Abstract
Representatives of only four well-characterized bacterial phyla were isolated from a pasture soil by using liquid serial dilution culture. In contrast, members of Acidobacteria, Verrucomicrobia, and Gemmatimonadetes and of other poorly represented bacterial lineages were isolated in earlier experiments with solidified versions of the same media. We conclude that, contrary to expectation, liquid serial dilution culture is inferior to culturing on solid media for isolating representatives of many bacterial phyla from soil.
Conn's description (12) of the inability of microbiologists to culture most soil microorganisms seems as relevant today as when it was published in 1918. The number of culturable cells in soil is often only about 1% of the total number of cells present. What has become apparent more recently is that this discrepancy is not just quantitative but also qualitative. Based on rRNA and rRNA gene abundance, groups of bacteria that are poorly represented by cultured representatives make up numerically significant populations within soil microbial communities (8, 20, 22, 24, 26). Serial liquid dilution culture has been used successfully to improve culturability (6, 9, 10) and facilitate the isolation of molecularly detected bacteria and archaea from freshwater, marine, and anoxic soil systems (11, 14, 25). We postulated that liquid serial dilution should also allow us to isolate representatives of poorly studied groups of bacteria from a nonsaturated soil.
Nine soil cores (25 mm in diameter, 100 mm long) were collected from one sample site at the Dairy Research Institute, Ellinbank, Victoria, Australia (18, 19, 28, 29) in May 1997 (one core), March and April 1998 (1 core each), and April, May, June, and July 1999 (two, one, one, and three cores, respectively). Cores were used individually, and only the lower 70 mm of each was used. Cultivation experiments, preparations for total cell counts, and dry-weight determinations were initiated within 3 h of collection as described elsewhere (18). Total counts of bacteria in the nine soil cores, determined after staining with DAPI (4′,6′-diamidino-2-phenylindole) and counted by fluorescence microscopy on three to five subsamples each (18), were between 1.2 × 109 (standard deviation [SD] = 9 × 107) and 1.6 × 109 (SD = 5 × 107) cells per g of dry soil. The mean count for the nine cores was 1.4 × 109 cells per g of dry soil (SD = 1.4 × 108). There was therefore little variation in the total cell counts in soil samples collected at different times and in different years from the same sample site. Our results are generally reported in terms of culturability, which is the most probable number (MPN) of culturable cells expressed as a percentage of the microscopically determined cell count for the sample used for that particular experiment. Accurately weighed samples of about 1 g of freshly sieved soil were added to 100-ml aliquots of sterile distilled water in 150-ml conical flasks and dispersed by stirring with Teflon-coated magnetic bars (8 mm diameter, 50 mm long) for 15 min at approximately 200 rpm. Three-tube MPN counting experiments (18) were prepared from these suspensions and incubated at 25°C in the dark. Growth in liquid cultures was scored as positive if visible turbidity developed. This was confirmed by microscopic examination of a sample of the culture. The MPN was calculated from the dry weight of soil, the dilution factor, and tables for three parallel dilution series (5).
Dilute nutrient broth (DNB) consisted of Difco nutrient broth (BD Diagnostic Systems, Sparks, Md.) at 0.08 g per liter of distilled water. The final pH was approximately 6.0. Medium VL55 contained 1.95 g of 2-[N-morpholino]ethanesulfonic acid, 0.2 mmol of MgSO4, 0.3 mmol of CaCl2, 0.2 mmol of (NH4)2HPO4, 1 ml of selenite-tungstate solution (31), and 1 ml of trace element solution SL-10 (32) per liter, and the pH was adjusted to 5.5 with a mixture of 200 mM NaOH plus 100 mM KOH. Medium VL70 contained 2.09 g of 3-[N-morpholino]propanesulfonic acid, 0.2 mmol of MgSO4, 0.3 mmol of CaCl2, 0.2 mmol of (NH4)2HPO4, 1 ml of selenite-tungstate solution, and 1 ml of trace element solution SL-10 per liter, and the pH was adjusted to 7.0 with a mixture of 200 mM NaOH plus 100 mM KOH. Media VL55 and VL70 were sterilized by autoclaving, and then the appropriate growth substrate was added from concentrated stocks that had been sterilized by autoclaving, in the case of polymers, or by passing them through Minisart sterile filters (Sartorius AG, Göttingen, Germany) with a pore size of 0.22 μm. The growth substrates and their final concentrations were 0.025% (wt/vol) arabic gum; 0.025% (wt/vol) cellulose powder; 0.025% (wt/vol) chitin; 0.025% (wt/vol) ghatti gum; 0.025% (wt/vol) karaya gum; 0.025% (wt/vol) pectin; 0.025% (wt/vol) sodium alginate; 0.025% (wt/vol) soluble starch; 0.025% (wt/vol) xanthan; 0.025% (wt/vol) xylan; 2 mM glucose; 2 mM methanol; 2 mM sodium acetate; 2 mM sodium benzoate; a mixture of 0.25 mM glucose, 0.5 mM glycine, 0.5 mM sodium acetate, 0.5 mM sodium glutamate, 0.5 mM sodium glycolate, and 0.5 mM sodium l-lactate; and an amino acid mixture (15) with the addition of 0.08 g of l-tryptophan per 100 ml of stock solution and added at 5 ml of stock solution per liter of medium. One milliliter of vitamin solution 1 (17) and 3 ml of vitamin solution 2 (17) were then added per liter of completed media VL55 and VL70.
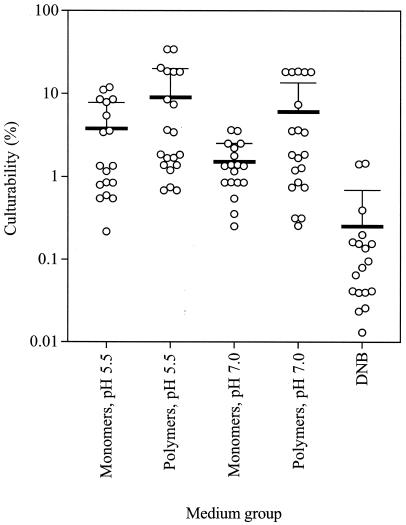
Thirteen of the 94 counting experiments, i.e., 14%, yielded counts that exceeded 10% of the total cell count, and 55 of the 94 counting experiments (59%) resulted in MPNs that exceeded 1% of the total cell count (Fig. 1). MPNs were variable, even when the same medium was used. For example, culturability varied over a 100-fold range with DNB (Fig. 1). The range of culturability on the different media was comparable to the plate counts obtained in earlier studies (18, 28), in which samples were also incubated for 12 weeks, using solid versions of the media used in this study. The highest count, obtained in two of the experiments, was 5.2 × 108 culturable units per g of dry soil, which corresponds to a culturability of 34%. DNB yielded significantly lower culturabilities (P = 2 × 10−5 to 5 × 10−4 in two-tailed Student's t test) than the other media grouped by pH and substrate class (polymers or monomers) (Fig. 1). Sugar polymers as growth substrates yielded significantly higher culturabilities than did different monomers or mixes of monomers (P = 3 × 10−3). It is possible that the transfer of bacteria from the low-nutrient soil environment to high-nutrient laboratory media results in substrate-accelerated death (23, 30), especially when monomers are used as growth substrates. Initially, there are only very low concentrations of available substrates in the media with polymers, and regulatory changes in metabolism can occur as the available substrate concentration increases when the polymers are gradually hydrolyzed by growing bacteria. Similar increases in MPN using polymeric sugar growth substrates instead of readily utilizable simpler sugars have been noted earlier (11).
FIG. 1.
Culturability of bacteria obtained with polymeric and monomeric growth substrates in media at pH 5.5 and 7.0 and DNB as the growth medium. The thick horizontal lines indicate the means, and the error bars represent 1 SD from the mean.
Medium VL55, with a pH of 5.5, which is similar to that of soil (pH 5.3 to 5.5) (29), yielded higher average culturabilities (P = 0.13) (Fig. 1) than medium VL70, which had a pH of 7.0. We repeated this experiment using the five growth substrates that resulted in high MPNs (alginate, arabic gum, pectin, xanthan, xylan; three counting experiments with each substrate at each of the two pHs). Again, media with a pH of 5.5 resulted in higher average culturabilities (P = 0.06) than media with a pH of 7.0.
Eighty-nine isolates were cultured from the terminal growth-positive tubes of serial liquid dilution cultures by streaking the material taken up in an inoculating loop onto the surface of an agar-solidified plate of the same medium as that used for that MPN experiment. Solid media were prepared as described elsewhere (18, 19), so that the final substrate concentration was twice that of the corresponding liquid medium. After incubation at 25°C, visibly different colonies were selected, restreaked, and subcultured until microscopically pure cultures were obtained. These isolates originated from the samples collected in May 1997, April 1998, and April 1999. Cultures could not be obtained on subculturing from all of the turbid growth-positive terminal tubes, even though microscopy revealed that microbial cells were present in the original liquid cultures. In other cases, more than one colony type developed on the plates inoculated with material from a single terminal tube, and so multiple isolates were obtained. The greatest number of isolates from any one of the replicates was four, and these belonged to four different families (data not shown). The isolates were identified by comparative analysis of near-complete 16S rRNA genes, determined as described elsewhere (28) (GenBank accession no. AF408943 to AF409031), initially on the basis of BLAST comparisons (2). Sequences that could not be clearly assigned to a family-level group (i.e., <97% identical to full-length 16S rRNA gene sequences with unambiguous family assignments) were analyzed more rigorously (28). Family-level assignments were based on monophyly of the isolate sequences with existing families, orders, classes, and phyla. If the sequences were not reproducibly affiliated with recognized families (13) within a higher-level grouping, novel families were proposed as the most conservative estimate of a novel group with a higher-level ranking than species and genus.
Many of the isolates obtained from the terminal positive cultures of the serial liquid dilution experiments were members of genera that are typically thought of as soil-inhabiting groups (1, 3). Members of the phylum Actinobacteria accounted for 53 of the 89 isolates (60%), and all were members of the well-characterized subclass Actinobacteridae. These could be assigned to 17 different families within the suborders Micrococcineae (28 isolates), Corynebacterineae (10 isolates), Propionibacterineae (7 isolates), Frankineae (5 isolates), Streptomycineae (2 isolates), and Pseudonocardineae (1 isolate). Eight of the 53 isolates were members of the genus Arthrobacter, and seven were members of the genus Mycobacterium. Four of the Actinobacteridae isolates represent four as-yet-unnamed novel families in this subclass. A further 19 of the 89 isolates (21%) were members of the class Alphaproteobacteria of the phylum Proteobacteria and could be assigned to six different families. Eleven of these 19 isolates were members of the genus Bradyrhizobium or closely related genera in the family “Bradyrhizobiaceae.” Another 11 of the 89 isolates (12%) were members of the class Betaproteobacteria, and these belonged to just two genera, with 10 of the 11 being members of the genus Burkholderia. The remaining six of the 89 isolates (7%) were members of the class Gammaproteobacteria (3 isolates), of the class Sphingobacteria in the phylum Bacteroidetes (2 isolates), and of the class “Bacilli” of the phylum Firmicutes (1 isolate). Two members of the class Gammaproteobacteria belong to one as-yet-unnamed family.
Simpson's and Shannon's indices (4) of family-level diversity, and the number of families relative to collection size, were similar for the collection of isolates obtained in this study and in three comparable studies conducted on soil from the Ellinbank sample site (18, 19, 28) using solid media (Table 1). Therefore, we conclude that the present collection of isolates is no less diverse at the family level than the three collections obtained with solid media. However, at the phylum (division) level, the collection of isolates obtained from serial liquid dilution was not as representative of soil bacterial diversity as the collections obtained on solid media. All liquid culture isolates belong to only four phyla, the Proteobacteria, Firmicutes, Bacteroidetes, and Actinobacteria (subclass Actinobacteridae). In contrast, the three solid-medium cultivation studies resulted in the isolation of members of five additional bacterial phyla and of basal lineages in the Actinobacteria with few cultivated representatives: Acidobacteria, Verrucomicrobia, Planctomycetes, Gemmatimonadetes, “Deinococcus-Thermus,” and the subclasses Acidimicrobidae and Rubrobacteridae of the Actinobacteria (16 to 17% of isolates) (Table 1). Culture-independent studies suggest that these additional rarely isolated lineages are not trivial components of the soil microbiota. The Acidobacteria, Verrucomicrobia, Gemmatimonadetes, Acidimicrobidae, and Rubrobacteridae are common and abundant in geographically diverse soils (8, 16, 20, 22, 24), including the Ellinbank site (L. Schoenborn and P. H. Janssen, unpublished data).
TABLE 1.
Comparison of the diversity of bacterial isolates obtained from a pasture soil using liquid and solid media
| Growth medium | No. of isolates | % of isolates affiliated with
|
No. of families | Diversity index at family level
|
Reference | ||
|---|---|---|---|---|---|---|---|
| Well-represented groupsa | Poorly represented groupsb | Shannon's | Simpson's | ||||
| Liquid; range of media | 89 | 100 | 0 | 29 | 3.0 | 14.5 | This study |
| Solid; DNB | 30 | 83.3 | 16.7 | 16 | 2.6 | 11.0 | 18 |
| Solid; VL55 + xylan | 71 | 83.1 | 16.9 | 27 | 3.0 | 15.5 | 28 |
| Solid; range of media | 350 | 83.8 | 16.2 | 60 | 3.4 | 15.5 | 19 |
Members of the subclass Actinobacteridae of the phylum Actinobacteria and of the phyla Proteobacteria, Bacteroidetes, and Firmicutes.
Members of the phyla Acidobacteria, “Deinococcus-Thermus,” Gemmatimonadetes, Planctomycetes, and Verrucomicrobia and of the subclasses Acidimicrobidae and Rubrobacteridae of the phylum Actinobacteria.
We conclude that solid media are more useful for isolating soil bacteria belonging to little-studied phyla than are liquid media. There are a number of possible explanations for this unanticipated finding. Members of poorly represented groups simply may not grow in liquid culture, although this is unlikely since representatives of the Acidobacteria, Verrucomicrobia, Rubrobacteridae, and Acidimicrobidae, originally isolated on solid media (18, 19, 28), have been successfully grown in liquid media (X. Chen, M. Sait, P. Sangwan, S. J. Joseph, and P. H. Janssen, unpublished data). The spatial separation of bacteria on solid media (overcoming negative interactions such as lysis by coinoculated bacteriophage or antibiotic production) may have a greater overall influence on culturability of poorly studied groups than the provision for possible positive interactions in liquid culture, such as signals to initiate growth (21). Members of the rarely cultured groups may not grow rapidly enough to establish themselves in the liquid cultures to ensure their subsequent isolation. Based on calculations described by Button et al. (10) and Roussanov et al. (27), analysis of our MPN data (not shown) suggests that the majority (71%) of cultures originated from liquid cultures inoculated with multiple viable cells. If these cells represent more than one species, then only the fastest-growing species will be isolated. This selection on the basis of growth rate will not be so apparent where spatially separated cells can independently develop into detectable cultures, such as on solid media (18, 19, 28) or by using gel encapsulation (33) or automated cell separation (7, 10).
Acknowledgments
We thank Cameron Gourley and Sharon Aarons (Dairy Research Institute, Ellinbank) for help with access to the sampling site.
This work was supported by a grant from the Australian Research Council.
REFERENCES
- 1.Alexander, M. 1977. Introduction to soil microbiology. John Wiley and Sons, New York, N.Y.
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Balows, A, H. G. Trüper, M. Dworkin, W. Harder, and K.-H. Schleifer (ed.). 1992. The prokaryotes. A handbook on the biology of bacteria: ecophysiology, isolation, identification, applications, 2nd ed. Springer, New York, N.Y.
- 4.Begon, M., J. L. Harper, and C. R. Townsend. 1996. Ecology, 3rd ed. Blackwell Science, Oxford, United Kingdom.
- 5.Beliaeff, B., and J. Y. Mary. 1993. The “most probable number” estimate and its confidence limits. Water Res. 27:799-805. [Google Scholar]
- 6.Bruns, A., H. Cypionka, and J. Overmann. 2002. Cyclic AMP and acyl homoserine lactones increase the cultivation efficiency of heterotrophic bacteria from the central Baltic Sea. Appl. Environ. Microbiol. 68:3978-3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bruns, A., H. Hoffelner, and J. Overmann. 2003. A novel approach for high throughput cultivation assays and the isolation of planktonic bacteria. FEMS Microbiol. Ecol. 45:161-171. [DOI] [PubMed] [Google Scholar]
- 8.Buckley, D. H., and T. M. Schmidt. 2002. Exploring the biodiversity of soil—a microbial rain forest, p. 183-208. In J. T. Staley and A.-L. Reysenbach (ed.), Biodiversity of microbial life: foundation of the Earth's biosphere. Wiley-Liss, Inc., New York, N.Y.
- 9.Bussmann, I., B. Philipp, and B. Schink. 2001. Factors influencing the cultivability of lake water bacteria. J. Microbiol. Methods 47:41-50. [DOI] [PubMed] [Google Scholar]
- 10.Button, D. K., F. Schut, P. Quang, R. Martin, and B. R. Robertson. 1993. Viability and isolation of typical marine oligotrophic bacteria by dilution culture: theory, procedures and initial results. Appl. Environ. Microbiol. 59:881-891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chin, K.-J., D. Hahn, U. Hengstmann, W. Liesack, and P. H. Janssen. 1999. Characterization and identification of numerically abundant culturable bacteria from the anoxic bulk soil of rice paddy microcosms. Appl. Environ. Microbiol. 65:5042-5049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Conn, H. J. 1918. The microscopic study of bacteria and fungi in soil. N. Y. Agric. Exp. Sta. Tech. Bull. 64:3-20. [Google Scholar]
- 13.Garrity, G. M., M. Winters, and D. B. Searles. 2001. Taxonomic outline of the procaryotic genera, release 1.0. Springer, New York, N.Y.
- 14.Groβkopf, R., P. H. Janssen, and W. Liesack. 1998. Diversity and structure of the methanogenic community in anoxic rice paddy soil microcosms as examined by cultivation and direct 16S rRNA gene sequence retrieval. Appl. Environ. Microbiol. 64:960-969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hudson, J. A., K. A. Schofield, H. W. Morgan, and R. M. Daniel. 1989. Thermonema lapsum gen. nov., sp. nov., a thermophilic gliding bacterium. Int. J. Syst. Bacteriol. 39:485-487. [Google Scholar]
- 16.Hugenholtz, P., B. M. Goebel, and N. R. Pace. 1998. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J. Bacteriol. 180:4765-4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Janssen, P. H., A. Schuhmann, E. Mörschel, and F. A. Rainey. 1997. Novel anaerobic ultramicrobacteria belonging to the Verrucomicrobiales lineage of bacterial descent isolated by dilution culture from anoxic rice paddy soil. Appl. Environ. Microbiol. 63:1382-1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Janssen, P. H., P. S. Yates, B. E. Grinton, P. M. Taylor, and M. Sait. 2002. Improved culturability of soil bacteria and isolation in pure culture of novel members of the divisions Acidobacteria, Actinobacteria, Proteobacteria, and Verrucomicrobia. Appl. Environ. Microbiol. 68:2391-2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Joseph, S. J., P. Hugenholtz, P. Sangwan, C. A. Osborne, and P. H. Janssen. 2003. Laboratory cultivation of widespread and previously uncultured soil bacteria. Appl. Environ. Microbiol. 69:7210-7215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liesack, W., and P. F. Dunfield. 2002. Biodiversity in soils: use of molecular methods for its characterization, p. 528-544. In G. Bitton (ed.), Encyclopedia of environmental microbiology. John Wiley and Sons, New York, N.Y.
- 21.Miller, M. B., and B. L. Bassler. 2001. Quorum sensing in bacteria. Annu. Rev. Microbiol. 55:165-199. [DOI] [PubMed] [Google Scholar]
- 22.Mummey, D. L., and P. D. Stahl. 2003. Candidate division BD: phylogeny, distribution and abundance in soil ecosystems. Syst. Appl. Microbiol. 26:228-235. [DOI] [PubMed] [Google Scholar]
- 23.Postgate, J. R., and J. R. Hunter. 1964. Accelerated death of Aerobacter aerogenes starved in the presence of growth limiting substrates. J. Gen. Microbiol. 34:459-473. [DOI] [PubMed] [Google Scholar]
- 24.Rappé, M. S., and S. J. Giovannoni. 2003. The uncultured microbial majority. Annu. Rev. Microbiol. 57:369-398. [DOI] [PubMed] [Google Scholar]
- 25.Rappé, M. S., S. A. Connon, K. L. Vergen, and S. J. Giovannoni. 2002. Cultivation of the ubiquitous SAR11 marine bacterioplankton clade. Nature 418:630-633. [DOI] [PubMed] [Google Scholar]
- 26.Rheims, H., A. Felske, S. Seufert, and E. Stackebrandt. 1999. Molecular monitoring of an uncultured group of the class Actinobacteria in two terrestrial environments. J. Microbiol. Methods 36:65-75. [DOI] [PubMed] [Google Scholar]
- 27.Roussanov, B., D. M. Hawkins, and S. R. Tatini. 1996. Estimating bacterial density from tube dilution data by a Bayesian method. Food Microbiol. 13:341-363. [Google Scholar]
- 28.Sait, M., P. Hugenholtz, and P. H. Janssen. 2002. Cultivation of globally-distributed soil bacteria from phylogenetic lineages previously only detected in cultivation-independent surveys. Environ. Microbiol. 4:654-666. [DOI] [PubMed] [Google Scholar]
- 29.Singh, D. K., P. W. G. Sale, C. J. P. Gourley, and C. Hasthorpe. 1999. High phosphorus supply increases persistence and growth of white clover in grazed dairy pastures during dry summer conditions. Aust. J. Exp. Agric. 39:579-585. [Google Scholar]
- 30.Straskrabová, V. 1983. The effect of substrate shock on populations of starving aquatic bacteria. J. Appl. Bacteriol. 54:217-224. [Google Scholar]
- 31.Tschech, A., and N. Pfennig. 1984. Growth yield increase linked to caffeate reduction in Acetobacterium woodii. Arch. Microbiol. 137:163-167. [Google Scholar]
- 32.Widdel, F., G. W. Kohring, and F. Mayer. 1983. Studies on dissimilatory sulfate-reducing bacteria that decompose fatty acids. III. Characterization of the filamentous gliding Desulfonema limicola gen. nov. sp. nov., and Desulfonema magnum sp. nov. Arch. Microbiol. 134:286-294. [Google Scholar]
- 33.Zengler, K., G. Toledo, M. Rappé, J. Elkins, E. J. Mathur, J. M. Short, and M. Keller. 2002. Cultivating the uncultured. Proc. Natl. Acad. Sci. USA 99:15681-15686. [DOI] [PMC free article] [PubMed] [Google Scholar]