Abstract
Hydraulic flush waste removal systems coupled to solid/liquid separators and circulated treatment lagoons are commonly utilized to manage the large amounts of animal waste produced on high-intensity dairy farms. Although these systems are common, little is known about the microbial populations that inhabit them or how they change as they traverse the system. Using culture-based and non-culture-based methods, we characterized the microbial community structure of manure, water from the separator pit, and water from the circulated treatment lagoon from a large dairy in the San Joaquin Valley of California. Our results show that both total bacterial numbers and bacterial diversity are highest in manure, followed by the separator pit water and the lagoon water. The most prevalent phylum in all locations was the Firmicutes (low-G+C, gram-positive bacteria). The most commonly occurring operational taxonomic unit (OTU) had a 16S rRNA gene (rDNA) sequence 96 to 99% similar to that of Clostridium lituseburense and represented approximately 6% of the manure derived sequences, 14% of the separator pit-derived sequences and 20% of the lagoon-derived sequences. Also highly prevalent was an OTU with a 16S rDNA sequence 97 to 100% similar to that of Eubacterium tenue, comprising approximately 3% of the manure-derived sequences, 6% of the separator pit-derived sequences and 9% of the lagoon-derived sequences. Taken together, these sequences represent approximately one-third of the total organisms in the lagoon waters, suggesting that they are well adapted to this environment.
Modern dairy farms generate large amounts of animal waste that must be managed to ensure good animal, as well as human, health. It is common for dairies to house more than 1,000 cows, each producing approximately 35 kg of manure a day (35); thus, a 1,000-cow farm will produce approximately 35,000 kg of manure per day or more than 12,000,000 kg per year. In modern dairies, cows spend a majority of their productive time residing in free stalls where they defecate and urinate on cement floors. To remove the animal waste, water is flushed across the cement floor and the waste is carried into separator pits, where the solids and liquids are segregated. The solid portion is removed and used as soil amendment for surrounding crop fields or as animal bedding material. The liquid portion can be treated by pumping the wastewater through artificially constructed wetlands, which reduce chemical and biological oxygen demand, the concentration of total nitrogen, ammonia, phosphate, suspended solids, and bacteria (22). However, the cost of constructing and maintaining artificial wetlands is considerable, and thus, the wastewater is more commonly treated by pumping it into storage lagoons that range in holding capacity from 10 million to 60 million liters. The lagoon waters may be circulated by mechanical mixers, which enhance oxygen transfer into the wastewater, prevent stratification of the lagoons, and are believed to enhance bacterial degradation of organic compounds. The water from the lagoons is reused to flush the waste from the free stall floors, and thus, the cycle is repeated. This type of system has many advantages, including low labor costs for removing waste from the free stall floors, low construction and maintenance costs, little additional water being needed once the system is functioning, and the fact that the water can be used as an inexpensive fertilizer that can be efficiently pumped into surrounding fields that produce crops destined for both animal and human consumption.
However, cow manure may contain bacteria that are pathogenic to both humans and livestock, such as Escherichia coli O157:H7 (16), Campylobacter spp. (54), Salmonella spp. (52), and Mycobacterium spp. (33, 39). Because flush water animal waste treatment systems are in effect “closed loops,” there is concern that they may concentrate pathogens that could adversely affect the health of livestock or humans who consume crops fertilized with the wastewater (6, 36, 41; P. R. Cieslak, T. J. Barrett, P. M. Griffin, K. F. Gensheimer, G. Beckett, J. Buffington, and M. G. Smith, Letter, Lancet 342:367, 1993). It has also been suggested that bacteria and other contaminants can percolate through the soil and contaminate groundwater (12, 20). Little is known about the bacterial populations that inhabit these ecosystems or how their community structures change as they transit these wastewater treatment systems. In this study we characterized the chemical and bacterial composition of animal waste on the free stall floor, wastewater from the separator pit, and the circulated holding lagoon water from a large (>1,000-cow) dairy farm in the San Joaquin Valley of California using both culture-based (plate counting) and non-culture-based (16S rRNA gene [rDNA] sequence analysis and pathogen-specific PCR assays) methods.
MATERIALS AND METHODS
Sample collection and preparation.
Manure and wastewater samples were taken from a large dairy farm (>1,000 cows), located in the San Joaquin Valley of California, between November 2002 and July 2003. One-liter water samples were taken from the separator pit and from the circulated holding lagoon using a 5-m pole with a 2-liter bucket attached to the end of it by submersing the bucket into the water to a depth of approximately 1 m. The temperature of the water at the time of sampling was between 12 and 16°C. Manure samples of approximately 1 g were taken from 25 locations around the free stall floor and combined in a sterile 50-ml collection tube. Samples were transported to the laboratory, stored overnight at 4°C, and processed the next day. Bacteria were collected from wastewater by high-speed centrifugation. Ten 0.25-g samples of the resulting pellet or solid manure samples were transferred to 2-ml tubes that were frozen at −80°C until processed further as described under DNA extraction.
Viable counts of bacteria in manure and wastewater.
Viable bacteria in manure and waste waters were enumerated by performing serial dilutions in phosphate-buffered saline which was vortex agitated for 1 min prior to plating onto brain heart infusion agar plates, which were incubated at 30°C under normal atmospheric conditions or in an anaerobic chamber. To determine the number of coliform bacteria, samples were diluted as described above but plated onto MacConkey agar plates and incubated at 37°C. CFU in all plates were enumerated after 24 h. All media were purchased as dehydrated powders from Difco (Detroit, Mich.).
Chemical analysis of water samples.
Water samples were packed on ice and sent by next-day delivery to A&L Western Agricultural Labs (Modesto, Calif.), a California accredited environmental testing lab, for chemical analysis. For specific information on testing procedures, see http://www.al-labs-west.com/environmental.htm.
DNA extraction from wastewaters and manure.
DNA was extracted from manure or wastewater pellets using a modification of the MoBio UltraClean fecal DNA isolation kit (MoBio, Solano Beach, Calif.). Briefly, manure or bacterial pellets were thawed at room temperature and mixed with the kit-supplied beads as well as an equal amount of 0.1-mm silica-zirconium beads. The cells were disrupted by placing the tubes in a mini-bead beater (Biospec Products, Bartlesville, Okla.) and shaking three times for 60 s. The rest of the protocol was preformed as described by the manufacturer. For a given sample, 10 extractions were performed and the DNA was pooled, precipitated, and suspended at a concentration of approximately 50 ng/μl in sterile water.
PCR amplification of 16S rDNA sequences and library construction.
PCR amplification of 16S rDNA sequences was carried out using the eubacterium-specific primers 27f (5′AGAGTTTGATCCTGGCTCAG3′) and 1392r (5′GACGGGCGGTGTGTAC3′) (3). PCRs were performed as recommended by Polz and Cavanaugh (42) to reduce bias in amplification. Briefly, 50-μl reaction volumes contained 200 μM deoxynucleoside triphosphates, 100 ng of genomic DNA, 2 U of Expand High Fidelity enzyme mix (Roche, Nutley, N.J.), 5 μl of 10× Expand High Fidelity buffer with 15 mM MgCl2, and 1 μM (each) primer. PCRs were carried out in a GeneAmp PCR System 9700 thermocycler (Applied Biosystems, Foster City, Calif.) under the following conditions: one cycle of 95°C for 5 min, 15 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 1.5 min, one cycle of 5 min at 72°C, and holding at 4°C. The PCR products were purified using the QIAquick PCR purification kit (QIAGEN, Valencia Calif.). PCR products were cloned using the QIAGEN PCR cloning kit as per the manufacturer's instructions and transformed into E. coli TOP10F′ cells (Invitrogen, Carlsbad, Calif.) by heat shock (42°C for 30 s). Clones were plated on Luria-Bertani agar plates containing kanamycin (50 μg/ml), isopropyl-β-D-thiogalactopyranoside (20 mM), and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (80 μg/ml), and white colonies were selected and grown in 96-well plates in Luria-Bertani kanamycin broth. A total of 7 PCRs were performed for each sample, and 96 clones were picked from each PCR to minimize potential PCR bias (42).
Template preparation and sequencing.
DNA sequence templates were prepared in 96-well format using the Templiphi 100 amplification kit (Amersham Biosciences, Sunnyvale, Calif.) as per the manufacturer's instructions. Sequencing reactions were performed using the BigDye Terminator, version 3.1, cycle sequencing kit (Applied Biosystems) as described by the manufacturer, using the primer 1392r. Cycle sequencing reactions were purified by using the DyeEx 96 kit (QIAGEN); electrophoresis and readout were performed by using an Applied Biosystems 3100 Genetic Analyzer.
DNA sequence analysis, rarefaction analysis, dendrogram construction, and statistical methods.
DNA sequences were edited manually to correct falsely identified bases and trimmed to remove unreadable sequence at the 3′ and 5′ ends using the Chromas software (Technelysium Pty. Ltd., Helensvale, Australia). Only sequences with reads longer than 500 bases were used, and they averaged approximately 600 bp. The small-subunit ribosomal sequences were downloaded from the Ribosomal Database Project II (http://rdp.cme.msu.edu/download/SSU_rRNA/unaligned/), release 8.1, which contained 34,531 sequences (7). A BLASTable 16S rDNA nucleotide database was constructed using the FORMATDB program (ftp://ftp.ncbi.nih.gov/toolbox/ncbi_tools/). The predicted 16S rDNA nucleotide sequences from this study were compared to the 16S rDNA sequences in the database using a FASTA-formatted file containing the 16S rDNA sequences and the BLASTALL program (ftp://ftp.ncbi.nih.gov/toolbox/ncbi_tools/). Operational taxonomic units (OTUs) were defined as clones that shared 97% or greater sequence similarity. For dendrogram construction, 10 partial 16S rDNA sequences representing the most prevalent OTUs from each environment (manure, separator pit, and lagoon water) were aligned using CLUSTALW. Also included in the alignment were the 16S rDNA sequences most similar (>88% nucleotide identity) to each OTU, in the National Center for Biotechnology Information nr database (see Table 3). The 16S rDNA sequences from the homologs were first trimmed to approximate the start point and length of the OTU rDNA sequences. Phylogenetic and molecular evolutionary analyses were conducted using MEGA, version 2.1 (24). The dendrogram was constructed using the neighbor-joining algorithm and the Jukes-Cantor distance estimation method.
TABLE 3.
Ten most abundant OTUs from each library
| Source and OTU | Organism with or accession no. of most similar sequence in GenBank | % Similarity | No. of clonesa |
|---|---|---|---|
| Manure | |||
| M1 | Clostridium lituseburense | 97-99 | 31 |
| M2 | AF129864 | 95-97 | 26 |
| M3 | Ruminococcus bromii | 96-98 | 15 |
| M4 | AF018563 | 92-94 | 15 |
| M5 | AF018563 | 96-98 | 14 |
| M6 | Acinetobacter johnsonii | 98-100 | 14 |
| M7 | U81762 | 95-96 | 14 |
| M8 | Bacillus silvestris | 97 | 12 |
| M9 | AB009169 | 94-97 | 13 |
| M10 | Eubacterium tenue | 99 | 13 |
| Separator pit | |||
| SP1 | Clostridium lituseburense | 96-99 | 69 |
| SP2 | Eubacterium tenue | 97-100 | 31 |
| SP3 | Turicibacter sanguinis | 99-100 | 18 |
| SP4 | AF129864 | 95-96 | 15 |
| SP5 | Salinicoccus roseus | 96-99 | 14 |
| SP6 | Alkalibacterium olivoapovliticus | 92-93 | 13 |
| SP7 | Bacillus thermocloacae | 91-92 | 12 |
| SP8 | Alkalibacterium olivoapovliticus | 94-96 | 10 |
| SP9 | Clostridium celatum | 96-98 | 8 |
| SP10 | Lactosphaera pasteurii | 95-98 | 8 |
| Lagoon | |||
| L1 | Clostridium lituseburense | 97-99 | 99 |
| L2 | Eubacterium tenue | 98-100 | 47 |
| L3 | U81676 | 94-97 | 35 |
| L4 | AF087054 | 89 | 17 |
| L5 | Turicibacter sanguinis | 99-100 | 13 |
| L6 | Clostridium aminobutyricum | 93-94 | 13 |
| L7 | Streptococcus suis | 92-94 | 11 |
| L8 | Thiocapsa rosea | 95-96 | 11 |
| L9 | Achromatium sp. HK15 | 93-94 | 10 |
| L10 | U81715 | 94-96 | 9 |
Total numbers of clones in the top 10 for each location were as follows: 167 (33% of total) for manure, 198 (39% of total) for the separator pit, and 265 (63% of total) for the lagoon.
Diversity within the libraries was analyzed by rarefaction using the analytical approximation algorithm of Hurlbert (21), with 95% confidence intervals estimated as described by Heck (18), using the freeware program aRarefactWin by Holland (19). Coverage values were calculated by the equation C = 1 − (n/N) × 100, where n is the number of OTUs, N is the number of clones examined, and C is the percent coverage.
PCR detection of pathogens.
DNA extracted from manure or wastewater pellets was analyzed by sequence-specific PCR to detect the presence of Salmonella (29), E. coli O157 (34), Campylobacter spp. (9, 15), Yersinia spp. (49), Listeria spp. (4), pathogenic E. coli (10, 40), toxigenic staphylococci (23), and Clostridium perfringens (57). The specific genes and PCR primers are shown in Table 1. PCR was performed as described previously (31). Each PCR was performed with one primer set using annealing conditions based on the midpoint temperature (Tm) of the specific primer set. Clinical isolates from the veterinary diagnostic laboratory served as positive controls for the PCR assays.
TABLE 1.
PCR primers used for the detection of pathogen-specific gene markers in DNA extracted from manure, first separator pit water, and lagoon water
| Organism | PCR targeta | Primer sequenceb |
|---|---|---|
| Campylobacter jejuni | ceuE | F: CCTGCTACGGTGAAAGTTTTGC |
| R: GATCTTTTTGTTTGTGCTGC | ||
| Campylobacter coli | 23S rDNA | F: TATTCCAATACCAACATTAGT |
| R: TACAGCGTGGACTACCAGGGT | ||
| Clostridium perfringens | α toxin | F: GTTGATAGCGCAGGACATGTTAAG |
| R: CATGTAGTCATCTGTTCCAGCATC | ||
| β toxin | F: ACTATACAGACAGATCATTCAACC | |
| R: TTAGGAGCAGTTAGAACTACAGAC | ||
| ɛ toxin | F: ACTGCAACTACTACTCATACTGTG | |
| R: CTGGTGCCTTAATAGAAAGACTCC | ||
| ι toxin | F: GCGATGAAAAGCCTACACCACTAC | |
| R: GGTATATCCTCCACGCATATAGTC | ||
| E. coli | O157 rfb | F: CGTGATGATGTTGAGTTG |
| R: AGATTGGTTGGCATTACTG | ||
| LT1 | F: TGGATTCATCATGCACCACAAGG | |
| R: CCATTTCTCTTTTGCCTGCCATC | ||
| VT1 (stx1) | F: ACGTTACAGCGTGTTGCTGGGATC | |
| R: TGTGGCTGGGTTCGTTAATACGGC | ||
| VT2 (stx2) | F: TGTTGGCTGGGTTCGTTAATACGG | |
| R: TCCGTTGTCATGGAAACCGTTGTC | ||
| STI | F: TTTCCCCTCTTTTAGTCAGTCAACTG | |
| R: GGCAGGATTACAACAAAGTTCACAG | ||
| STII | F: CCCCCTCTCTTTTGCACTTCTTTCC | |
| R: TGCTCCAGCAGTACCATCTCTAACCC | ||
| CNF1 | F: GGCGACAAATGCAGTATTGCTTGG | |
| R: GACGTTGGTTGCGGTAATTTTGGG | ||
| eaeA | F: TGAGCGGCTGGCATGAGTCATAC | |
| R: TCGATCCCCATCGTCACCAGAGG | ||
| K99 | F: TATTATCTTAGGTGGTATGG | |
| R: GGTATCCTTTAGCAGCAGTATTTC | ||
| F41 | F: GCATCAGCGGCAGTATCT | |
| R: GTCCCTAGCTCAGTATTATCACCT | ||
| Listeria spp. | iap | F: ATGAATATGAAAAAGCAAC |
| R: TTATACGCCACCGAAGCCAAC | ||
| M. avium subsp. paratuberculosis | IS900 | F: GAAGGGTGTTCGGGGCCGTCGCTTAGG |
| R: GGCGTTGAGGTCGATCGCCCACGTGAC | ||
| Salmonella sp. | inv | F: CTGTTGAACAACCCATTTGT |
| R: CGGATCTCATTAATCAACAAT | ||
| Staphylococcus aureus | sea | F: TTGGAAACGGTTAAAACGAA |
| R: GAACCTTCCCATCAAAAACA | ||
| seb | F: TCGCATCAAACTGACAAACG | |
| R: GCAGGTACTCTATAAGTGCC | ||
| sec | F: GACATAAAAGCTAGGAATTT | |
| R: AAATCGGATTAACATTATCC | ||
| sed | F: CTAGTTTGGTAATATCTCCT | |
| R: TAATGCTATATCTTATAGG | ||
| tsst1 | F: ATGGCAGCATCAGCTTGATA | |
| R: TTTCCAATAACCACCCGTT | ||
| Yersinia spp. | 16S rDNA | F: GCGGCAGCGGGAAGTAGTTA |
| R: TACAGCGTGGACTACCAGGGT |
Limits of detection were as follows: E. coli and C. perfringens, 10 pg; Yersina spp. and Salmonella sp., 10 fg (10 pg correlates to 104 cells, and 10 fg correlates to 10 cells).
F, forward; R, reverse.
Nucleotide sequence accession numbers.
DNA sequences representative of each OTU were deposited in GenBank under the accession numbers AY438422 to AY438481 and AY438706 to AY438974.
RESULTS
Cultural and chemical analysis of manure, separator pit water, and lagoon water.
Total bacterial numbers based on brain heart infusion aerobic plate counts were estimated to be 2.1 × 109 CFU/g in manure, 1.9 × 106 CFU/ml in the separator pit water, and 2.8 × 105 CFU/ml in the lagoon water (Table 2). Anaerobic plate counts revealed approximately threefold more bacteria in each location with 6.9 × 109 CFU/g in manure, 5.5 × 106 CFU/ml in the separator pit water, and 6.7 × 105 CFU/ml in the lagoon water (Table 2). Coliform bacteria as measured on MacConkey agar plates represented only a small fraction of these bacteria, with 2.4 × 106 CFU/g in manure, 1.3 × 103 CFU/ml in the separator pit water, and 1.1 × 102 CFU/ml in the lagoon water (Table 2). The concentrations of several chemical species were assayed and found to decrease from the separator pit water to the lagoon water (Table 2). The concentration of total nitrogen decreased by more than twofold from the separator pit to the lagoon. The concentration of ammonia, the predominant nitrogen species (representing 83% of the separator pit water nitrogen and 70% of the lagoon water nitrogen) decreased from 36 mM in the separator pit water to 14 mM in the lagoon water, a 2.6-fold reduction. Nitrate and nitrite were below detectable limits (<38 and <43 μM, respectively). Sulfate levels decreased from 4.3 mM in the separator pit water to 0.7 mM in the lagoon water, a >6-fold decrease. The concentration of iron and sodium decreased by 3.4- and 1.6-fold, respectively. The pH of the water increased slightly as it traversed the system, starting at 7.65 in the separator pit water and reaching 7.95 in the lagoon water.
TABLE 2.
Chemical and cultural analysis of wastewaters and manure
| Parameter | Value for source
|
||
|---|---|---|---|
| Manure | Separator pit water | Lagoon water | |
| APCa | 2.1 × 109 CFU/g | 1.9 × 106 CFU/ml | 2.8 × 105 CFU/ml |
| AnPCb | 6.9 × 109 CFU/g | 5.5 × 106 CFU/ml | 6.7 × 105 CFU/ml |
| CPCc | 2.4 × 106 CFU/g | 1.3 × 103 CFU/ml | 1.1 × 102 CFU/ml |
| Total Nd | NDe | 774 mg/liter (55 mM) | 348 mg/liter (24 mM) |
| Ammonia concn | ND | 644 mg/liter (36 mM) | 245 mg/liter (14 mM) |
| Nitrate concn | ND | <2 mg/liter (<38 μM) | <2 mg/liter (<38 μM) |
| Nitrite concn | ND | <2 mg/liter (<43 μM) | <2 mg/liter (<43 μM) |
| Sulfate concn | ND | 419 mg/liter (4.3 mM) | 65 mg/liter (0.7 mM) |
| Fe concn | ND | 4.7 mg/liter (247 μM) | 1.4 mg/liter (73 μM) |
| Na concn | ND | 400 mg/liter (17 mM) | 257 mg/liter (11 mM) |
| pH | ND | 7.65 | 7.95 |
Aerobic plate counts.
Anaerobic plate counts.
Coliform plate counts.
Total Kjeldahl nitrogen.
ND, not determined.
Analysis of 16S rDNA sequences from manure, separator pit water, and lagoon water.
We constructed 16S rDNA libraries derived from DNA extracted from manure, the separator pit water, and the lagoon water containing 500 sequences each (for a complete list, see http://www.pw.usda.gov/wrrcpagedoc/PUBSUPPORT/PUBLICATION_SUPPORT.HTML). The sequences were BLAST analyzed against a 16S rDNA database containing more than 33,000 sequences (7). Each library contained multiple sequences that were most similar to 16S rDNA sequences from as of yet uncultured organisms, and thus, no information about these organisms is available. These sequences represented 45% of the manure-derived library, 21% of the separator pit water-derived library, and 35% of the lagoon water-derived library. Of the remaining sequences, for which information is available, the majority from all locations were most similar to those of gram-positive organisms: 87% from manure, 90% from the separator pit water, and 80% from the lagoon water.
Our analysis of the sequences derived from manure identified six phyla. Within these six phyla we identified 85 OTUs, as defined by 97% or greater sequence similarity, and 277 clones. The Firmicutes (low-G+C, gram-positive bacteria) were the most predominant phyla, representing 77% of the total isolates with 49 OTUs and 214 clones. The Actinobacteria (high-G+C, gram-positive bacteria) represented 9% of the total clones with 14 OTUs and 26 clones. The Bacteroidetes represented 7% of the total isolates with 9 OTUs and 19 clones. The Proteobacteria represented 5% of the total isolates with 10 OTUs and 15 clones, followed by the Cyanobacteria, representing 0.7% of the total clones (2 OTUs and 2 clones), and the Spirochaetes with 0.4% (1 OTU and 1 clone).
The sequences derived from the separator pit water comprised 5 phyla, with 153 OTUs and 342 clones. Similar to the manure-derived library, the phylum Firmicutes represented the most predominant phylum, representing 80% of the isolates with 63 OTUs and 274 clones. The phylum Actinobacteria represented 10% of the total clones, with 24 OTUs and 35 clones, followed by the Proteobacteria, representing 5% of the total clones (10 OTUs and 18 clones), the Bacteroidetes, containing 4% of the total (7 OTUs and 14 clones), and the Chloroflexi, with 0.03% of the clones (1 OTU and 1 clone).
The sequences obtained from the lagoon water represented organisms within 6 phyla, comprising 72 OTUs and 324 clones. Again the phylum Firmicutes predominated, with 77% of the total clones, with 41 OTUs and 251 clones. However, in contrast to the manure- and separator pit water-derived libraries, the second most abundant phylum in the lagoon water-derived library was the Proteobacteria, which comprised 15% of the clones and contained 16 OTUs and 50 clones, followed by the Actinobacteria with 3% of the total clones (7 OTUs and 9 clones), the Bacteroidetes with 3% of the total clones (5 OTUs and 9 clones), the Planctomycetes with 1% of the total clones (2 OTUs and 4 clones), and the Chloroflexi with 0.003% of the total clones (1 OTU and 1 clone).
Estimates of diversity, coverage analysis, and identification of the 10 most prevalent species from each location.
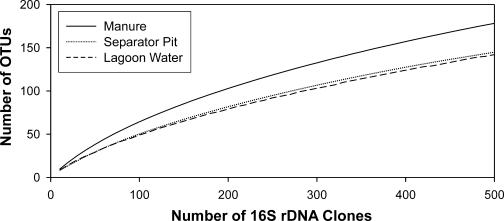
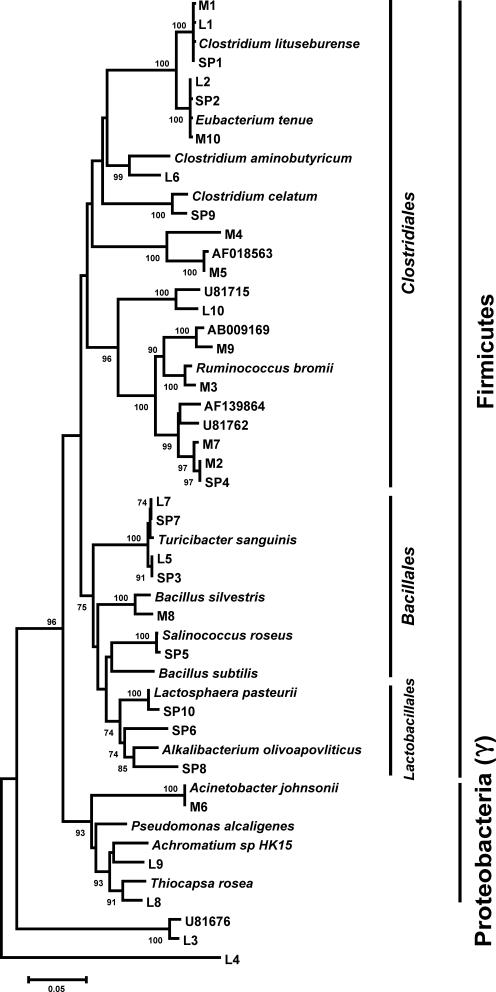
We applied rarefaction analysis to determine if the 500 clone libraries were sufficient to estimate the diversity in our samples (Fig. 1). The rarefaction curves did not reach saturation, suggesting that further cloning would have revealed more OTUs in each sample; however, the slopes of these curves do decrease towards the end points, indicating that the most prevalent bacterial groups were likely identified. Indeed, coverage analysis estimate the level of coverage at 64% for the manure-derived library, 69% for the separator pit water-derived library, and 72% for the lagoon water-derived library. To further characterize the level of diversity within each library, we examined the evenness within each library by calculating what percentage of the total population was represented by the 10 most prevalent OTUs (Table 3 and Fig. 2). For the manure-derived library, the 10 most numerous OTUs accounted for only 33% of the total OTUs; however, for the separator pit water- and lagoon water-derived libraries, that number increased to 40 and 53%, respectively.
FIG. 1.
Rarefaction curves for 16S rDNA clone libraries derived from DNA extracted from manure, the separator pit water, and the lagoon water using the Rarefaction Calculator (located at http://www.uga.edu/∼strata/software/) by Holland. Sequences were grouped into OTUs based on 97% or greater sequence similarity. Solid lines refer to the manure-derived library, dotted lines refer to the separator pit water-derived library, and dashed lines refer to the lagoon water-derived library.
FIG. 2.
Phylogenetic relationship of the OTUs isolated from manure (M), the separator pit (SP), and lagoon water (L). The dendrogram was constructed using the neighbor-joining algorithm and the Jukes-Cantor distance estimation method. Bootstrap values (>70%) generated from 1,000 replicates are shown at the nodes. The scale bar represents substitutions per site. Phylum and order designations are indicated on the right. Due to space limitations, the two orders represented within the γ-Proteobacteria are not labeled. Acinetobacter johnsonii, M6, and Pseudomonas alcaligenes are members of the order Pseudomonadales; Achromatium, L8, L9, and Thiocapsa rosea are members of the order Chromatiales. Labels were added in Corel Draw 11 (Corel, Ottawa, Canada).
Detection of 16S rDNA sequences similar to those of pathogenic microorganisms.
The 16S rDNA cloning method detected sequences with 97% or greater sequence similarity to pathogenic bacteria. Of the 500 sequences derived from manure, we identified one sequence that was 97% similar to that of Clostridium difficile. Of the 500 sequences derived from the separator pit water, we detected two sequences with 98 to 99% similarity to that of Clostridium septicum and one sequence 97% similar to that of Dermatophilus congolensis. However, none of these sequences was detected in the lagoon water. To increase the sensitivity of detection for pathogenic microorganisms, we performed pathogen-specific PCR on DNA isolated from each location, using primers specific for genetic markers found in Salmonella, pathogenic E. coli, Staphylococcus aureus, Campylobacter jejuni, Campylobacter coli, Clostridium perfringens, Listeria spp., Mycobacterium avium subsp. paratuberculosis, and Yersinia spp. (Table 1). No DNA fragments were amplified by these primer sets, except for the Yersinia spp. PCR from manure and the separator pit water derived DNA; however, further evaluation revealed that the fragments did not represent pathogenic Yersinia spp. (e.g., Yersinia pestis, Yersinia pseudotuberculosis, or Yersinia enterocolitica).
DISCUSSION
The use of hydraulic flush systems to remove animal waste has multiple advantages. However, little information is known about the microbial populations that inhabit these ecosystems, and there is concern that they may be environmental reservoirs for bacteria that are pathogenic to animals and humans. In order to gain more information about these systems, we enumerated the number of bacteria per gram in the manure entering the system and the number of bacteria per milliliter in the separator pit and in the storage lagoons (Table 2). We observed that the number of bacteria, as measured by aerobic and anaerobic plate counts, decreased from manure to the separator pit water to the lagoon water; likewise, the number of coliform bacteria decreased as they traversed the system. Although cultural techniques such as these are inherently inaccurate due to cultural bias, these data imply that the system efficiently reduces the bacterial load of the water as it transits the system.
The concentrations of several chemicals, such as NH3, SO4, and Fe, were also reduced from the separator pit water to the lagoon water (Table 2). The majority of nitrogen in both the separator pit water and the lagoon water was in the form of NH3, likely the result of bacterial mineralization of organic nitrogen-containing compounds, such as peptides and urea. The levels of NO3 and NO2 were undetectable in either the lagoon or the separator pit water. We hypothesize that their absence is the result of the lack of nitrification due to the anaerobic nature of these systems and the depletion of any existing NO3 and NO2 by microbial anaerobic respiration as well as assimilation of these chemicals by microorganisms. The loss of SO4 from the separator pit to the lagoon is also likely due to its reduction to SO3 and eventually to sulfide by microbial anaerobic respiration, as well as assimilatory sulfate reduction.
To better define the bacterial populations that inhabit these systems, we adopted a strategy that does not rely on cultivation. We constructed 16S rDNA libraries with DNA amplified from manure, the separator pit water, and lagoon water and sequenced a total of 1,500 clones. Although we used methods previously shown to reduce PCR bias, such as high DNA template concentration (100 ng/reaction), avoidance of primer degeneracies, and few amplification cycles (15 cycles) (42), we cannot be sure that the number of a particular 16S rDNA sequence in our libraries correlates to its abundance in the environment. Approximately one-third of the sequences isolated from each location were most similar to sequences of uncultured organisms recovered in previous studies using 16S rDNA cloning methods (2, 47, 53). Since these 16S rDNA sequences represent as of yet uncultured organisms, little information can be derived about their physiology. Of the sequences that are closely related to 16S rDNA sequences of cultured organisms, the majority in all locations were most similar to gram-positive organisms, with the phylum Firmicutes (low-G+C, gram-positive bacteria) representing approximately half of the clones from each location. This was not surprising, since many ruminant bacteria, including the genera Clostridium, Bacillus, and Ruminococcus, reside within this group. Indeed, sequences 96 to 99% similar to that of Clostridium lituseburense were the most commonly occurring clones in each library and represented approximately 6% of the manure clones, 14% of the separator pit clones, and 20% of the lagoon clones. Clones with sequences 97 to 100% similar to that of Eubacterium tenue were also frequently isolated, comprising approximately 3% of the manure clones, 6% of the separator pit clones, and 9% of the lagoon clones. These organisms appear to be well adapted to both the host and wastewater environments, since their sequences predominate among the manure library clones and increase in abundance by approximately threefold in the lagoon water library. Furthermore, because these two sequences account for approximately one-third of the lagoon water-derived clones, these organisms may be of value as indicators of dairy wastewater contamination and source tracking.
Three OTUs that have a 97% or greater homology to known pathogenic bacteria were detected and included: C. difficile, C. septicum, and D. congolensis. C. difficile is an etiological agent of diarrhea in humans and swine (55, 56). C. septicum causes pseudo-blackleg in cattle and sheep, an infection of skeletal and heart muscle that results in the inflammation of these tissues and is often fatal (37). In humans, C. septicum can cause gas gangrene (5) and myonecrosis, a rare but often life-threatening infection (1). D. congolensis is the etiological agent of dermatophilosis, an exudative dermatitis of ruminants that can damage hides and decrease milk production (30). D. congolensis has also been reported to cause pitted keratolysis in humans (13). Although these OTUs have high sequence similarity to pathogens, we cannot be absolutely sure, based on 16S rDNA sequence alone, that these organisms are pathogenic. Based on previous reports, we were surprised that we did not detect any of the common livestock-associated pathogens, such as Salmonella, pathogenic E. coli, S. aureus, C. jejuni, C. coli, C. perfringens, Listeria spp., M. avium subsp. paratuberculosis, or Yersinia spp. It is likely that some of these species were present but below the detectable level of the PCR assays, which ranges from 104 to 10 cells.
Ibekwe et al. (22) analyzed the microbial community structure of dairy waste effluent from a constructed wetland using denaturing gradient gel electrophoresis (DGGE). Although wetland systems are different from the separator pit/lagoon system we studied and appear to select for ammonia-oxidizing bacteria, such as Nitrosospira- and Nitrosomonas-like organisms, that we did not detect, we did find some similarities. For example, the most abundant 16S rDNA sequence from our system, which represented Clostridium lituseburense, was 97% similar to their DGGE band 4430. We also observed that DGGE bands 3214 and 212 were 96 and 94% similar to sequences identified in our study but represented low-abundance clones.
Interestingly, the bacterial community structure of dairy waste lagoon water is in some ways similar to that of broiler chicken litter (31) and liquid swine manure (27). Lu et al. (31) observed that broiler chicken litter contained approximately 82% gram-positive bacteria, which is very similar to what we observed (86% for manure, 90% for the separator pit water, and 80% for the lagoon water). Their observed distribution of 62% Firmicutes, 24% Actinobacteria, and 12% Proteobacteria is also in general agreement with our observations. The bacterial community structure of liquid swine manure reported by Leung and Topp (27) also shares similarities to what we observed. For example, they observed a predominant DGGE band corresponding to a 16S rDNA sequence that was most similar to U81676, the third-most-abundant clone isolated from the lagoon water. They also observed DGGE bands corresponding to 16S rDNA sequences of AF001778, U81735, and UBA400574, which were also observed in our study at low frequencies.
Many of the 16S rDNA sequences isolated in our study have been previously characterized as representing bacteria associated with animals. For example, 16S rDNA sequences similar to those of Clostridium lituseburense (11), Clostridium celatum (17), Ruminococcus bromii (51), E. tenue (45), Turicibacter sanguinis (8), and AF371945 (26) have been isolated from the gastrointestinal tracts of humans and animals. In addition, 16S rDNA sequences similar to those of Acinetobacter johnsonii (32), Bacillus thermocloacae (25), Lactosphaera pasteurii (28), AF129864 (14), U81762 (14), U81676 (14), and AF087054 (46) have been isolated from anaerobic digestors and activated sludge, while others similar to those of Bacillus silvestris, Bacillus marinus (43), Salinococcus roseus (50), Thiocapsa rosea (48), and Alkalibacterium olivoapovliticus (38) appear to represent environmental organisms associated with plant rhizosphere, marine sediment, and both salt and fresh water.
The microbial composition of diary wastewater treatment systems is complex and is in need of further evaluation. Although this study sequenced more than 1,500 bacterial 16S rDNA clones isolated from various locations in the treatment system and has given new insights as to the bacterial populations that inhabit these systems, more information is needed. For instance, we were not able to identify any of the common pathogens associated with dairy waste, such as pathogenic E. coli, which have caused outbreaks in close proximity to dairy farms and have been blamed on the seepage of waste into groundwater (44). If dairy farms were to blame, was this an isolated incident or are there pathogens residing in undetectably low numbers in dairy farm environments that are able to grow under favorable conditions? Furthermore, different dairies use different waste management strategies, such as artificial wetlands, aerated versus nonaerated holding lagoons, solid/liquid separation versus bulk flow methods; could one of these methods be more susceptible to pathogen persistence? If so, what conditions favor regrowth? Future studies will address these and other questions.
Acknowledgments
We thank Anna Korn from the USDA/ARS/FCR for her assistance throughout this project, Jenn Brofft from the Skidaway Institute of Oceanography for insightful discussion and technical reading of the manuscript, Mark Mackiewicz from the University of Georgia, Center for Applied Genetic Technologies, for help with sequencing, and George Robertson from the USDA/ARS/BCE for web page development.
This work was funded by the U.S. Department of Agriculture, Agricultural Research Service, and National Programs 108 (Food Safety) and 206 (Manure and By-Product Utilization).
REFERENCES
- 1.Abella, B. S., P. Kuchinic, T. Hiraoka, and D. S. Howes. 2003. Atraumatic clostridial myonecrosis: case report and literature review. J. Emerg. Med. 24:401-405. [DOI] [PubMed] [Google Scholar]
- 2.Bowman, J. P., S. M. Rea, S. A. McCammon, and T. A. McMeekin. 2000. Diversity and community structure within anoxic sediment from marine salinity meromictic lakes and a coastal meromictic marine basin, Vestfold Hilds, Eastern Antarctica. Environ. Microbiol. 2:227-237. [DOI] [PubMed] [Google Scholar]
- 3.Brofft, J. E., J. V. McArthur, and L. J. Shimkets. 2002. Recovery of novel bacterial diversity from a forested wetland impacted by reject coal. Environ. Microbiol. 4:764-769. [DOI] [PubMed] [Google Scholar]
- 4.Bubert, A., I. Hein, M. Rauch, A. Lehner, B. Yoon, W. Goebel, and M. Wagner. 1999. Detection and differentiation of Listeria spp. by a single reaction based on multiplex PCR. Appl. Environ. Microbiol. 65:4688-4692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carron, P., and D. Tagan. 2003. Fulminant spontaneous Clostridium septicum gas gangrene. Ann. Chir. 128:391-393. (In French.) [DOI] [PubMed] [Google Scholar]
- 6.Chapman, P. A., C. A. Siddons, J. Manning, and C. Cheetham. 1997. An outbreak of infection due to verocytotoxin-producing Escherichia coli O157 in four families: the influence of laboratory methods on the outcome of the investigation. Epidemiol. Infect. 119:113-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cole, J. R., B. Chai, T. L. Marsh, R. J. Farris, Q. Wang, S. A. Kulam, S. Chandra, D. M. McGarrell, T. M. Schmidt, G. M. Garrity, and J. M. Tiedje. 2003. The Ribosomal Database Project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res. 31:442-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Egert, M., B. Wagner, T. Lemke, A. Brune, and M. W. Friedrich. 2003. Microbial community structure in midgut and hindgut of the humus-feeding larva of Pachnoda ephippiata (Coleoptera: Scarabaeidae). Appl. Environ. Microbiol. 69:6659-6668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eyers, M., S. Chapelle, G. Van Camp, H. Goossens, and R. De Wachter. 1993. Discrimination among thermophilic Campylobacter species by polymerase chain reaction amplification of 23S rRNA gene fragments. J. Clin. Microbiol. 31:3340-3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Franck, S. M., B. T. Bosworth, and H. W. Moon. 1998. Multiplex PCR for enterotoxigenic, attaching and effacing, and Shiga toxin-producing Escherichia coli strains from calves. J. Clin. Microbiol. 36:1795-1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Franks, A. H., H. J. Harmsen, G. C. Raangs, G. J. Jansen, F. Schut, and G. W. Welling. 1998. Variations of bacterial populations in human feces measured by fluorescent in situ hybridization with group-specific 16S rRNA-targeted oligonucleotide probes. Appl. Environ. Microbiol. 64:3336-3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gagliardi, J. V., and J. S. Karns. 2000. Leaching of Escherichia coli O157:H7 in diverse soils under various agricultural management practices. Appl. Environ. Microbiol. 66:877-883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gillum, R. L., S. M. Qadri, M. N. Al-Ahdal, D. H. Connor, and A. J. Strano. 1988. Pitted keratolysis: a manifestation of human dermatophilosis. Dermatologica 177:305-308. [DOI] [PubMed] [Google Scholar]
- 14.Godon, J. J., E. Zumstein, P. Dabert, F. Habouzit, and R. Moletta. 1997. Molecular microbial diversity of an anaerobic digestor as determined by small-subunit rDNA sequence analysis. Appl. Environ. Microbiol. 63:2802-2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gonzalez, I., K. A. Grant, P. T. Richardson, S. F. Park, and M. D. Collins. 1997. Specific identification of the enteropathogens Campylobacter jejuni and Campylobacter coli by using a PCR test based on the ceuE gene encoding a putative virulence determinant. J. Clin. Microbiol. 35:759-763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hancock, D. D., T. E. Besser, and D. H. Rice. 1998. Ecology of E. coli 0157:H7 in cattle and impact of management practices. ASM Press, Washington D.C.
- 17.Hauschild, A. H. W., and L. V. Holdeman. 1974. Clostridium celatum sp. nov., isolated from normal human feces. Int. J. Syst. Bacteriol. 24:478-481. [Google Scholar]
- 18.Heck, K. L., Jr., G. Van Belle, and D. Simberloff. 1975. Explicit calculation of the rarefaction diversity measurement and the determination of sufficient sample size. Ecology 56:1459-1461. [Google Scholar]
- 19.Holland, S. 1998. aRarefactWin. University of Georgia, Athens.
- 20.Howell, J. M., M. S. Coyne, and P. Cornelius. 1995. Fecal bacteria in agricultural waters of the bluegrass region of Kentucky. J. Environ. Qual. 24:1-14. [Google Scholar]
- 21.Hurlbert, S. H. 1971. The nonconcept of species diversity: a critique and alternative parameters. Ecology 52:577-586. [DOI] [PubMed] [Google Scholar]
- 22.Ibekwe, A. M., C. M. Grieve, and S. R. Lyon. 2003. Characterization of microbial communities and composition in constructed dairy wetland wastewater effluent. Appl. Environ. Microbiol. 69:5060-5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson, W. M., S. D. Tyler, E. P. Ewan, F. E. Ashton, D. R. Pollard, and K. R. Rozee. 1991. Detection of genes for enterotoxins, exfoliative toxins, and toxic shock syndrome toxin 1 in Staphylococcus aureus by the polymerase chain reaction. J. Clin. Microbiol. 29:426-430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 25.Kurisu, F., H. Satoh, T. Mino, and T. Matsuo. 2002. Microbial community analysis of thermophilic contact oxidation process by using ribosomal RNA approaches and the quinone profile method. Water Res. 36:429-438. [DOI] [PubMed] [Google Scholar]
- 26.Leser, T. D., J. Z. Amenuvor, T. K. Jensen, R. H. Lindecrona, M. Boye, and K. Moller. 2002. Culture-independent analysis of gut bacteria: the pig gastrointestinal tract microbiota revisited. Appl. Environ. Microbiol. 68:673-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leung, K., and E. Topp. 2001. Bacterial community dynamics in liquid swine manure during storage: molecular analysis using DGGE/PCR of 16S rDNA. FEMS Microbiol. Ecol. 38:169-177. [Google Scholar]
- 28.Liu, J. R., P. Burrell, E. M. Seviour, J. A. Soddell, L. L. Blackall, and R. J. Seviour. 2000. The filamentous bacterial morphotype ‘Nostocoida limicola' I contains at least two previously described genera in the low G+C gram positive bacteria. Syst. Appl. Microbiol. 23:528-534. [DOI] [PubMed] [Google Scholar]
- 29.Liu, T., K. Liljebjelke, E. Bartlett, C. Hofacre, S. Sanchez, and J. J. Maurer. 2002. Application of nested polymerase chain reaction to detection of Salmonella in poultry environment. J. Food Prot. 65:1227-1232. [DOI] [PubMed] [Google Scholar]
- 30.Lloyd, D. H. 1976. The economic effects of bovine streptothricosis, p. 274-291. In D. H. Lloyd and K. C. Sellers (ed.), Dermatophilus infection in animals and man. Academic Press, London, United Kingdom.
- 31.Lu, J., S. Sanchez, C. Hofacre, J. J. Maurer, B. G. Harmon, and M. D. Lee. 2003. Evaluation of broiler litter with reference to the microbial composition as assessed by using 16S rRNA and functional gene markers. Appl. Environ. Microbiol. 69:901-908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Malik, A., M. Sakamoto, S. Hanazaki, M. Osawa, T. Suzuki, M. Tochigi, and K. Kakii. 2003. Coaggregation among nonflocculating bacteria isolated from activated sludge. Appl. Environ. Microbiol. 69:6056-6063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Manning, E. J. 2001. Mycobacterium avium subspecies paratuberculosis: a review of current knowledge. J. Zool. Wildl. Med. 32:293-304. [DOI] [PubMed] [Google Scholar]
- 34.Maurer, J. J., D. Schmidt, P. Petrosko, S. Sanchez, L. Bolton, and M. D. Lee. 1999. Development of primers to O-antigen biosynthesis genes for specific detection of Escherichia coli O157 by PCR. Appl. Environ. Microbiol. 65:2954-2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miner, R. J., F. J. Humenik, and M. R. Overcash. 2000. Managing livestock wastes to preserve environmental quality, 1st ed. Iowa State University Press, Ames.
- 36.Natvig, E. E., S. C. Ingham, B. H. Ingham, L. R. Cooperband, and T. R. Roper. 2002. Salmonella enterica serovar Typhimurium and Escherichia coli contamination of root and leaf vegetables grown in soils with incorporated bovine manure. Appl. Environ. Microbiol. 68:2737-2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nervig, R. M., S. E. Maloy, K. D. Claus, and D. R. Kolbe. 1981. Clostridium septicum infection in cattle in the United States. J. Am. Vet. Med. Assoc. 179:479. [PubMed] [Google Scholar]
- 38.Ntougias, S., and N. J. Russell. 2001. Alkalibacterium olivoapovliticus gen. nov., sp. nov., a new obligately alkaliphilic bacterium isolated from edible-olive wash-waters. Int. J. Syst. Evol. Microbiol. 51:1161-1170. [DOI] [PubMed] [Google Scholar]
- 39.O'Reilly, L. M., and C. J. Daborn. 1995. The epidemiology of Mycobacterium bovis infections in animals and man: a review. Tuber. Lung Dis. 76(Suppl. 1):1-46. [DOI] [PubMed] [Google Scholar]
- 40.Pass, M. A., R. Odedra, and R. M. Batt. 2000. Multiplex PCRs for identification of Escherichia coli virulence genes. J. Clin. Microbiol. 38:2001-2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pell, A. N. 1997. Manure and microbes: public and animal health problem? J. Dairy Sci. 80:2673-2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Polz, M. F., and C. M. Cavanaugh. 1998. Bias in template-to-product ratios in multitemplate PCR. Appl. Environ. Microbiol. 64:3724-3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pukall, R., I. Kramer, M. Rohde, and E. Stackebrandt. 2001. Microbial diversity of cultivatable bacteria associated with the North Sea bryozoan Flustra foliacea. Syst. Appl. Microbiol. 24:623-633. [DOI] [PubMed] [Google Scholar]
- 44.Ritter, L., K. Solomon, P. Sibley, K. Hall, P. Keen, G. Mattu, and B. Linton. 2002. Sources, pathways, and relative risks of contaminants in surface water and groundwater: a perspective prepared for the Walkerton inquiry. J. Toxicol. Environ. Health A 65:1-142. [DOI] [PubMed] [Google Scholar]
- 45.Schwiertz, A., G. Le Blay, and M. Blaut. 2000. Quantification of different Eubacterium spp. in human fecal samples with species-specific 16S rRNA-targeted oligonucleotide probes. Appl. Environ. Microbiol. 66:375-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Snaidr, J., B. Fuchs, G. Wallner, M. Wagner, K. H. Schleifer, and R. Amann. 1999. Phylogeny and in situ identification of a morphologically conspicuous bacterium, Candidatus Magnospira bakii, present at very low frequency in activated sludge. Environ. Microbiol. 1:125-135. [DOI] [PubMed] [Google Scholar]
- 47.Tajima, K., R. I. Aminov, T. Nagamine, K. Ogata, M. Nakamura, H. Natsui, and Y. Benno. 1999. Rumen bacterial diversity as determined by sequence analysis of 16S rDNA libraries. FEMS Microbiol. Ecol. 29:159-169. [Google Scholar]
- 48.Tonolla, M., A. Demarta, R. Peduzzi, and D. Hahn. 1999. In situ analysis of phototrophic sulfur bacteria in the chemocline of meromictic Lake Cadagno (Switzerland). Appl. Environ. Microbiol. 65:1325-1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Trebesius, K., D. Harmsen, A. Rakin, J. Schmelz, and J. Heesemann. 1998. Development of rRNA-targeted PCR and in situ hybridization with fluorescently labelled oligonucleotides for detection of Yersinia species. J. Clin. Microbiol. 36:2557-2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ventosa, A., J. J. Nieto, and A. Oren. 1998. Biology of moderately halophilic aerobic bacteria. Microbiol. Mol. Biol. Rev. 62:504-544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang, R. F., W. W. Cao, and C. E. Cerniglia. 1997. PCR detection of Ruminococcus spp. in human and animal faecal samples. Mol. Cell Probes. 11:259-265. [DOI] [PubMed] [Google Scholar]
- 52.Warnick, L. D., L. M. Crofton, K. D. Pelzer, and M. J. Hawkins. 2001. Risk factors for clinical salmonellosis in Virginia, USA cattle herds. Prev. Vet. Med. 49:259-275. [DOI] [PubMed] [Google Scholar]
- 53.Watanabe, K., M. Teramoto, and S. Harayama. 1999. An outbreak of nonflocculating catabolic populations caused the breakdown of a phenol-digesting activated-sludge process. Appl. Environ. Microbiol. 65:2813-2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wesley, I. V., S. J. Wells, K. M. Harmon, A. Green, L. Schroeder-Tucker, M. Glover, and I. Siddique. 2000. Fecal shedding of Campylobacter and Arcobacter spp. in dairy cattle. Appl. Environ. Microbiol. 66:1994-2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yablon, S. A., R. Krotenberg, and K. Fruhmann. 1992. Diarrhea in hospitalized patients. Am. J. Phys. Med. Rehabil. 71:102-107. [DOI] [PubMed] [Google Scholar]
- 56.Yaeger, M., N. Funk, and L. Hoffman. 2002. A survey of agents associated with neonatal diarrhea in Iowa swine including Clostridium difficile and porcine reproductive and respiratory syndrome virus. J. Vet. Diagn. Investig. 14:281-287. [DOI] [PubMed] [Google Scholar]
- 57.Yoo, H. S., S. U. Lee, K. Y. Park, and Y. H. Park. 1997. Molecular typing and epidemiological survey of prevalence of Clostridium perfringens types by multiplex PCR. J. Clin. Microbiol. 35:228-232. [DOI] [PMC free article] [PubMed] [Google Scholar]