On the contrary, slight changes in NFIA expression were observed in the bins that corresponded to the cortical plate where PAX6 is not expressed but the NFI transcription factors are strongly expressed.
Figure 3. NFI protein expression is not reduced in the ventricular zone of E13 and E15 Pax6Sey/Sey neocortex.
Representative immunofluorescence images of rostral, middle and caudal cortical regions stained for NFIA (A; red) or NFIB (B; red) and co-stained for DAPI (blue) in E13 wildtype (WT) and Pax6Sey/Sey mutant (SEY) horizontal brain sections representing the middle of the dorsal telencephalon. Fluorescence intensity histograms represent the mean fluorescence intensity of NFIA (A) or NFIB (B) expression, divided into 20 equally spaced bins from the ventricular to pial surfaces and normalized to fluorescence intensity of DAPI within each bin (n = 6 per genotype). Mean fluorescence intensity for Pax6Sey/Sey is represented in red with wildtype in blue. NFIA and NFIB showed no significant change in expression in any of the bins that represented the ventricular zone of Pax6Sey/Sey mutants. (C D). Similar analyses were performed using E15 Pax6Sey/Sey and wildtype brains sectioned coronally, with matched rostral and caudal sections analyzed independently. Similar to E13, NFIA (C) and NFIB (D) expression were not significantly changed in bins corresponding to the ventricular zone, but NFI expression was higher in the cortical plate of the Pax6Sey/Sey mutant. Error bars represent standard error of the mean. VZ = ventricular zone, MZ = marginal zone, D = dorsal, M = medial. Scale bar represents 50 μm.
We conducted similar analyses on E15 Pax6Sey/Sey mutant brains sectioned coronally, and independently analyzed matched rostral and caudal sections. Again, no significant decrease in NFI expression was detected in bins that correspond to the VZ (Fig. 3C,D). In the bins that represented the cortical plate, we observed that peak NFIA expression shifted dorsally towards the pial surface in the Pax6Sey/Sey mutants as compared to wildtype controls. Nevertheless, as the VZ cells that express PAX6 and the NFI transcription factors showed no mis-regulation of NFI expression, PAX6 does not function to activate Nfi transcription in radial glial cells of the developing neocortex.
Discussion
The identification of transcriptional regulatory cascades is integral to determining whether similar neurodevelopmental disorders are the result of interconnected or distinct causes. In the developing telencephalon, the NFI and PAX6 transcription factors are involved in proliferation and neurogenesis2,3,4,5,6,7,8,27,29,34,35,36,37. Haploinsufficiency of these transcription factors in humans is similarly implicated with agenesis of the corpus callosum9,10,11,12,13,14,15,16,38,39. As PAX6 expression precedes NFI expression in the neocortex1,18 and deletion of Pax6 results in reduced Nfi expression in the Pax6Sey/Sey neocortex17, PAX6 was hypothesized to directly regulate Nfi expression during neocortical development.
In the current study, we present several lines of evidence that contradict this hypothesis. Whilst gene expression analyses of Pax6Sey/Sey neocortex validate the reduction of Nfi expression reported previously17,27,29,30, our immunohistochemistry analyses to investigate NFI expression, and the inconsistencies in Nfi expression observed amongst the Pax6 mouse models suggest possible discrepancies27,32. Significantly, we report from our analyses as well as the data published by Price and colleagues34 that Nfia and Nfib are not mis-regulated at the mRNA or protein levels in the Pax6Sey/Sey VZ where PAX6 may autonomously regulate these transcription factors. This suggests that, despite the reduction of Nfi expression observed in the Pax6Sey/Sey neocortex, PAX6 is not a direct transcriptional activator of Nfi expression in the developing neocortex.
Interestingly, we observed increased NFIA expression in the cortical plate of Pax6Sey/Sey mutants (Fig. 3C). As PAX6 is not expressed in the developing cortical plate, PAX6 is unlikely to transcriptionally activate Nfia expression in this region. Rather, it is likely that the previously described precocious neurogenesis in the Pax6Sey/Sey mutant results in a thinner cortical plate that is disproportionately populated by deep layer neurons35,37,40,41,42. As NFIA is more highly expressed in the deep layer neurons as compared to upper layer neurons1,3, this could account for the increased NFI protein expression observed within this region.
Our findings in this study are supported by previous independent chromatin immunoprecipitation experiments suggesting that Nfia and Nfib are not direct targets of PAX6 during neocortical development. Chromatin immunoprecipitation for PAX6 using E12.5 neocortical and E15.5 forebrain tissues does not detect binding of this transcription factor to the Nfi promoter regions27,43. Interestingly, binding of PAX6 at the Nfia and Nfib promoter regions in mice was detected for the pancreatic βT3 cells43 and the adult olfactory bulb44, respectively.
Similarly, our study is also supported by the phenotypes observed upon deletion of Nfi in mice, or NFI haploinsufficiency in humans as compared to that observed with PAX6 deficiency. For instance, PAX6 deficiency in humans and mice is associated with microcephaly45,46,47. In contrast, NFI reduction is implicated in ventriculomegaly and macrocephaly9,10,11,12. These opposite phenotypes suggest that if PAX6 were to regulate Nfi expression, it would likely function as a transcriptional repressor. However, no increase in Nfi expression was observed in the VZ of the Pax6Sey/Sey neocortex by mRNA expression (Fig. 2B) or immunohistochemistry analyses (Fig. 3). Taken together, this evidence indicates that PAX6 is not a direct regulator of Nfi expression during neocortical development.
While our study demonstrates that down-regulation of expression does not necessarily imply a direct transcriptional relationship, similar precautions are required to prevent over-interpretation of our findings. Specifically, binding of PAX6 to the Nfi promoter in pancreatic βT3 cells43 and the adult olfactory bulb44 suggest that PAX6 may regulate Nfi expression within other tissue types or perhaps even within the neocortex at ages not examined. Interestingly, we note that within our analyses of E13 Nfib heterozygous mice, high expression of both PAX6 and β-galactosidase was present within the hippocampal primordia (Fig. 2E,F). However, as our analyses were strictly restricted to the developing neocortex, whether PAX6 regulates Nfi transcription during hippocampal development necessitates further investigation.
Although we show that PAX6 is not a direct transcriptional activator of Nfi expression in the neocortex, an interesting area of future study would be to determine the mechanism underlying the reduced Nfi expression observed in the Pax6Sey/Sey neocortex. We propose an alternative hypothesis for this based on previous studies of the Pax6Sey/Sey mutant. As PAX6 is involved in establishing the boundary between the dorsal and ventral telencephalon22,48,49,50,51, and is required for normal cell cycle exit and cortical cell identity specification35,37,40, the reduced Nfi expression observed in the Pax6Sey/Sey mutant may represent a change in the cellular composition of the tissue analyzed in gene expression analyses. For example, Nfi expression may be indirectly reduced as radial glial cells that lacked functional PAX6 expression may differentiate prematurely or transition towards a ventral telencephalic progenitor cell fate22,35,37,40,48,49,50,51. This in turn results in the thinner cortical plate observed in Pax6Sey/Sey mutants35,37,40,41,42. As NFIA and NFIB are expressed in the VZ and the cortical plate of the developing neocortex, these changes in cellular composition may account for the reduced expression of these transcription factors.
To conclude, by analyzing NFI expression in the wildtype and Pax6Sey/Sey neocortices, we determined that PAX6 does not directly regulate Nfi expression during neocortical development. Our findings demonstrate the importance of careful phenotypic analyses when interpreting transcriptional regulatory cascades predicted from mixed cellular populations.
Methods
Animal breeding and tissue collection
All mice used in this study were bred at The University of Queensland with approval from the institutional Animal Ethics Committee and all experiments were performed in accordance with the approved guidelines. Nfib knockout mice33 and Pax6Sey-neu (referred to as Pax6Sey)19 (original breeding stocks were a kind gift from Professor Grant Mastick, University of Nevada, Reno) were maintained on the C57Bl/6J and 102/El x C3H/Rl x C57Bl/6J backgrounds respectively. To generate timed-pregnant females, male and female mice were placed together overnight and checked the following day for vaginal plugs. This day was designated as E0 if a vaginal plug was present. Embryos were removed from dams that were placed under anesthesia using sodium pentobarbital (Abbott Laboratories), and were either drop fixed (E13 embryos) or transcardially perfused (E15 embryos) with 0.9% (w/w) saline, followed by 4% (w/w) paraformaldehyde in 1x phosphate-buffered saline (PBS; pH 7.4) for further analyses. For mRNA expression analyses of E13 neocortical tissue, the dorsolateral cortex (excluding the medial cortex) was microdissected in ice-cold sterile PBS and immediately snap frozen for mRNA isolation.
Immunohistochemistry
Brains of paraformaldehyde-fixed embryos were removed from the skull and sectioned coronally, sagittally or horizontally at 50 μm using a vibratome. Chromogenic immunohistochemistry (IHC) or fluorescence immunohistochemistry (IF) was conducted as previously described with minor modifications1. Primary antibodies used for immunohistochemistry were as follows: rabbit anti-NFIA (1:500; ARP32714, Aviva Systems Biology (IF) or 1:10,000; HPA008884, Sigma (IHC)), rabbit anti-NFIB (1:500; ARP32716, Aviva Systems Biology or 1:10,000; HPA003956, Sigma), chicken anti-β-galactosidase (1:500; ab9361, Abcam (IF)) and rabbit anti-PAX6 (1:500; ab2237, Abcam (IF)). For IHC, a biotinylated goat anti-rabbit secondary antibody (1:500; BA-5000, Vector Laboratories) was used, followed by incubation with the VECTASTAIN elite ABC kit (Vector Laboratories) and nickel DAB staining as previously described1. For IF, Alexa Fluor 488-conjugated goat anti-chicken (1:500; A-11039, Invitrogen) and Alexa Fluor 555-conjugated goat anti-rabbit (1:500; A-11034, Invitrogen) secondary antibodies were used for detection. Immunofluorescent sections were counterstained with 4`,6-diamidino-2-phenylindole, dihydrochloride (DAPI, Invitrogen) to label cell nuclei and coverslipped using ProLong Gold anti-fade reagent (Invitrogen). To minimize variability for IF quantification, matched sections that were to be compared between Pax6Sey/Sey and wildtype littermates were mounted on the same slide to ensure identical staining conditions.
Imaging and data analysis
Brightfield and wide-field fluorescence imaging was performed with a Zeiss upright Axio-Imager Z1 microscope fitted with Axio- Cam HRc and HRm cameras and Apotome. Images were acquired with AxioVision software (Carl Zeiss) or ZEN blue (Carl Zeiss). For IF, matched sections from 6 brains were imaged per condition using identical acquisition settings to allow for quantification. Following this, matched neocortical regions of 100 μm width were cropped from each section in Photoshop (Adobe) and imported into ImageJ software (NIH). To generate fluorescence intensity histograms, cropped sections were divided into 20 bins of equal size across the ventricular to pial surfaces and fluorescence intensity corresponding to NFIA or NFIB was normalized to DAPI intensity within each bin. Statistical significance was determined using one-way ANOVA, with p-values below 0.05 considered significant. All values are presented as the mean, with error bars representing the standard error of the mean. Images in figures are representative images that have been identically cropped, enhanced for contrast and brightness, and pseudo-colored to permit overlay using Adobe Photoshop software.
Quantification of Nfi mRNA expression
Quantitative PCR was performed as previously described52. Briefly, RNA of snap-frozen E13 neocortex of Pax6Sey mutant or wildtype littermates of 3 litters was isolated using TRIzol reagent (Invitrogen) and 1 μg of RNA was reverse transcribed using SuperScript III First-Strand Synthesis SuperMix (Invitrogen) and Oligo(dT) primers (Invitrogen) as per the manufacturer’s instructions. Diluted samples were used for real-time qPCR with Platinum SYBR Green qPCR SuperMix (Invitrogen) and 0.25 μM forward and reverse primers. The qPCR reaction was carried out on a Rotor-Gene 3000 (Corbett Life Science) with the following thermocycler conditions: 10 minutes at 95 °C followed by 40 cycles with 15 seconds denaturation at 95 °C, 20 seconds annealing at 60 °C, and 30 seconds extension at 72 °C. Relative expression was determined using the ΔΔCt method with the housekeeping gene beta-2-microglobulin used as a relative standard. Statistical analyses were performed using one-way ANOVA. Error bars represent the standard error of the mean. Primers were as previously described28: B2MM 5`-agactgatacatacgcctgcag-3`, B2MMC 5`-gcaggttcaaatgaatcttcag-3`, mNFI-AE23 5`-ggcatactttgtacatgcagc-3`, mNFI-AE4C2 5`-acctgatgtgacaaagctgtcc-3`, mNFI-BE2 5`-gtttttggcatactacgtgcagg-3` and mNFI-BE3C 5`-ctctgatacattgaagactccg-3`.
Dual-luciferase reporter assays
The U251 MG human glioblastoma53 and NE-4C mouse neuroepithelial54 cell lines were obtained from American Type Culture Collection (ATCC) and cultured in DMEM medium (Invitrogen), supplemented with 10% fetal bovine serum, 100 U/mL penicillin and 100 μg/mL streptomycin (Invitrogen) at 37 °C in a humidified atmosphere containing 5% CO2. The mouse Nfia (2,329 base pairs) and Nfib (2,484 base pairs) promoters were cloned into the pGL4.23 luciferase construct (Promega) to drive firefly luciferase expression, using primers with added NheI (forward primer) and BglII (reverse primer) restrictions sites; pNfia_F 5`-ttacatgttggttccctcagc-3`, pNfia_R 5`-tgcttacctgggtgagacag-3` pNfib_F 5`-ccaggctcttggtttacagg-3`, pNfib_R 5`-tcgccttaaaacgcactttc-3’. The mouse PAX6 coding sequence was cloned into pCAGIG (Addgene) from pMXIG-Pax655, kindly provided by Professor Magdalena Gotz. Cells were seeded into a 96-well plate 24 hours prior to transfection at a density of 3,000 cells per well. pNfia-Luc, pNfib-Luc or the control pGL4.23 plasmid were co-transfected with either pCAGIG-PAX6 or pCAGIG into seeded cells using FuGENE HD (Promega). Renilla luciferase (pRL-SV40, Promega) was co-transfected with all transfections as an internal control for normalization of firefly luciferase activity. Luciferase activity was assayed 48 hours after transfection using the Dual-Luciferase Reporter Assay System (Promega). All experimental conditions were performed as three independent experiments of triplicates, of which one is represented. Statistical significance was determined using a one-way ANOVA.
Microarray expression data analysis
Expression data of Pax6Sey mutant E12.5 neocortical radial glial cells (GSE38703)34, Pax6Sey, Pax6Leca2 and Pax6Leca4 mutant E14 rostral cortex (GSE35260)29 and Pax6Sey mutant E14.5 neocortex (GSE32271)30 were obtained from the Gene Expression Omnibus database. The processed data in 2log scale were converted to absolute scale and the difference in expression of Nfia or Nfib as compared to the wildtype embryos was calculated and represented as a percentage of the expression in wildtype controls. When more than 3 samples were available, statistical significance was determined using one-way ANOVA. Expression in human cortex at 12 and 13 weeks post-conception was obtained from the GSE25219 dataset24 and analyzed and visualized using the R2: microarray analysis and visualization platform ( http://r2.amc.nl) as previously described56.
Additional Information
How to cite this article: Bunt, J. et al. PAX6 does not regulate Nfia and Nfib expression during neocortical development. Sci. Rep. 5, 10668; doi: 10.1038/srep10668 (2015).
Supplementary Material
Acknowledgments
We are grateful to Rowan Tweedale and Laura Fenlon for critical comments on the manuscript. We also thank Thomas Pollak and Gonzalo Almarza for technical assistance with immunohistochemistry and image acquisition. This work was supported by grants from the Australian Research Council (DP150104748) and the National Health and Medical Research Council (NHMRC), Australia (GNT560504) to LJR. LJR is supported by an NHMRC Principal Research Fellowship.
Footnotes
Author Contributions J.B. and L.J.R. designed the experiments. J.B., L.Z. and S.M. performed the experiments. J.B., J.W.C.L., L.Z. and S.M. analyzed the data and prepared the figures. J.B., J.W.C.L. and L.J.R. wrote the manuscript.
References
- Plachez C. et al. Nuclear factor I gene expression in the developing forebrain. J. Comp. Neurol. 508, 385–401 (2008). [DOI] [PubMed] [Google Scholar]
- Barry G. et al. Specific glial populations regulate hippocampal morphogenesis. J. Neurosci 28, 12328–12340 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betancourt J., Katzman S. & Chen B. Nuclear factor one B regulates neural stem cell differentiation and axonal projection of corticofugal neurons. J. Comp. Neurol. 522, 6–35, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell C. E. et al. The transcription factor Nfix is essential for normal brain development. BMC Dev. Biol. 8, 52, 10.1186/1471-213X-8-52 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- das Neves L. et al. Disruption of the murine nuclear factor I-A gene (Nfia) results in perinatal lethality, hydrocephalus, and agenesis of the corpus callosum. Proc. Natl. Acad Sci. 96, 11946–11951 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heng Y. H. et al. NFIX regulates neural progenitor cell differentiation during hippocampal morphogenesis. Cerebr Cortex 24, 261–79 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper M. et al. Multiple non-cell-autonomous defects underlie neocortical callosal dysgenesis in Nfib-deficient mice. Neural Dev. 4, 43 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu T., Butz K. G., Plachez C., Gronostajski R. M. & Richards L. J. Abnormal development of forebrain midline glia and commissural projections in Nfia knock-out mice. J. Neurosci 23, 203–212 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C. P. et al. Chromosome 1p32-p31 deletion syndrome: prenatal diagnosis by array comparative genomic hybridization using uncultured amniocytes and association with NFIA haploinsufficiency, ventriculomegaly, corpus callosum hypogenesis, abnormal external genitalia, and intrauterine growth restriction. Taiwan J. Obstet Gynecol 50, 345–352, (2011). [DOI] [PubMed] [Google Scholar]
- Koehler U. et al. A novel 1p31.3p32.2 deletion involving the NFIA gene detected by array CGH in a patient with macrocephaly and hypoplasia of the corpus callosum. Eur. J. Pediatr 169, 463–468 (2010). [DOI] [PubMed] [Google Scholar]
- Lu W. et al. NFIA haploinsufficiency is associated with a CNS malformation syndrome and urinary tract defects. PLoS Genet 3, e80, 10.1371/journal.pgen.0030080 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao A. et al. An intragenic deletion of the NFIA gene in a patient with a hypoplastic corpus callosum, craniofacial abnormalities and urinary tract defects. Eur. J. Med. Genet 57, 65–70 (2014). [DOI] [PubMed] [Google Scholar]
- Klaassens M. et al. Malan syndrome: Sotos-like overgrowth with de novo NFIX sequence variants and deletions in six new patients and a review of the literature. Eur. J. Hum. Genet, 10.1038/ejhg.2014.162 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malan V. et al. Distinct effects of allelic NFIX mutations on nonsense-mediated mRNA decay engender either a Sotos-like or a Marshall-Smith syndrome. Am. J. Hum. Genet 87, 189–198 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priolo M. et al. A peculiar mutation in the DNA-binding/dimerization domain of NFIX causes Sotos-like overgrowth syndrome: a new case. Gene 511, 103–105 (2012). [DOI] [PubMed] [Google Scholar]
- Yoneda Y. et al. Missense mutations in the DNA-binding/dimerization domain of NFIX cause Sotos-like features. J. Hum. Genet 57, 207–211 (2012). [DOI] [PubMed] [Google Scholar]
- Holm P. C. et al. Loss- and gain-of-function analyses reveal targets of Pax6 in the developing mouse telencephalon. Mol. Cell. Neurosci 34, 99–119 (2007). [DOI] [PubMed] [Google Scholar]
- Walther C. & Gruss P. Pax-6, a murine paired box gene, is expressed in the developing CNS. Development 113, 1435–1449 (1991). [DOI] [PubMed] [Google Scholar]
- Favor J., Neuhauser-Klaus A. & Ehling U. H. The effect of dose fractionation on the frequency of ethylnitrosourea-induced dominant cataract and recessive specific locus mutations in germ cells of the mouse. Mutat Res. 198, 269–275 (1988). [DOI] [PubMed] [Google Scholar]
- Hill R. E. et al. Mouse small eye results from mutations in a paired-like homeobox-containing gene. Nature 354, 522–525 (1991). [DOI] [PubMed] [Google Scholar]
- Roberts R. Small eyes-a new dominan eye mutant in the mouse. Genetics Res. Camb. 9, 121–122 (1967). [Google Scholar]
- Stoykova A. & Gruss P. Roles of Pax-genes in developing and adult brain as suggested by expression patterns. J. Neurosci 14, 1395–1412 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen Institute for Brain Science. Allen Developing Mouse Brain Atlas [Internet]. (2013) Available at : http://developingmouse.brain-map.org. (Accessed: 1st November 2014). [Google Scholar]
- Kang H. J. et al. Spatio-temporal transcriptome of the human brain. Nature 478, 483–489 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Ameele J., Tiberi L., Vanderhaeghen P. & Espuny-Camacho I. Thinking out of the dish: what to learn about cortical development using pluripotent stem cells. Trends Neurosci 37, 334–342 (2014). [DOI] [PubMed] [Google Scholar]
- Machon O. et al. Lens morphogenesis is dependent on Pax6-mediated inhibition of the canonical Wnt/beta-catenin signaling in the lens surface ectoderm. Genesis 48, 86–95 (2010). [DOI] [PubMed] [Google Scholar]
- Sansom S. N. et al. The level of the transcription factor Pax6 is essential for controlling the balance between neural stem cell self-renewal and neurogenesis. PLoS Genet 5, e1000511, 10.1371/journal.pgen.1000511 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messina G. et al. Nfix regulates fetal-specific transcription in developing skeletal muscle. Cell 140, 554–566 (2010). [DOI] [PubMed] [Google Scholar]
- Walcher T. et al. Functional dissection of the paired domain of Pax6 reveals molecular mechanisms of coordinating neurogenesis and proliferation. Development 140, 1123–1136 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boppana S., Scheglov A., Geffers R. & Tarabykin V. Cellular retinaldehyde-binding protein (CRALBP) is a direct downstream target of transcription factor Pax6. Biochim. Biophys. Acta 1820, 151–156 (2012). [DOI] [PubMed] [Google Scholar]
- Thaung C. et al. Novel ENU-induced eye mutations in the mouse: models for human eye disease. Hum Mol. Genet 11, 755–767 (2002). [DOI] [PubMed] [Google Scholar]
- Xie Q., Ung D., Khafizov K., Fiser A. & Cvekl A. Gene regulation by PAX6: structural-functional correlations of missense mutants and transcriptional control of Trpm3/miR-204. Mol. Vis. 20, 270–282 (2014). [PMC free article] [PubMed] [Google Scholar]
- Steele-Perkins G. et al. The transcription factor gene Nfib is essential for both lung maturation and brain development. Mol. Cell. Biol. 25, 685–698 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi D. et al. Pax6 exerts regional control of cortical progenitor proliferation via direct repression of Cdk6 and hypophosphorylation of pRb. Neuron 78, 269–284 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estivill-Torrus G., Pearson H., van Heyningen V., Price D. J. & Rashbass P. Pax6 is required to regulate the cell cycle and the rate of progression from symmetrical to asymmetrical division in mammalian cortical progenitors. Development 129, 455–466 (2002). [DOI] [PubMed] [Google Scholar]
- Gotz M., Stoykova A. & Gruss P. Pax6 controls radial glia differentiation in the cerebral cortex. Neuron 21, 1031–1044 (1998). [DOI] [PubMed] [Google Scholar]
- Quinn J. C. et al. Pax6 controls cerebral cortical cell number by regulating exit from the cell cycle and specifies cortical cell identity by a cell autonomous mechanism. Dev. Biol. 302, 50–65 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards T. J., Sherr E. H., Barkovich A. J. & Richards L. J. Clinical, genetic and imaging findings identify new causes for corpus callosum development syndromes. Brain 137, 1579–1613 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abouzeid H. et al. PAX6 aniridia and interhemispheric brain anomalies. Mol. Vis. 15, 2074–2083 (2009). [PMC free article] [PubMed] [Google Scholar]
- Asami M. et al. The role of Pax6 in regulating the orientation and mode of cell division of progenitors in the mouse cerebral cortex. Development 138, 5067–5078, (2011). [DOI] [PubMed] [Google Scholar]
- Pinon M. C., Tuoc T. C., Ashery-Padan R., Molnar Z. & Stoykova A. Altered molecular regionalization and normal thalamocortical connections in cortex-specific Pax6 knock-out mice. J. Neurosci 28, 8724–8734 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones L., Lopez-Bendito G., Gruss P., Stoykova A. & Molnar Z. Pax6 is required for the normal development of the forebrain axonal connections. Development 129, 5041–5052 (2002). [DOI] [PubMed] [Google Scholar]
- Xie Q. et al. Pax6 interactions with chromatin and identification of its novel direct target genes in lens and forebrain. PLoS One 8, e54507, 10.1371/journal.pone.0054507 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninkovic J. et al. The BAF complex interacts with Pax6 in adult neural progenitors to establish a neurogenic cross-regulatory transcriptional network. Cell Stem Cell. 13, 403–418 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoykova A., Fritsch R., Walther C. & Gruss P. Forebrain patterning defects in Small eye mutant mice. Development 122, 3453–3465 (1996). [DOI] [PubMed] [Google Scholar]
- Glaser T. et al. Absence of PAX6 gene mutations in Gillespie syndrome (partial aniridia, cerebellar ataxia, and mental retardation). Genomics 19, 145–148 (1994). [DOI] [PubMed] [Google Scholar]
- Solomon B. D. et al. Compound heterozygosity for mutations in PAX6 in a patient with complex brain anomaly, neonatal diabetes mellitus, and microophthalmia. Am. J. Med. Genet A 149A, 2543–2546 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoykova A., Treichel D., Hallonet M. & Gruss P. Pax6 modulates the dorsoventral patterning of the mammalian telencephalon. J Neurosci 20, 8042–8050 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun K., Potter S. & Rubenstein J. L. Gsh2 and Pax6 play complementary roles in dorsoventral patterning of the mammalian telencephalon. Development 128, 193–205 (2001). [DOI] [PubMed] [Google Scholar]
- Toresson H., Potter S. S. & Campbell K. Genetic control of dorsal-ventral identity in the telencephalon: opposing roles for Pax6 and Gsh2. Development 127, 4361–4371 (2000). [DOI] [PubMed] [Google Scholar]
- Stenman J., Yu R. T., Evans R. M. & Campbell K. Tlx and Pax6 co-operate genetically to establish the pallio-subpallial boundary in the embryonic mouse telencephalon. Development 130, 1113–1122 (2003). [DOI] [PubMed] [Google Scholar]
- Piper M. et al. NFIA controls telencephalic progenitor cell differentiation through repression of the Notch effector Hes1. J. Neurosci 30, 9127–9139 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponten J. & Macintyre E. H. Long term culture of normal and neoplastic human glia. Acta Pathol Microbiol Scand 74, 465–486 (1968). [DOI] [PubMed] [Google Scholar]
- Schlett K. & Madarasz E. Retinoic acid induced neural differentiation in a neuroectodermal cell line immortalized by p53 deficiency. J. Neurosci Res. 47, 405–415 (1997). [PubMed] [Google Scholar]
- Ninkovic J. et al. The transcription factor Pax6 regulates survival of dopaminergic olfactory bulb neurons via crystallin alphaA. Neuron 68, 682–694 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunt J. et al. Regulation of cell cycle genes and induction of senescence by overexpression of OTX2 in medulloblastoma cell lines. Mol. Cancer Res. 8, 1344–1357 (2010). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.