Abstract
Termites are an important component of tropical soil communities and have a significant effect on the structure and nutrient content of soil. Digestion in termites is related to gut structure, gut physicochemical conditions, and gut symbiotic microbiota. Here we describe the use of 16S rRNA gene sequencing and terminal-restriction fragment length polymorphism (T-RFLP) analysis to examine methanogenic archaea (MA) in the guts and food-soil of the soil-feeder Cubitermes fungifaber Sjostedt across a range of soil types. If these MA are strictly vertically inherited, then the MA in guts should be the same in all individuals even if the soils differ across sites. In contrast, gut MA should reflect what is present in soil if populations are merely a reflection of what is ingested as the insects forage. We show clear differences between the euryarchaeal communities in termite guts and in food-soils from five different sites. Analysis of 16S rRNA gene clones indicated little overlap between the gut and soil communities. Gut clones were related to a termite-derived Methanomicrobiales cluster, to Methanobrevibacter and, surprisingly, to the haloalkaliphile Natronococcus. Soil clones clustered with Methanosarcina, Methanomicrococcus, or rice cluster I. T-RFLP analysis indicated that the archaeal communities in the soil samples differed from site to site, whereas those in termite guts were similar between sites. There was some overlap between the gut and soil communities, but these may represent transient populations in either guts or soil. Our data do not support the hypothesis that termite gut MA are derived from their food-soil but also do not support a purely vertical transmission of gut microflora.
Termites are an extremely important component of tropical soil decomposer communities. The abundance of termites, coupled with their consumption and digestion of plant-derived material, means they have a major influence on soil structure, plant decomposition, carbon mineralization, and nutrient availability (3, 22). Members of the six families of lower termites feed on wood or grass, but those of the Termitidae family, the higher termite family that includes ∼70% of all known termite species, feed on a wide range of plant material at different stages of decomposition (12, 15). The majority of species in the Termitidae feed on highly humified plant material in soil, and these soil-feeding termites are especially diverse and abundant in tropical forest soils (11).
Digestion in termites is closely related to gut structure, the physicochemical conditions in different gut regions, and the symbiotic microbiota found within their guts, without which the insects cannot survive (5, 8). The microbiota include taxa drawn from all three domains of life (19), although the flagellate protists that feature prominently in textbook treatments of termite symbioses are absent from the higher termites. This absence of gut flagellates in the Termitidae appears to be correlated with their evolution and diversification in feeding habits. The degree to which gut prokaryotes vary between termites is far from clear, but differences in host diet have been correlated with differences in microbial community processing of fermentation-generated H2. In anaerobic gut regions of both lower and higher wood-feeding termites, bacterial acetogenesis outcompetes methanogenesis for H2, whereas the reverse is true in the guts of soil-feeders (4, 7, 39). Since methanogenesis is restricted to members of the Archaea, it is not surprising that archaeal 16S rRNA is relatively more abundant in the guts of soil-feeders than in those of other termites (6). Although 16S rRNA gene sequences of other Archaea have been reported from guts of Cubitermes orthognathus (18), methanogens are the only Archaea to have been detected in significant numbers and the only ones known to contribute to anaerobic decomposition in termite guts.
The genus Cubitermes has been extensively used as a model for investigating soil-feeding in termites. Their guts are highly structured and compartmentalized environments characterized by steep gradients of O2, H2, and pH both between and within segments (9, 20, 34) with structured microbial communities (18, 35, 36). The third and fourth proctodeal gut segments contain virtually all of the methanogenic activity of the hindgut, although both methanogens and acetogens are located there. It has been suggested that either the acetogens utilize a non-H2 substrate or else they rely on cross-epithelial H2 transfer from the anterior gut regions (39).
The relationship between termites and their gut microbes, especially the methanogenic Archaea, are important in understanding the function of termites in the global ecosystem. In addition, termite gut microbial communities are an ideal model for asking questions about the relationship between microbial biodiversity and function in situ (19). However, analyzing such microbial communities is a complex and difficult task. A number of methods have been used, but many, such as the cloning and sequencing of PCR-amplified 16S rRNA gene sequences, are laborious, time-consuming, and cannot easily be applied to a large number of samples. Rapid methods of profiling communities do exist, but most of these offer only a snapshot of the community as a whole, without the resolution of identifying and tracking individual species, phylotypes, etc. Terminal-restriction fragment length polymorphism (T-RFLP) analysis, however, is a rapid method that can identify and track individual taxa and has already been used to study the archaeal microflora of termite guts (18).
In T-RFLP analysis specific groups of organisms have terminal restriction fragments (T-RFs) of a characteristic size, and so the composition of a whole community, or a subgroup of the community can be identified (23, 28). However, when used to analyze entire, complex microbial communities such as those in termite guts, T-RFLP can give a large number of peaks that make accurate peak identification and interpretation difficult. In contrast, when used to identify and track a subset of the community, such as the methanogens, this method can be used to a high degree of resolution on a large number of samples.
We describe here the use of 16S rRNA gene sequencing and T-RFLP analysis to examine methanogenic archaea in the guts and food-soil of Cubitermes fungifaber, a soil-feeding termite that is widely distributed in areas of disturbed tropical forests, where it encounters and ingests a variety of soil types. If methanogens in guts are strictly vertically inherited, then the methanogen communities should be the same in all individual termites, even if the archaea present in the soil differ from site to site. The reverse should be true (methanogens in guts should reflect what is present in the soil) if new populations are constantly ingested as the insects forage. T-RFLP profiles should allow us to distinguish between the two hypotheses.
MATERIALS AND METHODS
Sample collection.
We collected samples from nine sites in southern Cameroon, West Africa. All sites were closed-canopy forest but included a range of soil types. These soils were characterized by pH (measured from a 1:1 soil-deionized water suspension) (40), qualitative mineralogy (13), and color (Munsell soil color charts) (1).
We identified the characteristic mushroom-shaped termite mounds of C. fungifaber and gathered soil samples from directly beneath the mounds but away from any termite tunnels or galleries. We removed the guts of worker termites and stored them in liquid N2 within 3 to 4 h of collection. We also stored voucher specimens in 100% ethanol in order to confirm termite identification. Samples were replicated at one site to evaluate intrasite variability. Upon our return to the United Kingdom, we stored samples at −70°C until DNA was extracted.
DNA extraction from soils and termite guts.
We extracted DNA from the soil (∼0.5 g [wet weight]) and whole termite guts (in batches of 10) by using the hydroxyapatite spin-column method (30). Initially, this did not give DNA that we could amplify consistently using PCR, and so the method was revised. Prior to the Sephadex G-75 desalting step the eluted DNA was passed through a 0.5-ml polyvinylpolypyrrolidone spin column (2). This step significantly reduced humic contamination, although it also resulted in a reduction in DNA yield. We visualized extracted DNA by ethidium bromide-stained agarose gel electrophoresis (1.4% (wt/vol) agarose gel in 1× Tris-acetate-EDTA (TAE) buffer (33). We excised gel slices containing DNA fragments of >5,000 bp and extracted the DNA by using the QiaQuick gel extraction kit (Qiagen, Crawley, West Sussex, United Kingdom) and stored it in 10 mM Tris buffer (pH 7.5) at −20°C until use. We determined whether the DNA could be used in PCR by amplification with the general bacterial primers Epsilon and 1541R as described previously (32).
Amplification and analysis of euryarchaeal 16S rRNA genes in termite guts and soils.
In order to specifically target the methanogen community in the termite guts and soils, we used euryarchaeal specific primers that target all of the known MA diversity to amplify the DNA (16, 26). This approach requires a heminested PCR with the primers 1Af and 1404R in a touchdown PCR in the first round. We purified this PCR product by using the Qiagen QiaQuick system. In the second round of PCR we amplified this purified product with 1Af and 1100R for the clone library and 1Af and 1100R-FAM for T-RF analysis (for details, see M. A. Munson et al. [26]), and the products were purified as described above. In both rounds we used multiple PCRs that were bulked prior to purification to avoid stochastic biases in individual PCRs (42). The labeled primer (1100R-FAM) is some 180 bp farther along the 16S rRNA gene than other archaeal primers used in T-RFLP analysis (17, 18).
To identity the archaeal communities in the termites and soils and to identify peaks detected in our T-RFLP analysis, a 16S rRNA gene clone library was produced from the termite guts and soil from site B, Kribi. We cloned the purified PCR product from the secondary PCR into pGEM-T Easy (Promega) and then blue-white screened after transformation into DH5α maximum-efficiency competent cells (Life Technologies). We checked putatively positive clones by amplification with the vector-based primers M13f and M13R. Clones containing a fragment of the correct size we partly sequenced by using M13R. We aligned the resulting sequences (50 clones from the guts and 71 from the soil) with reference taxa and available environmental clones within the Genetics Database Environment that is distributed by the Ribosome Database Project (25) and analyzed them by using PAUP (38). We generated a neighbor-joining tree, rooted by using the sequence from Thermoplasma acidophilum, based on about 300 nucleotides for both forward and reverse directions and grouped the clones. From this tree, we chose 85 representative clones (34 from the termite guts and 51 from the soil) and fully sequenced them. We aligned the complete sequences, reference taxa, and available environmental clones, particularly those from a previous study on Archaea in C. orthognathus (18), in the ARB sequence analysis environment (www.arb-home.de). The shorter length (∼800 bp) of the C. orthognathus (18) and rice soil clones (10) reduced the number of bases we could analyze, and so we used 718 aligned positions from 76 taxa for further analysis. We calculated pairwise distances for all alignable sites by using the Logdet/Paralinear distances method (21, 24) as described previously (31). Logdet/Paralinear distances assume all sites can vary, which is not true for most datasets. Thus, we estimated the number of variable sites in the alignment by using a two-state (variable and invariable) maximum-likelihood model in PAUP, and subsequent phylogenetic analysis was limited to only variable positions (63% of sites). We evaluated the tree by bootstrap analysis (100 replicates). We also calculated Logdet/Paralinear distances trees for a smaller data set by utilizing the full length of our clones (53 taxa, 1,005 positions [60% of which were analyzed]) and a data set including methanogen-related clones from other termites (89 taxa, 434 positions of which 60% were analyzed) in order to compare trees to determine whether any topological changes occurred. Once we had identified clones by phylogenetic analysis we determined the clones' resultant T-RF by in silico analysis and verified these computer-based analyses on representative clones by T-RFLP analysis.
We quantified fluorescently labeled PCR products from the termite guts and soil samples by comparison with size markers of known quantities (BioLine, London, United Kingdom) and digested ca. 60 ng of total DNA by using the 4-bp cutter TaqI (Promega, Southampton, United Kingdom) as described by the manufacturers. We ran a portion of the cut PCR product from each sample on a 4% NuSieve 3:1 agarose gel (Cambrex Bioscience, Wokingham, United Kingdom) in 1× TAE at 6 V cm−1 for 2 h to determine whether the product had cut effectively and to compare restriction patterns between termite guts and soils from the five sites. We ran about 2 ng of the restricted PCR product on an ABI 377 automatic sequencer (Sequencing Facility, Natural History Museum, London, United Kingdom) with Genescan 2500 ROX specific size markers (ABI, Warrington, United Kingdom). We identified T-RF fragments with sizes of from 50 to 1,060 bp and peak heights of ≥100 fluorescence units by using the Genescan and Genotyper software (ABI). We analyzed the T-RFLP patterns detected and determined the relationships between the termite gut and soil archaeal communities as described by Dunbar et al. (14). This involved standardizing the total peak area detected to the smallest sample. The presence of peaks exceeding a minimum peak area were then used in a nearest neighbor cluster analysis with a distance matrix derived by using the Jaccard coefficient to determine T-RFLP pattern similarities on a purely qualitative basis.
Nucleotide sequence accession numbers.
16S rRNA gene sequence data has been deposited in GenBank under accession numbers AY487177 to AY487207.
RESULTS
Sample collection and soil typing.
We collected C. fungifaber termites and associated soil samples from nine different sites throughout south eastern Cameroon (a region covering ca. 10,000 km2) and determined soil pH, texture, and color. We chose paired samples of termites and soil from five of these sites, spanning the full range of soil properties, for further study, including three replicate samples collected from site D (at Nkometou) (Table 1).
TABLE 1.
Soil type and characteristics for the samples collected and used in this study
| Site | Soil
|
||
|---|---|---|---|
| pH | Texture | Munsell soil colora | |
| A (Bitylli) | 3.9 | Silty clay | 10YR 4/4 (dark yellowish brown) |
| B (Kribi) | 4.0 | Sandy loam | 10YR 4/4 (dark yellowish brown) |
| C (Ekok) | 4.1 | Clay | 10YR 3/4 (dark yellowish brown) |
| D-1 (Nkometou) | 4.6 | Sandy clay | 5YR 3/2 (dark reddish brown) |
| D-2 (Nkometou) | 5.4 | Sandy clay | 5YR 3/2 (dark reddish brown) |
| D-3 (Nkometou) | 4.4 | Sandy clay | 5YR 3/2 (dark reddish brown) |
| E (Mbalmayo) | 5.2 | Clay loam | 5YR 3/1 (very dark gray) |
See reference 1.
DNA extraction and PCR amplification of 16S rRNA gene sequences.
We successfully extracted DNA from all termite gut and soil samples. Extraction yields (determined by visual analysis in comparison to DNA size markers [Bioline]) were similar in all cases, giving ca. 6 μg of DNA g (wet weight) of soil−1 and 1 μg of DNA per 10 termite guts.
Phylogenetic analysis of cloned archaeal 16S rRNA genes.
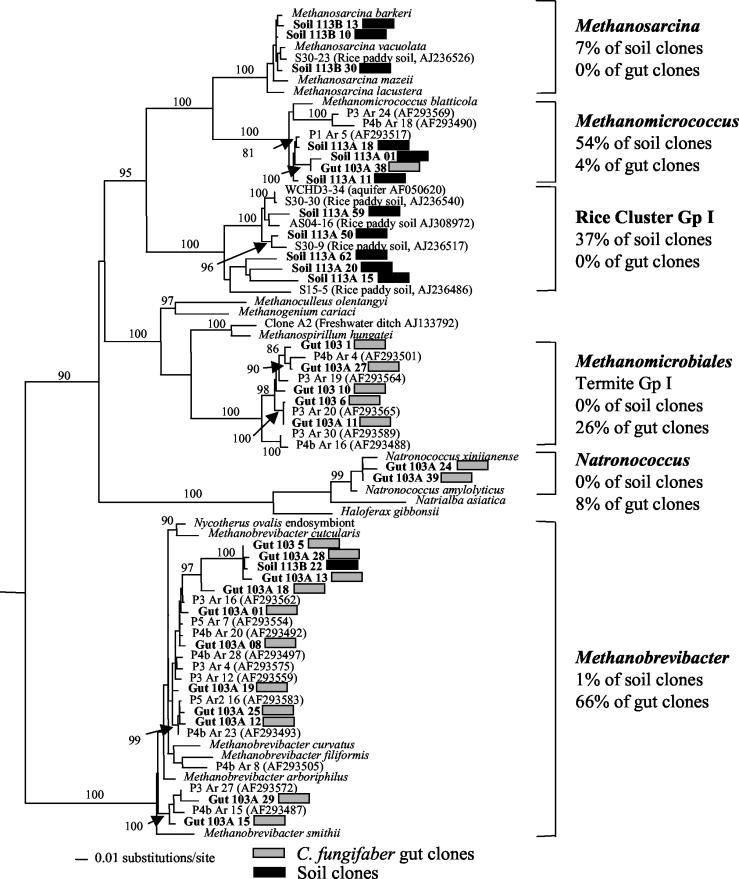
We obtained 121 clones containing 16S rRNA gene sequences from guts and soil from site B (Kribi), 50 from C. fungifaber guts, and the rest from soil. Phylogenetic analysis of ∼300 nucleotide positions of each clone indicated that some of the sequences were very similar. We obtained complete sequences for 85 representative clones (34 from guts [18 shown in Fig. 1 ] and 51 from soil [12 shown in Fig. 1]). The identities, proportions, and T-RF values are shown for these clones in Table 2. The phylogenetic relationships of representative 16S rRNA gene sequences from C. fungifaber guts and food-soil to those of cultivated organisms and other environmental clones are shown in Fig. 1. The LogDet/Paralinear distances method used to infer this tree work optimally if all sequences are the same length. Although our sequences were all >1,000 bp, we limited the analysis to 718 nucleotide positions in order to include relevant shorter sequences from other studies (10, 18). Phylogenetic relationships inferred from analysis of our full-length data, as well those based on a shorter data set that included methanogen sequences from other termites, were essentially identical to those in Fig. 1 (data not shown).
FIG. 1.
Inferred phylogenetic relationships between archaeal C. fungifaber gut and soil sequences from site B (Kribi) reference taxa and other environmental clones. This is a Logdet/Paralinear distance tree based on 61% (the estimated number of variable sites) of 718 alignable nucleotides. Bootstrap (100 replicates) values of >70% are shown at the nodes. Gp, group.
TABLE 2.
Identification of clones from the termite guts and soils from site B (Kribi)
| Clone(s) in treea | T-RF | No. of clones | Closest isolated relative | % Similarity | Cluster identity (% of clone library)b |
|---|---|---|---|---|---|
| Guts (All gut 103) | |||||
| A08, A19 (plus others) | 994-996 | 13 | Methanobrevibacter filiformis | 92-95 | |
| 5, A13, A18, A28 | 993-1005 | 4 | Methanobrevibacter filiformis | 92-95 | |
| A12, A25 | 269 | 2 | Methanobrevibacter filiformis | 92-95 | Methanobrevibacter spp. (66) |
| A01 | 571 | 1 | Methanobrevibacter filiformis | 92-95 | |
| A15, A29 (plus others) | 994-998 | 13 | Methanobrevibacter smithii | 94 | |
| 6, 10, A11 (plus others) | 569-571 | 6 | Methanospirillum hungatei | 89 | Termite archaeal group I (22) |
| 1, A27 (plus others) | 569-570 | 5 | Methanospirillum hungatei | 89 | |
| A24, A39 (plus others) | 118-119 | 4 | Natronococcus amyloticus | 97 | Natronococcus (8) |
| A38 (plus one other) | 1008 | 2 | Methanomicrococcus blatticola | 97 | Methanomicrococcus (4) |
| Soil (All soil 113) | |||||
| A11, A18 (plus others) | 1010-1013 | 20 | Methanomicrococcus blatticola | 96 | Methanomicrococcus (46) |
| A01 (plus others) | 1011 | 12 | Methanomicrococcus blatticola | 96 | |
| A15, A20 (plus others) | 118 | 22 | Methanosarcina lacustera | 82-84 | |
| A50, A59 (plus others) | 118 | 9 | Methanosarcina lacustera | 82-84 | Rice cluster I (45) |
| A62 | 570 | 1 | Methanosarcina lacustera | 82-84 | |
| B10, B13 (plus others) | 363 | 5 | Methanosarcina barkeri | 98 | Methanosarcina (8) |
| B30 | No cut | 1 | Methanosarcina vacuolata | 98 | |
| B22 | 999 | 1 | Methanobrevibacter filiformis | 92 | Methanobrevibacter (1) |
See Fig. 1.
For guts, 50 clones; for soil, 71 clones.
A wide diversity of euryarchaeal 16S rRNA gene sequences were detected from both guts and soil, but there was almost no overlap between the two clone libraries (Fig. 1). All but one of the soil clone sequences were related to the Methanosarcinales. In addition to clones specifically related to Methanosarcina, this clade included environmental sequences from “rice cluster I” (RC I [17]) and clones related to Methanomicrococcus blatticola, an isolate from the gut of the cockroach Periplaneta americana (37). Two clones from the guts of C. fungifaber (represented by gut 103A 38 in Fig. 1) were also related to M. blatticola. Almost all of the other 16S rRNA gene sequences recovered from the guts of C. fungifaber were either related to Methanomicrobiales or to the genus Methanobrevibacter. An exception was the surprising and close relationship between four gut clones and Natronococcus, an obligately extreme haloalkaliphile. The C. fungifaber gut clones related to Methanomicrobiales sequences were part of an exclusively termite-related clade that included clones from the P3 and P4 segments of C. orthognathus termite guts, as well as clones from Nasutitermes takasogoensis and Pericapritermes nitobei guts (27). We henceforth refer to this cluster as termite archaeal group I. Similarly, C. fungifaber clones related to Methanobrevibacter species clustered with C. orthognathus clones from gut sections P3, P4, and P5 (18). There was a single soil clone (soil 113B 22) that was also related to the Methanobrevibacter.
T-RFLP analysis of Euryarchaea in termite guts and soils.
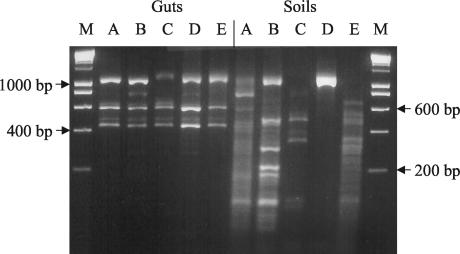
PCR product from each gut and soil sample was digested with TaqI and an aliquot run on an ethidium bromide-stained agarose gel (Fig. 2). The restriction patterns for the termite gut samples show a remarkable similarity, whereas the soil samples are all distinctly different.
FIG. 2.
TaqI restriction analysis of euryarchaeal PCR products from C. fungifaber gut and soil samples from five different tropical soils on a 4% NuSieve (3:1) ethidium bromide-stained agarose gel in 1× TAE. M, Hyperladder I (Bioline). Sites are labeled A to E as in Table 1.
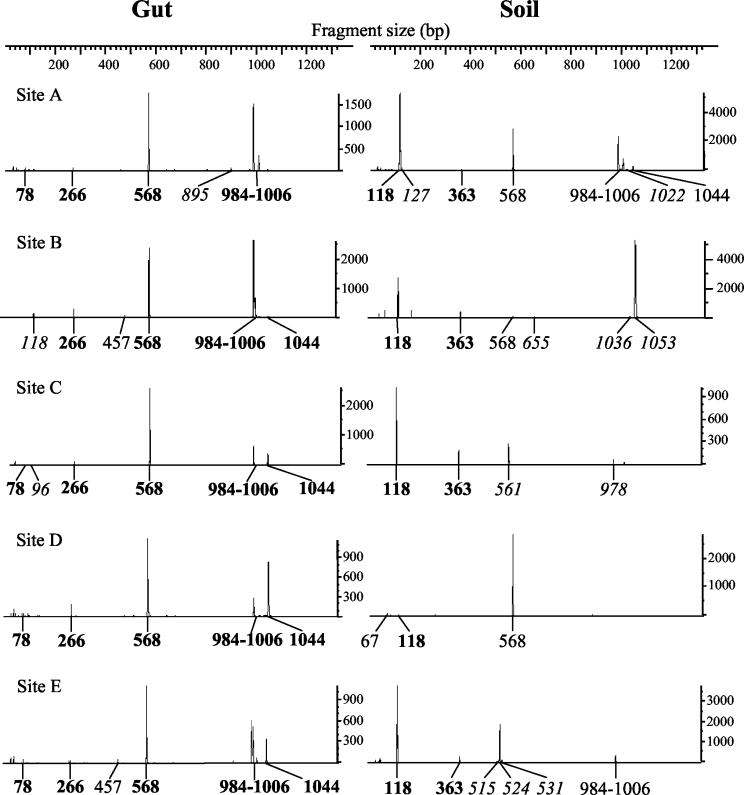
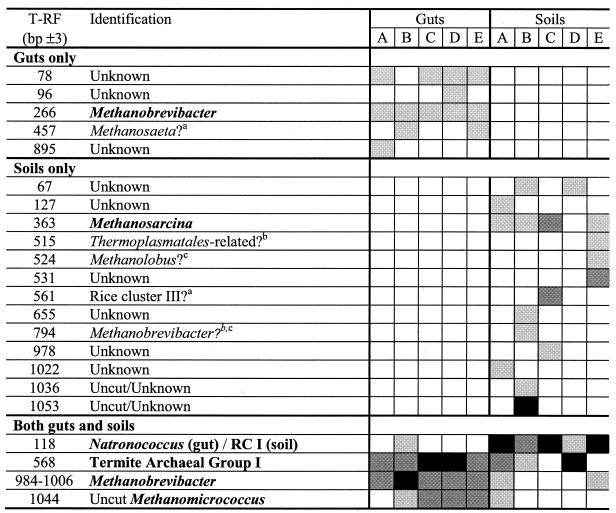
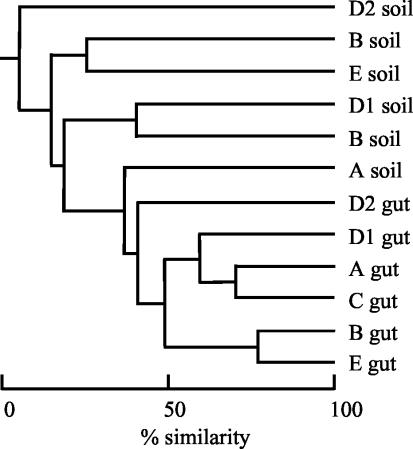
T-RFLP analysis of the gut and soil samples indicated that the samples are diverse with numerous peaks in almost all samples tested (Fig. 3). After peak area standardization by the method of Dunbar et al. (14) and comparison to in silico restriction digestion of our clones sequences, we identified 22 T-RFLP groups (T-RF peaks) that were used in subsequent analysis (Table 3). Of these peaks, 9 were found in termite guts and 17 were found in soil samples. Five peaks (T-RF 266, Methanobrevibacter; T-RF 568, termite archaeal group I; T-RF 984-1006, Methanobrevibacter; unidentified T-RF 78; and uncut PCR product 1044) were detected in gut samples from at least four of the five sites. The majority of the peaks found exclusively in soil samples were only detected once (11 of 13). Only two peaks were detected in at least four soil samples (T-RF 118 [RC I/Natronococcus] and T-RF 363 [Methanosarcina]). Four peaks were found in both gut and soil samples, but in two cases the peak was detected in only a single gut or soil sample. Cluster analysis of the T-RFLPs showed the gut profiles from the five sites forming a single cluster, whereas the soil profiles were not clearly related to each other (Fig. 4). In summary, T-RFLP profiles of the termite gut samples were less diverse and showed greater similarity than those from the soil samples.
FIG. 3.
T-RFLP profiles from gut and soil samples from five sites. T-RFs in boldface are those found in ≥4 of the 5 gut or soil samples; those in italics are unique to that sample. T-RF 984-1006 is a combined group since these all appear to be related to Methanobrevibacter.
TABLE 3.
Representation of the T-RFLP data from all five sitesa
Identifications in boldface represent T- RFs identified in our sequence analysis; others are tentatively based on T-RF values quoted in the literature or from in silico analysis of sequence data. Lightly shaded boxes are T-RFs that represent <10%, medium shaded boxes are T-RFs that represent 10 to 50%, and dark-shaded boxes are T-RFs that represent >50% of the total area measured in each analysis. The primers we used produce T-RFs that are approximately 180 bp longer than those in Friedrich et al. (18). Tentative identifications were based on T-RF data from Fey and Conrad (17) (superscript a), Friedrich et al. (18) (superscript b), or in silico analysis of sequence data (superscript c).
FIG. 4.
Jaccard cluster analysis (group average link) of T-RFLP analysis data from soil and termite guts samples from the five sites analyzed. The data were analyzed as described by Dunbar et al. (14).
Half of the detected peaks were either identified by our cloning analysis (indicated in boldface in Table 3) or have been identified previously (10, 17) and are tentatively identified here. Interestingly, the peak detected in all of the soil samples, T-RF 118, was identified as the novel Methanosarcinales family RC I (10, 17) by our sequence analysis. With the primer set more generally used for T-RFLP analysis of Archaea (18), this group would have given a T-RF of 389, coinciding with the termite archaeal group I peak in the termite guts (T-RF 388 [our T-RF of 568 minus 180 bp]), and we would have failed to separate the two groups.
DISCUSSION
Methanogen community composition in termite guts and in soil.
Our intention at the onset was to test the hypotheses that the methanogen community in the guts of C. fungifaber was either derived from their food-soil or vertically transmitted from termite to termite. In our analysis, there is a distinct difference between the methanogen community in the guts of termites and from the soil in all five sites tested. Thus, C. fungifaber maintains a gut methanogen community that is different from its food-soil and does not even appear to be a substantial subset of the soil methanogen diversity. We can now speculate further about the relationship between termites and methanogens. Is the methanogen community in the termite guts vertically transmitted between generations and is there support for coevolution of the termites and methanogens? Although there were strong similarities between termite gut contents from the five sites, our analysis did not reveal identical gut microbiota in these termites that would have been suggestive of coevolution between the host and its symbionts. Thus, neither of the two extreme explanations of the origin of methanogens in C. fungifaber guts, that of purely environmental or that of a purely coevolved community, appear to be true. The most likely solution is a mix of the two with some coevolved and some environmental groups. The apparently termite-specific termite archaeal group I is the clade that, on the basis of data presented here, is the best candidate for a termite-specific subset of the community, and this putative evolutionary relationship will be investigated in future studies.
Analysis of the methanogen communities in termite guts and food-soils.
In analyses of communities in situ, it is vital that the methodology utilized is robust and accurately reflects the community under study. There is strong support in the literature to suggest that T-RFLP analysis is such a method, but the interpretation of results can be difficult (28). T-RFLP analysis of the termite guts and soils suggested some overlap between the guts and soils. However, on further analysis the overlaps between the guts and soils may be less significant than is at first apparent. Of the four T-RF peaks that we found in both guts and soils, two are represented in only a single gut or soil sample. T-RF 118 is a diphyletic fragment that represents both a number of obligately halophilic archaea, including Natronococcus spp., and also the soil group RC I. Clone-based identification suggested that the T-RF detected in the site B gut represented Natronococcus spp. and that detected in the soil samples was from soil group RC I, which would separate this T-RF into gut- and soil-specific groups. T-RF 1044 represents uncut PCR product, and thus any identification must be considered very tentative. The other two T-RFs, 568 and 984-1006, could genuinely represent methanogens that are present in both soil and guts. Both of these peaks represent important members of the termite gut community. Due to the sheer abundance of soil-feeding termites in this habitat, it is likely that the soil had been frequently modified by termites. Thus, organisms that are usually resident in termite guts may be transiently present in termite-digested soil. Similarly, the detection of clones related to Methanomicrococcus in the termite gut clone library (Fig. 1) may be derived from soil simply passing through the gut. However, there is an alternative explanation in the case of T-RF 568. One clone, 113 A62, was found in the soil from site B that had an aberrant T-RF. This clone clusters in RC I and should have a T-RF of 118; instead, it has a T-RF of 570, placing it within the termite specific T-RF 568 cluster. Analysis of this clone's sequence revealed a 2-bp change (GA to TC) at the Taq1 restriction site that would have produced a T-RF of 118. Such a change is unlikely to be due to PCR or sequencing errors and thus may represent a real RC I community with a T-RF of 568. However, if these gut-associated groups are genuinely active in soil, this would indicate that these two groups represent an environmental pool of methanogens available to termites.
We detected a number of T-RFs that we could not identify from our clone libraries but that were consistent with those identified previous studies (Table 3). An unidentified peak (457 bp) in the site A and C gut samples corresponds to the T-RF of the obligate-acetate-utilizing methanogen Methanosaeta (17). The peak at 524 bp in soil from site A may be associated with the C1 compound-utilizing methanogen Methanolobus and the 561-bp peak matches the T-RF of a rice paddy soil T-RF named RC III by Fey and Conrad (17). However, we have no confirmation that these peaks do correspond to these groups, and these identifications must be considered to be very tentative and need to be confirmed by cloning and sequencing.
We also detected a number of T-RFs that we could not identify. Most of these (six of nine) were in the soil samples (Table 3). The most prevalent unidentified peak was T-RF 78, which was detected in four of the five gut samples. Without an identification we cannot speculate on the identity or function of the organisms from which these peaks were derived except to say their 16S rRNA genes were amplifiable with primers specific for Archaea.
We detected a T-RF in soil from site E that may be from the Thermoplasmatales-related group detected by Friedrich et al. in C. orthognathus (18). This Thermoplasmatales-related group has been detected in a variety of different environments (41), including estuarine sediments, with our primer set (26), and so we could have reasonably expected to detect this group in the guts of C. fungifaber with our primers. Therefore, the most likely explanation of our inability to detect clones related to this group in our gut samples is that it represents a difference in archaeal communities between the two Cubitermes species.
The power of using T-RFLP in the analysis of methanogens in termite guts can be clearly seen in the overlap between our analyses and those of Friedrich et al. with C. orthognathus (18). Our analysis, with a primer set different from that of Friedrich et al., recovered all of the methanogen groups they detected, as well as several clones related to the obligately halophilic Natronococcus spp. We extracted DNA from whole guts and recovered all of the euryarchaeal groups that Friedrich et al. detected in their analysis of separate gut sections, with the exception of the group distantly related to the Thermoplasmatales. This suggests that our more-complex, heminested PCR approach is an effective method for the analysis of whole termite guts.
A novel microbe in invertebrate guts.
An interesting finding of the present study was the detection of clones closely related to the obligately haloalkaliphilic Natronococcus in the clone library from the termite guts. The T-RF for this group (118 bp) was detected in the gut samples from site B. (The detection of the same T-RF in all five soil samples was probably due to the presence of RC I-related taxa rather than Natronococcus [based on our clone data].) Halophilic Archaea are obligate extreme halophiles and often do not survive when salinities fall to <10% (wt/vol; 1.5 M NaCl). Some parts of the Cubitermes gut reach pH 12 (9), which may well favor alkaliphilic organisms, but there is no indication that the guts are extremely saline as well. The finding of these clones, then, is a conundrum. Halophile related clones have been detected in estuarine sediments (26), and members of the halophile community in these estuarine sediments have been isolated and appear to grow at seawater salinities (29). Thus, it may be that some organisms within the halophiles' radiation are capable of growth and survival under nonextreme conditions, and this may be the case here. We are trying to bring these organisms into culture in order to show that they genuinely exist within termite guts.
The case for coevolution between termites and their methanogen communities.
Despite minor differences detected in the gut archaeal communities between the savannah-dwelling east African C. orthognathus and the forest-dwelling west African C. fungifaber, there is a substantial amount of agreement between the two analyses. It therefore seems that methanogen communities within soil-feeding termite guts may be very stable across species. This is even more striking given that the methanogen community in the soils they feed on are so diverse. Our data do not support the hypothesis that methanogens in termite guts are derived from their food-soil, although it is possible that they are present in soil but below our detection limit. Whether such a community is active in soil or merely a transient termite-derived community is a moot point; the ability to survive and successfully inoculate a termite gut is the primary issue and is a matter beyond the scope of the present study. Our data also do not support a purely vertical transmission of gut microflora. However, it does appear that at least one group of methanogens (termite archaeal group I) is found only in termite guts, and these are probably prime candidates for a coevolutionary relationship with termites. It is also pertinent to ask, given the differences in the gut microflora of soil- and wood-feeding termites, what role the development of specialized methanogen assemblages plays in the evolution of soil-feeding in the higher termites.
Acknowledgments
This study was funded by a grant (NER/B/S/2000/00703) to P.E. and K.J.P. by the Natural Environment Research Council, United Kingdom. The sampling trip to Cameroon was funded by the Natural History Museum, London, United Kingdom, and the U.S. National Science Foundation.
Any opinion, findings and conclusions or recommendations expressed in this study are those of the authors and do not necessarily reflect the views of the National Science Foundation.
We thank the Sequencing Unit at the Natural History Museum, London, United Kingdom, and one anonymous reviewer for comments that clarified and improved the manuscript.
REFERENCES
- 1.Anonymous. 2000. Munsell soil colour charts. Gretag-Macbeth, New Windsor, N.Y.
- 2.Berthelet, M., L. G. Whyte, and C. W. Greer. 1996. Rapid, direct extraction of DNA from soils for PCR analysis using polyvinylpolypyrrolidone spin columns. FEMS Microbiol. Lett. 138:17-22. [DOI] [PubMed] [Google Scholar]
- 3.Bignell, D., and P. Eggleton. 2000. Termites in ecosystems, p. 363-387. In T. Abe, M. Higashi, and D. Bignell (ed.), Termites: evolution, sociality, symbiosis, ecology. Kluwer Academic Press, Dordrecht, The Netherlands.
- 4.Bignell, D. E., P. Eggleton, L. Nunes, and K. L. Thomas. 1997. Termites as mediators of carbon fluxes in tropical forest: budgets for carbon dioxide and methane emissions, p. 109-134. In A. D. Watt, N. E. Stork, and M. D. Hunter (ed.), Forests and insects. Chapman & Hall, Plc., London, United Kingdom.
- 5.Brauman, A. 2000. Effect of gut transit and mound deposit on soil organic matter transformations in the soil-feeding termite: a review. Eur. J. Soil Biol. 36:117-125. [Google Scholar]
- 6.Brauman, A., J. Dore, P. Eggleton, D. Bignell, J. A. Breznak, and M. D. Kane. 2001. Molecular phylogenetic profiling of prokaryotic communities in guts of termites with different feeding habits. FEMS Microbiol. Ecol. 35:27-36. [DOI] [PubMed] [Google Scholar]
- 7.Brauman, A., M. D. Kane, M. Labat, and J. A. Breznak. 1992. Genesis of acetate and methane by gut bacteria of nutritionally diverse termites. Science 257:1384-1387. [DOI] [PubMed] [Google Scholar]
- 8.Breznak, J. A. 2000. Ecology of prokaryotic microbes in the guts of woodland litter-feeding termites, p. 209-232. In T. Abe, M. Higashi, and D. Bignell (ed.), Termites: evolution, sociality, symbiosis, ecology. Kluwer Academic Press, Dordrecht, The Netherlands.
- 9.Brune, A., and M. Kuhl. 1996. pH profiles of the extremely alkaline hindguts of soil-feeding termites (Isoptera: Termitidae) determined with microelectrodes. J. Insect Physiol. 42:1121-1127. [Google Scholar]
- 10.Chin, K. J., T. Lukow, and R. Conrad. 1999. Effect of temperature on structure and function of the methanogenic archaeal community in an anoxic rice field soil. Appl. Environ. Microbiol. 65:2341-2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davies, R. G., P. Eggleton, D. T. Jones, F. J. Gathorne-Hardy, and L. M. Hernández. 2003. Evolution of termite functional diversity: analysis and synthesis of local ecological and regional influences on local species richness. J. Biogeogr. 30:847-877. [Google Scholar]
- 12.Donovan, S. E., P. Eggleton, and D. E. Bignell. 2001. Gut content analysis and a new feeding group classification of termites. Ecol. Entomol. 26:356-366. [Google Scholar]
- 13.Dubbin, W. E. 2001. Soils. Natural History Museum, London, England.
- 14.Dunbar, J., L. O. Ticknor, and C. R. Kuske. 2001. Phylogenetic specificity and reproducibility and new method for analysis of terminal restriction fragment profiles of 16S rRNA genes from bacterial communities. Appl. Environ. Microbiol. 67:190-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eggleton, P., and I. Tayasu. 2001. Feeding groups, lifetypes, and the global ecology of termites. Ecol. Res. 16:941-960. [Google Scholar]
- 16.Embley, T. M., B. J. Finlay, R. H. Thomas, and P. L. Dyal. 1992. The use of rRNA sequences and fluorescent probes to investigate the phylogenetic positions of the anaerobic ciliate Metopus palaeformis and its archaeobacterial endosymbiont. J. Gen. Microbiol. 138:1479-1487. [DOI] [PubMed] [Google Scholar]
- 17.Fey, A., and R. Conrad. 2000. Effect of temperature on carbon and electron flow and on the archaeal community in methanogenic rice field soil. Appl. Environ. Microbiol. 66:4790-4798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Friedrich, M. W., D. Schmitt-Wagner, T. Lueders, and A. Brune. 2001. Axial differences in community structure of Crenarchaeota and Euryarchaeota in the highly compartmentalized gut of the soil-feeding termite Cubitermes orthognathus. Appl. Environ. Microbiol. 67:4880-4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kane, M. D., and U. G. Mueller. 2002. Insights from insect-microbe symbioses, p. 289-313. In J. T. Staley and A.-L. Reysenbech (ed.), Biodiversity of microbial life: foundation of earth's biosphere. Wiley-Liss, Inc., New York, N.Y.
- 20.Kappler, A., and A. Brune. 1999. Influence of gut alkalinity and oxygen status on mobilization and size-class distribution of humic acids in the hindgut of soil-feeding termites. Appl. Soil Ecol. 13:219-229. [Google Scholar]
- 21.Lake, J. A. 1994. Reconstructing evolutionary trees from DNA and protein sequences: paralinear distances. Proc. Natl. Acad. Sci. USA 91:1455-1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lavelle, P., D. Bignell, M. Lepage, V. Wolters, P. Roger, P. Ineson, O. W. Heal, and S. Dhillion. 1997. Soil function in a changing world: the role of invertebrate ecosystem engineers. Eur. J. Soil Biol. 33:159-193. [Google Scholar]
- 23.Liu, W., T. L. Marsh, H. Cheng, and L. Forney. 1997. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl. Environ. Microbiol. 63:4516-4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lockhart, P. J., M. A. Steel, M. D. Hendy, and D. Penny. 1994. Recovering evolutionary trees under a more realistic model of sequence evolution. Mol. Biol. Evol. 11:605-612. [DOI] [PubMed] [Google Scholar]
- 25.Maidak, B. L., N. Larsen, J. McCaughey, R. Overbeek, G. J. Olsen, K. Fogel, J. Blandy, and C. R. Woese. 1994. The ribosomal database project. Nucleic Acids Res. 22:3485-3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Munson, M. A., D. B. Nedwell, and T. M. Embley. 1997. Phylogenetic diversity of Archaea in sediment samples from a coastal salt marsh. Appl. Environ. Microbiol. 63:4729-4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ohkuma, M., S. Noda, and T. Kudo. 1999. Phylogenetic relationships of symbiotic methanogens in diverse termites. FEMS Microbiol. Lett. 171:147-153. [DOI] [PubMed] [Google Scholar]
- 28.Osborn, A. M., E. R. B. Moore, and K. N. Timmis. 2000. An evaluation of terminal-restriction fragment length polymorphism (T-RFLP) analysis for the study of microbial community structure and dynamics. Environ. Microbiol. 2:39-50. [DOI] [PubMed] [Google Scholar]
- 29.Purdy, K. J., T. Cresswell-Maynard, D. B. Nedwell, T. J. McGenity, W. D. Grant, K. N. Timmis, and T. M. Embley. 2004. Isolation of haloarchaea that grow at low salinities. Environ. Microbiol. 591-595. 6: [DOI] [PubMed]
- 30.Purdy, K. J., T. M. Embley, S. Takii, and D. B. Nedwell. 1996. Rapid extraction of DNA and rRNA from sediments using a novel hydroxyapatite spin-column method. Appl. Environ. Microbiol. 62:3905-3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Purdy, K. J., M. A. Munson, D. B. Nedwell, and T. M. Embley. 2002. Comparison of the molecular diversity of the methanogenic community at the freshwater and marine ends of a UK estuary. FEMS Microbiol. Ecol. 39:17-21. [DOI] [PubMed] [Google Scholar]
- 32.Purdy, K. J., D. B. Nedwell, and T. M. Embley. 2003. Analysis of the sulfate-reducing bacterial and methanogenic archaeal populations in contrasting Antarctic sediments. Appl. Environ. Microbiol. 69:3181-3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 34.Schmitt-Wagner, D., and A. Brune. 1999. Hydrogen profiles and localization of methanogenic activities in the highly compartmentalized hindgut of soil-feeding higher termites (Cubitermes spp.). Appl. Environ. Microbiol. 65:4490-4496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmitt-Wagner, D., M. W. Friedrich, B. Wagner, and A. Brune. 2003. Axial dynamics, stability, and interspecies similarity of bacterial community structure in the highly compartmentalized gut of soil-feeding termites (Cubitermes spp.). Appl. Environ. Microbiol. 69:6018-6024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmitt-Wagner, D., M. W. Friedrich, B. Wagner, and A. Brune. 2003. Phylogenetic diversity, abundance, and axial distribution of bacteria in the intestinal tract of two soil-feeding termites (Cubitermes spp.). Appl. Environ. Microbiol. 69:6007-6017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sprenger, W. W., M. C. van Belzen, J. Rosenberg, J. H. Hackstein, and J. T. Keltjens. 2000. Methanomicrococcus blatticola gen. nov., sp. nov., a methanol and methylamine-reducing methanogen from the hindgut of a cockroach Periplaneta americana. Int. J. Syst. Evol. Microbiol. 50:1989-1999. [DOI] [PubMed] [Google Scholar]
- 38.Swofford, D. L. 1998. PAUP*: phylogenetic analysis using parsimony (*and other methods), 4.0b4a ed. Sinauer Associates, Sunderland, Mass.
- 39.Tholen, A., and A. Brune. 1999. Localization in in situ activities of homoacetogenic bacteria in the highly compartmentalized hindgut of soil-feeding higher termites (Cubitermes spp.). Appl. Environ. Microbiol. 65:4497-4505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thomas, G. W. 1996. Soil pH and acidity, p. 475-490. In D. L. Sparks (ed.), Methods of soil analysis. Part 3. Chemical methods. American Society of Agronomy, Madison, Wis.
- 41.Vetriani, C., A. L. Reysenbach, and J. Dore. 1998. Recovery and phylogenetic analysis of archaeal rRNA sequences from continental shelf sediments. FEMS Microbiol. Lett. 161:83-88. [DOI] [PubMed] [Google Scholar]
- 42.Wagner, A., N. Blackstone, P. Cartwright, M. Dick, B. Misof, P. Snow, G. P. Wagner, J. Bartels, M. Murtha, and J. Pendleton. 1994. Surveys of gene families using polymerase chain reaction: PCR selection and PCR drift. Syst. Biol. 43:250-261. [Google Scholar]