Abstract
Microbial communities within the human oral cavity are dynamic associations of more than 500 bacterial species that form biofilms on the soft and hard tissues of the mouth. Understanding the development and spatial organization of oral biofilms has been facilitated by the use of in vitro models. We used a saliva-conditioned flow cell, with saliva as the sole nutritional source, as a model to examine the development of multispecies biofilm communities from an inoculum containing the coaggregation partners Streptococcus gordonii, Actinomyces naeslundii, Veillonella atypica, and Fusobacterium nucleatum. Biofilms inoculated with individual species in a sequential order were compared with biofilms inoculated with coaggregates of the four species. Our results indicated that flow cells inoculated sequentially produced biofilms with larger biovolumes compared to those biofilms inoculated with coaggregates. Individual-species biovolumes within the four-species communities also differed between the two modes of inoculation. Fluorescence in situ hybridization with genus- and species-specific probes revealed that the majority of cells in both sequentially and coaggregate-inoculated biofilms were S. gordonii, regardless of the inoculation order. However, the representation of A. naeslundii and V. atypica was significantly higher in biofilms inoculated with coaggregates compared to sequentially inoculated biofilms. Thus, these results indicate that the development of multispecies biofilm communities is influenced by coaggregations preformed in planktonic phase. Coaggregating bacteria such as certain streptococci are especially adapted to primary colonization of saliva-conditioned surfaces independent of the mode of inoculation and order of addition in the multispecies inoculum. Preformed coaggregations favor other bacterial strains and may facilitate symbiotic relationships.
Within the human oral cavity, complex interactions of bacteria result in the formation on enamel surfaces of multispecies biofilms known as dental plaque. Dental plaque is comprised of more than 500 species of bacteria (17, 26, 31), more than half of which are not yet cultivated (31). By forming microbial biofilms, oral bacteria overcome many of the challenges associated with retention in the oral cavity, such as high salivary flow and low-nutrient conditions.
Critical to the formation and development of oral biofilms is cell-to-cell communication (15). Mechanisms by which bacteria have been shown to communicate in oral biofilms include physical interactions (35), genetic exchange (18), and diffusible signals (1, 24). Two forms of physical interactions thought to play an important role in biofilm formation are coaggregation and coadhesion (2, 5, 6, 9). Coaggregation is the cell-cell recognition between genetically distinct bacteria in a planktonic suspension (12), whereas coadhesion refers to the recognition between a planktonic cell and a surface-attached cell (3). Coaggregation and coadhesion are mediated by the same kinds of cell-cell interactions (5, 35). Although coaggregation and coadhesion interactions are highly specific between pairs of bacterial partners, these interactions are widespread among oral bacteria and have been observed among all bacteria isolated from the oral cavity (13) as well as between certain oral bacteria and Candida albicans (11). These two binding interactions also occur among microbes isolated from the mammalian gut, human urogenital track, activated sludge, and freshwater ecosystems (21, 22, 32). Coaggregation and coadhesion are thought to contribute to biofilm formation by fostering mutualistic interactions between juxtaposed cells (29) and by providing more diverse attachment sites for planktonic bacteria to adhere to the developing biofilm surface (15, 33).
To understand the spatial organization and development of oral biofilms it is useful to examine the succession of organisms in the growing biofilm. These temporal changes to the biofilm occur via the attachment and growth of different bacterial species to the substratum. Previous studies of bacterial successions using culture-dependent techniques postulated that additions to the bacterial population on the tooth surface occur in a sequential order (19, 27, 28). These studies documented that in the first 4 h after professional cleaning 60 to 90% of the primary colonizers of the tooth surface are streptococci (28). Other early colonizers of the tooth surface include Actinomyces spp., Capnocytophaga spp., Haemophilus spp., and Veillonella spp. (19, 27, 28). Once the nascent tooth surface is colonized, biofilm accretion continues by the adhesion of late colonizers such as Fusobacterium nucleatum, Treponema spp., Tannerella forsythensis (formerly Bacteroides forsythus), and Porphyromonas gingivalis (26, 34).
With more than 500 species known from the human oral cavity, in vitro models have been employed to simplify these complex associations. One such model, the flow cell, facilitates the in situ examination of undisturbed biofilm communities. In this study, saliva-conditioned flow cells with saliva as the sole nutrient were used to determine whether differences in biofilm architecture and composition occurred in flow cells that were inoculated either sequentially with individual species or simultaneously with coaggregates of mixed species. To determine the biofilm composition, fluorescence in situ hybridization (FISH) was used to examine the mixed-species communities in the flow cells without disruption of the growing biofilm.
Four bacterial species were chosen as the inocula for the mixed-species biofilms; three of these species have been identified as primary colonizers of the tooth surface. These early colonizers include the facultative anaerobes Streptococcus gordonii, Actinomyces naeslundii, and Veillonella atypica. The fourth species used in this study, F. nucleatum, an obligate anaerobe, is one of the most abundant gram-negative bacteria in subgingival oral biofilms (26). F. nucleatum is capable of coaggregating with all known oral isolates (16) and has been shown to associate clinically with late-colonizing pathogenic bacteria (34). Together, these four species exemplify a wide range of metabolic and physiological characteristics that serve to represent a diverse array of oral bacteria.
We investigated these strains to test the hypothesis that order of inoculation as well as preformed coaggregations may alter the outcome of community species diversity following growth on saliva in our in vitro biofilm model. In this study, we provide evidence that flow cells inoculated sequentially produce larger biofilm volumes overall than flow cells inoculated with coaggregates of mixed species. In addition, we demonstrate that preformed coaggregations alter the species representation by an apparent symbiosis within the biofilm community. Extrapolating to conditions in vivo in developing dental plaque biofilms, we propose that coaggregations preformed in saliva could have profound effects on the architecture and community composition of growing dental biofilms.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The coaggregating partners A. naeslundii T14V, F. nucleatum PK1594, S. gordonii DL1, and V. atypica PK1910 were used as inocula (16) for the formation of multispecies biofilms. A. naeslundii was grown in Todd-Hewitt broth (Difco Laboratories, Detroit, Mich.). F. nucleatum was grown in brain heart infusion (Difco Laboratories) broth supplemented with 0.25% l-glutamic acid. S. gordonii was grown in brain heart infusion broth and V. atypica was grown in Schaedler (Difco Laboratories) broth supplemented with 0.1 M sodium lactate. All species were grown overnight in a Bactron anaerobic environmental chamber (N2-CO2-H2, 90:5:5; Sheldon Manufacturing Inc., Cornelius, Oreg.) at 37°C. Overnight bacterial cultures were harvested by centrifugation and washed twice with 25% sterile human saliva. The cell density was normalized to 2.5 × 105 cells per ml of 25% saliva as determined by spectrophotometric measurement at A600 and by direct cell counts with a Petroff-Hausser cell counter.
Flow cell preparation.
Two flow tracks (each track was 40 mm long, 3 mm wide, and 2 mm deep) were milled into a high-density polyethylene block, resulting in two chambers each with a 240-μl volume. A glass coverslip, which serves as the attachment site for the growing biofilm, was secured to the reusable flow cells with a silicone adhesive. The flow cells were cleaned overnight with 0.1 M HCl and rinsed with 5 ml of distilled water. To disinfect the flow cells, bleach was injected into the flow cells and allowed to incubate for 2 h, followed by continuous rinsing with sterile distilled water for 5 min using a peristaltic pump. The flow cells were then treated with 25% sterile human saliva for 15 min at 34°C to condition the glass surface with salivary components.
Saliva preparation.
Stimulated saliva was collected from at least six healthy individuals, pooled, and treated with 2.5 mM dithiothreitol for 10 min to reduce salivary protein aggregation. The saliva was then centrifuged and processed as previously described (8, 29). Briefly, the supernatant was diluted with distilled water to produce 25% saliva and then was filtered through a 0.22-μm-pore-size SFCA low-protein-binding filter (Nalge Nunc International, Rochester, N.Y.) and stored at −20°C. Prior to use, saliva was thawed and centrifuged to remove any precipitate that resulted from freezing and thawing.
Biofilm growth conditions.
Flow cells were inoculated with the four bacterial strains either independently in a sequential order or mixed together as a coaggregate. In sequentially inoculated flow cells, the flow cells were injected four times each with 500 μl of 25% saliva containing 1.25 × 105 cells of a single strain. The flow cells were inverted and the cells were allowed 15 min to adhere to the glass surface in the absence of flow. Between each injection, the flow cells were washed for 15 min by pumping sterile saliva through the flow cell. Unless otherwise indicated, the order in which the bacteria were added to the flow cells was as follows: S. gordonii, A. naeslundii, V. atypica, and F. nucleatum. All flow cells were maintained in an aerobic incubator at 34°C.
To inoculate flow cells with coaggregates of mixed species, 1.25 × 105 cells of each strain were added to 500 μl of 25% saliva and vortexed for 15 s (see Fig. 1). The resulting mixture was then injected into the flow cell. The flow cell was inverted for 15 min and maintained statically without flow. The flow cell was then washed for 15 min by pumping saliva though the flow cell. After washing, 25% saliva was pumped through the flow cells for either 1 or 14 h at a rate of 200 μl per min (7). Each experimental condition was independently repeated in triplicate. Saliva was used as the sole nutrient source for all flow cell experiments.
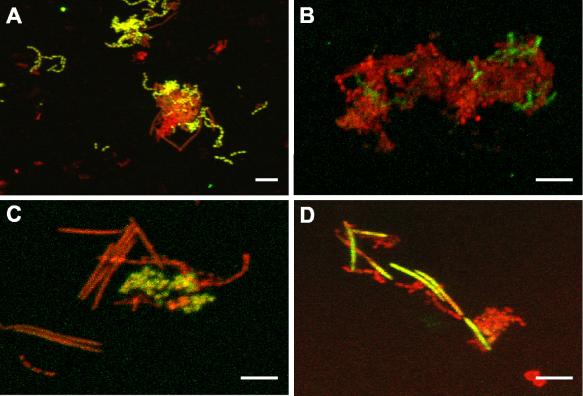
FIG. 1.
Confocal micrographs of planktonic cultures containing coaggregates of S. gordonii, A. naeslundii, V. atypica, and F. nucleatum processed for FISH with fluorescein isothiocyanate-labeled probes (green) and counterstained with general nucleic acid stain Syto 59 (red). Colocalization of both fluorescent markers appears yellow to yellow-green. (A) Streptococcus-specific probe targeting S. gordonii. (B) Species-specific probe labeling short rod-shaped A. naeslundii cells. (C) Clustered V. atypica cells hybridized with Veillonella-specific probe. (D) Long slender rod-shaped F. nucleatum cells labeled with F. nucleatum-specific probe. Bar, 5 μm.
Biofilm staining.
The resulting biofilms were labeled with either (i) a 5 μM solution of the general nucleic acid stain Syto 59 (red fluorescence) (Molecular Probes, Eugene, Oreg.), (ii) a LIVE/DEAD viability kit containing a solution of 1.67 μM Syto 9 (green fluorescence) and 10 μM propidium iodide (red fluorescence) (Molecular Probes), or (iii) by FISH (for conditions see below). After staining, all biofilms were examined in situ through the glass coverslip with a Leica (Exton, Pa.) TCS-SP2 confocal laser scanning microscope.
Oligonucleotide probe design and synthesis.
Oligonucleotide probes used in this study were designed to target the 16S rRNA and are listed in Table 1. The probes used in this study were either generated as previously described (30) or developed by using the probe design function of the ARB program (www.arb-home.de). To ensure probe specificity, probe JF201, designed to target A. naeslundii, was tested and demonstrated not to hybridize to a closely related species (Actinomyces denticolens ATCC 43322) that contained one mismatch. Probe JF3 matched only F. nucleatum subspecies nucleatum, vincentii, and animalis. The closest match for probe JF3 that was not a strain of F. nucleatum contained four mismatches and was isolated from a hydrothermal vent community. Probe analyses were conducted using the Probe Match subroutine of the ARB program in conjunction with the Ribosomal Database Project (20). All probes were synthesized and 5′-end-labeled (QIAGEN, Alameda, Calif.) for fluorescence with either fluorescein isothiocyanate or Cy3 (Amersham Life Sciences, Piscataway, N.J.).
TABLE 1.
Oligonucleotide probes used in this study
FISH.
To identify S. gordonii and A. naeslundii within the biofilm, flow cells were injected with 100% ethanol for 15 min at 34°C. For the identification of V. atypica and F. nucleatum, biofilms were fixed with 4% paraformaldehyde in phosphate-buffered saline (1.7 mM KH2PO4-5 mM Na2HPO4 with 0.15 M sodium chloride, pH 7.2) for 12 h at 4°C. After fixation, all biofilms were washed with phosphate-buffered saline and then exposed to a solution containing 10 mg of lysozyme per ml of 0.1 M Tris-HCl-0.05 M EDTA, pH 7.2, for 30 min at room temperature in order to permeabilize cells. After permeabilization, biofilms were dehydrated with a series of ethanol washes containing 50, 80, and 95% ethanol for 3 min each rinse. Biofilms were then incubated with 40 ng of oligonucleotide probe per μl of hybridization buffer (0.9 M NaCl, 0.1 M Tris-HCl [pH 7.2], 20% formamide [vol/vol], 1% blocking reagent [wt/vol; Roche, Indianapolis, Ind.], 0.0005% sodium dodecyl sulfate [wt/vol]). Biofilms exposed to probes E72 or JF201 were incubated at 46°C for 12 h, whereas biofilms labeled with JF3 or E79 were incubated at 46°C for 4 h. Following probe hybridization, biofilms were washed for 15 min using a peristaltic pump at a rate of 200 μl/min with a wash buffer containing 0.1 M Tris-HCl (pH 7.2), 0.18 M NaCl, 0.05 M EDTA, and 0.005% sodium dodecyl sulfate (wt/vol). After washing, the labeled biofilms were counterstained for 5 min with a general nucleic acid stain solution containing either 1.67 μM Syto 9 or 5 μM Syto 59 (Molecular Probes). Biofilms were then washed for 10 min with wash buffer and examined through the glass coverslip with confocal laser scanning microscopy.
Image and statistical analyses.
To quantify the biomass of the oral biofilms, the total fluorescent staining of confocal micrographs was analyzed using the image analysis program Imaris 3.3.2 (Bitplane AG, Zürich, Switzerland). The Imaris program calculates the biofilm volumes (biovolumes) from stacks of two-channel (red and green) images by measuring voxel intensities. Fluorescence intensity thresholds were set manually for red and green pixels, and a voxel size of 0.6 μm was used for the x, y, and z directions. The resulting biovolumes from the red and green fluorescence channels were then analyzed for statistical significance. At least eight micrographs were analyzed for each experimental group, and the mean and standard deviation were calculated for each group. Either one-way analysis of variance (ANOVA) or two-sample t tests were used to determine whether significant differences existed between different experimental groups. To determine which means were statistically different from each other in the ANOVA test a nonparametric Tukey's pairwise comparison test was also used. All tests were conducted at the 95% confidence level.
RESULTS
Probe specificity.
Mixed-species planktonic cultures of S. gordonii, A. naeslundii, V. atypica, and F. nucleatum were examined with FISH using genus- and species-specific oligonucleotide probes (Fig. 1). Each of the four species, S. gordonii (Fig. 1A), A. naeslundii (Fig. 1B), V. atypica (Fig. 1C), and F. nucleatum (Fig. 1D) were positively identified within the mixed community with low nonspecific binding of the other three species. Fluorescent labeling also revealed that all four species were able to coaggregate with the other members of the community.
Growth of oral bacteria in the flow cell.
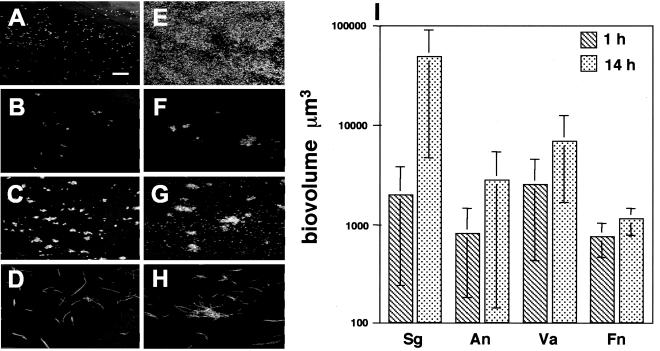
Prior to examining the growth of four-species biofilms, it was first necessary to determine whether each species was capable of independent biofilm growth in flow cells using saliva as the sole nutrient source. S. gordonii, A. naeslundii, V. atypica, and F. nucleatum monocultures were each incubated within flow cells for 1 and 14 h (Fig. 2). At 1 h the amount of cell attachment to the substratum was determined by fluorescent staining of the bacterial cells within each flow cell track. These results demonstrated that all four species attached to the saliva-conditioned flow cell within 1 h (Fig. 2A to D). Biofilms incubated for 14 h (Fig. 2E to H) showed statistically significant growth in only one of the four species, S. gordonii. The biovolume for each species was calculated after 1 and 14 h of salivary flow (Fig. 2I). Only S. gordonii demonstrated a statistically significant increase (P < 0.05) in biovolumes over the 14 h incubation. To determine whether the cells within the flow cells were viable after 14 h, cells were stained with the LIVE/DEAD viability stain, a fluorescent marker of membrane integrity. LIVE/DEAD staining revealed that most cells (>90%) in mono-species biofilms containing S. gordonii, A. naeslundii, or V. atypica appeared to have no membrane damage at 1 or 14 h (data not shown). In mono-species biofilms containing F. nucleatum, 17% of cells exhibited membrane damage at 1 h. However by 14 h, only 8% of F. nucleatum cells exhibited damage suggesting either plasma membrane recovery or growth (data not shown). These results suggest that the majority of all cells present within the flow cell were viable even though only S. gordonii grew as a monoculture.
FIG. 2.
Growth of mono-species biofilms. (A to D) Confocal micrographs of mono-species biofilms at 1 h stained with nucleic acid stain Syto 59. (A) S. gordonii (bar, 40 μm); (B) A. naeslundii; (C) V. atypica; (D) F. nucleatum. (E to H) Confocal micrographs of 14-h mono-species biofilms stained with Syto 59. (E) S. gordonii; (F) A. naeslundii; (G) V. atypica; (H) F. nucleatum. (I) Graph depicting biovolumes of mono-species biofilms. Abbreviations: Sg, S. gordonii; An, A. naeslundii; Va, V. atypica; Fn, F. nucleatum.
Sequentially or coaggregate-inoculated flow cells.
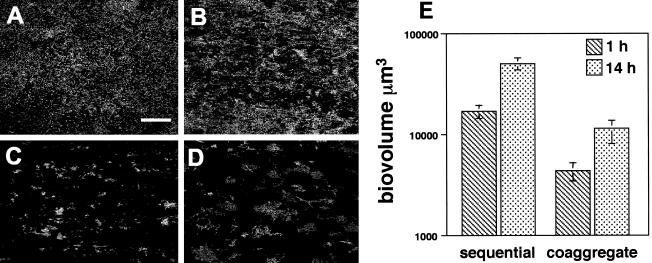
Biofilms inoculated sequentially with the four species were compared to those inoculated with coaggregates of the same four species. This comparison revealed inoculation-specific differences in the morphology and biovolumes of the mixed-species biofilms (Fig. 3). In flow cells inoculated with S. gordonii, A. naeslundii, V. atypica, and F. nucleatum in sequential order an extensive biofilm was visible after 1 h that covered much of the flow cell substratum (Fig. 3A) and increased in biovolume by 14 h (Fig. 3B). The four species are known to coaggregate strongly pairwise with each other (16), and they also formed multispecies coaggregates of various sizes (see Fig. 1). Flow cells inoculated simultaneously with coaggregated S. gordonii, A. naeslundii, V. atypica, and F. nucleatum also formed biofilms; however, areas of exposed substratum were visible after 1 h (Fig. 3C). By 14 h postinoculation with coaggregates of mixed species, spaces were still visible throughout the biofilm (Fig. 3D). The biofilm volumes are graphically represented in Fig. 3E. Statistical comparisons revealed that biovolumes were significantly larger at 14 h than at 1 h in sequentially inoculated biofilms (P < 0.0007) as well as in coaggregate-inoculated biofilms (P < 0.041). Significant differences were also observed when the total biovolumes of 1 and 14 h sequentially and coaggregate-inoculated biofilms were compared. Sequentially inoculated biofilms had significantly larger biovolumes than biofilms inoculated with coaggregates at 1 h (P < 0.002) and 14 h (P < 0.002).
FIG. 3.
Comparison of sequentially and coaggregate-inoculated biofilms. (A to D) Confocal micrographs of four-species biofilms containing S. gordonii, A. naeslundii, V. atypica, and F. nucleatum. Biofilms were stained with general nucleic acid stain Syto 59. (A) One-hour biofilms inoculated in a sequential order demonstrating the widespread coverage of flow cell surface. Bar, 40 μm. (B) Fourteen-hour sequentially inoculated biofilm. (C) One-hour coaggregate-inoculated biofilm. (D) Fourteen-hour biofilm inoculated with a coaggregate of the four species. (E) Biovolumes of sequentially and coaggregate-inoculated biofilms.
To examine whether additional adherence time would increase the biovolumes of coaggregate-inoculated biofilms, coaggregates were inoculated into flow cells and allowed to adhere to the saliva-conditioned substratum for 15, 30, and 60 min; washed; and incubated for 1 h with 25% sterile saliva. The resulting biovolumes were statistically compared using ANOVA and revealed no significant difference between the different incubation times (data not shown). These results demonstrate that increasing the time of initial adherence of coaggregates up to 60 min does not produce larger biovolumes.
Order of S. gordonii in sequentially inoculated biofilms.
To determine whether the order of species in sequentially inoculated flow cells contributed to biofilm development, biofilms inoculated in different orders with S. gordonii, A. naeslundii, V. atypica, and F. nucleatum were compared. S. gordonii was inoculated into the flow cell either first, second, third, or fourth, while the addition of three other species was not varied (i.e., A. naeslundii, V. atypica, and F. nucleatum). After inoculation of all four species, the biofilms were incubated with flowing saliva for 1 h then processed for FISH with a Streptococcus-specific probe to determine the amount of S. gordonii present within the mixed-species communities. S. gordonii appeared evenly distributed throughout the biofilm and most S. gordonii cells were associated with other members of the mixed-species community (data not shown). The total and S. gordonii biovolumes for each combination were calculated, and the results indicated that regardless of inoculation order, neither the total nor the S. gordonii biovolumes were significantly different from each other (data not shown). These data indicate that S. gordonii grows independently of the other three species and that it is the major contributing cell type in sequentially inoculated biofilms.
Biofilm species composition.
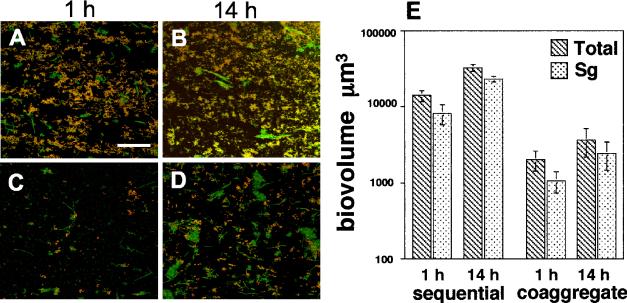
To examine the relative amounts of each species within the mixed-species communities, biofilms were grown for 1 and 14 h and examined by FISH using oligonucleotide probes. Biofilms inoculated sequentially were compared with those biofilms inoculated with coaggregates to determine whether differences in species composition occurred. In both sequentially and coaggregate-inoculated biofilms FISH revealed that the majority of the cells comprising 1- and 14-h biofilms were S. gordonii (Fig. 4A to D). Image analysis of the four-species biofilms indicated that S. gordonii ranged from 57 to 73% of the total biovolume of the four-species community at 1 and 14 h (Fig. 4E). As the biofilm grew, differences were detected in the S. gordonii biovolumes of sequentially and coaggregate-inoculated biofilms (Fig. 4E). S. gordonii biovolumes in sequentially inoculated biofilms were statistically higher than biofilms inoculated with coaggregates at both 1 h (P < 0.032) and 14 h (P < 0.007). Despite this difference, the S. gordonii biovolumes of both sequentially and coaggregate-inoculated biofilms remained constant over time with respect to the total biofilm indicating that the abundance of S. gordonii appeared to remain proportional to the total biovolume of the community (Fig. 4E).
FIG. 4.
Abundance of S. gordonii in four-species biofilm using FISH with Cy3-labled probes. (A to D) Confocal micrographs of mixed-species communities hybridized with Streptococcus-specific probe (red) and counterstained with nucleic acid stain Syto 9 (green). Colocalization of both fluorescent probes appears yellow. (A) One-hour biofilms inoculated in a sequential order. Bar, 40 μm. (B) Sequentially inoculated biofilm at 14 h. (C) One-hour biofilm inoculated with a coaggregate of mixed species. (D) Coaggregate-inoculated biofilm at 14 h. (E). Graph indicating the total biovolumes of the four-species biofilms (hatched bars) and biovolumes of S. gordonii (Sg) (stippled bars) in sequentially and coaggregate-inoculated biofilms at 1 and 14 h.
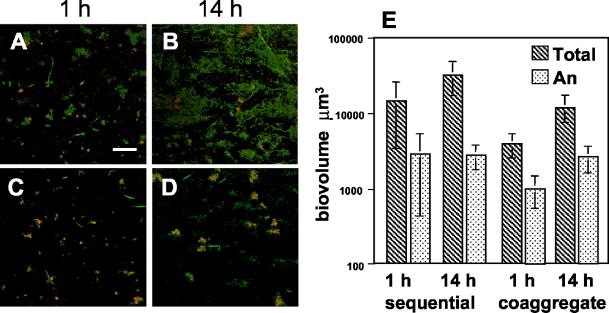
In biofilms hybridized with a species-specific probe for A. naeslundii, the biovolumes of A. naeslundii ranged from 8 to 28% of the total biofilm (Fig. 5). At 1 h, microcolonies of A. naeslundii were visibly small (approximately 5 μm diameter), and some, but not all, were associated with other species (Fig. 5A and C). At 14 h, the size of the A. naeslundii microcolonies increased (approximately 20 μm diameter), and all were visibly associated with other species in both sequentially and coaggregate-inoculated biofilms (Fig. 5B and D). Biovolumes calculated from confocal micrographs indicated that at 14 h, A. naeslundii accounted for between 8 (sequential) and 21% (coaggregate) of the total biovolume (Fig. 5E). Although sequentially inoculated biofilms contained a greater total biomass than biofilms inoculated with coaggregates (P < 0.031), A. naeslundii biovolumes were not statistically different between the two modes of inoculation at 1 h. At 14 h, there was no increase in A. naeslundii biovolumes in sequentially inoculated biofilms despite the increase in the total biovolume (P < 0.022). In contrast, at 14 h in coaggregate-inoculated biofilms there was a statistically significant increase in both the A. naeslundii biovolume (P < 0.010) and the total biovolume (P < 0.001). These results suggest that although the total biomass of biofilms was lower in coaggregate-inoculated biofilms compared to sequentially inoculated biofilms, A. naeslundii grew only when associated with mixed-species coaggregates during inoculation.
FIG. 5.
The presence of A. naeslundii in four-species biofilm using FISH with Cy3-labled probes. (A to D) Confocal micrographs of mixed-species biofilms hybridized with an A. naeslundii-specific probe (red) and counterstained with general nucleic acid stain Syto 9 (green). Colocalization of both probes appears yellow. (A) One-hour sequentially inoculated biofilm. Bar, 40 μm. (B) Fourteen-hour sequentially inoculated biofilm. (C) One-hour coaggregate-inoculated biofilm. (D) Fourteen-hour coaggregate-inoculated biofilm. (E) Graph indicating the total biovolumes of the four-species biofilms (hatched bars) and biovolumes of A. naeslundii (An) (stippled bars) in sequentially and coaggregate-inoculated biofilms.
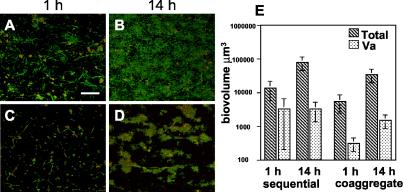
Biofilms hybridized with the Veillonella-specific probe revealed that V. atypica comprised between 5 to 12% of the total biovolume of the mixed-species community (Fig. 6). In sequentially inoculated biofilms, microcolonies of V. atypica were distributed throughout the biofilms at 1 h (Fig. 6A), and by 14 h the mixed-species community covered most of the flow cell substratum (Fig. 6B). In flow cells inoculated with coaggregates of the four species, V. atypica was visible throughout the biofilm at 1 and 14 h (Fig. 6C and D). As with A. naeslundii, by 14 h all V. atypica cells appeared to associate with other members of the mixed species community (Fig. 6D). Biovolumes of the total mixed-species community as well as V. atypica were calculated for each biofilm and compared (Fig. 6E). In sequentially inoculated biofilms, despite a statistically significant increase in the total biovolume of the community from 1 to 14 h (P < 0.003), there was no change in the biovolume of V. atypica. In coaggregate-inoculated biofilms, however, a significant increase was detected in both the total (P < 0.002) and V. atypica (P < 0.004) biovolumes between 1 and 14 h. As with A. naeslundii, the growth of V. atypica increased after 14 h only in coaggregate-inoculated biofilms despite an overall lower total biomass compared to sequentially inoculated biofilms.
FIG. 6.
Prevalence of V. atypica in four-species biofilm using FISH with Cy3-labled probes. (A to D) Confocal micrographs of four-species biofilms labeled with a Veillonella-specific probe (red) and counterstained with general nucleic acid stain Syto 9 (green). Areas of yellow indicate colocalization of both fluorescent markers. (A) One-hour sequentially inoculated biofilm. Bar, 40 μm. (B) Fourteen-hour sequentially inoculated biofilm. (C) One-hour coaggregate inoculated biofilm. (D) Fourteen-hour coaggregate-inoculated biofilm. (E) Graph indicating the total biovolumes of the four-species biofilm (hatched bars) and biovolumes of V. atypica (Va) (stippled bars) in sequentially and coaggregate-inoculated biofilms.
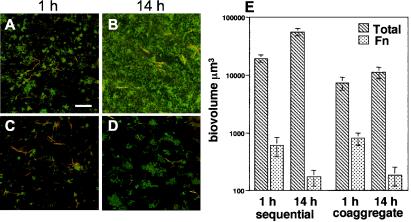
In mixed-species communities hybridized with an F. nucleatum-specific probe, F. nucleatum ranged from 3 to 11% of the total population (Fig. 7). As with the other four strains examined, F. nucleatum appeared evenly distributed throughout the biofilm and often associated with other species (Fig. 7A to D). Total and F. nucleatum biovolumes are graphically represented in Fig. 7E. Although a statistical difference was observed between the total biovolumes of sequentially and coaggregate-inoculated flow cells at 1 h (P < 0.002) and 14 h (P < 0.002), there was no statistical difference in the F. nucleatum biovolume between coaggregate- and sequentially inoculated biofilms at 1 h (P < 0.24) and 14 h (P < 0.082). By 14 h, however, the biovolumes of F. nucleatum had significantly decreased in both coaggregate- (P < 0.005) and sequentially (P < 0.03) inoculated biofilms.
FIG. 7.
Examination of F. nucleatum in a four-species biofilm using FISH with Cy3-labled probes. (A to D) Confocal micrographs of four-species biofilms labeled with a F. nucleatum-specific probe (red) and counterstained with general nucleic acid stain Syto 9 (green). Areas of yellow indicate colocalization of both fluorescent markers. (A) One-hour sequentially inoculated biofilm. Bar, 40 μm. (B) Fourteen-hour sequentially inoculated biofilm. (C) One-hour coaggregate-inoculated biofilm. (D) Fourteen-hour coaggregate-inoculated biofilm. (E) Graph indicating the total biovolumes of the four-species biofilm (hatched bars) and biovolumes of F. nucleatum (Fn) (stippled bars) in sequentially and coaggregate-inoculated biofilms.
DISCUSSION
The aim of this study was to examine the mechanisms of early oral biofilm formation using two modes of inoculation of a four-species community of oral bacteria under flowing conditions with saliva as the sole nutrient source. The results of this study provide evidence that (i) biofilms inoculated in a sequential order have higher total biovolumes than coaggregate-inoculated biofilms; (ii) monocultures of the four species all had the capacity to bind to the saliva-conditioned substratum independently, but only S. gordonii exhibited significant growth after 14 h; (iii) the major component of the mixed-species biofilms in both sequentially and coaggregate-inoculated biofilms was S. gordonii; and (iv) growth of A. naeslundii and V. atypica only occurred in coaggregate-inoculated biofilms and appeared as mixed-species clusters.
The comparison of mixed-species biofilms revealed that flow cells inoculated in a sequential order produced significantly more biomass than flow cells inoculated with coaggregates. Possible explanations for these differences include (i) an extended incubation time for sequentially inoculated biofilms due to the additional wash periods between inoculations, (ii) reduced adhesion of coaggregates to the substratum compared to the total collective adhesion of the four individual species, (iii) variations in the growth of each organism as a coaggregate compared to individual species, or (iv) selection of coadhesion partner and adherence site favors growth compared to growth in preformed coaggregates.
Although the same number of cells was inoculated under both experimental conditions, sequentially inoculated biofilms required an additional 60 min for rinsing. However, when biofilms incubated with coaggregates for 15 min were compared with coaggregate-inoculated biofilms incubated for an additional 60 min, no statistical difference was detected indicating that incubation time was not the cause of the larger biovolumes in sequentially inoculated biofilms. The results of these experiments do suggest, however, that coaggregates of multiple species were not able to bind as efficiently to the flow cell surface compared to the collective total coadhesion of individual species. This result was unexpected considering the additional binding possibilities potentially exhibited by four species collectively versus a single species.
In another model system consisting of saliva-conditioned hydroxyapatite disks and a five-membered oral bacterial biofilm consortium (10), growth was observed for actinomyces, fusobacteria, streptococci, and veillonellae over a period of 64 h. A mixture of equal amounts of saliva and a modified tryptone-yeast-extract-based fluid universal medium (mFUM) was the growth medium with either glucose or glucose plus sucrose as additional carbon sources. The hydroxyapatite disks were placed in sterile microtiter wells with mFUM under an anaerobic atmosphere. All five strains adhered strongly to the saliva-conditioned surface. While interspecies associations were observed at later stages of biofilm formation, the authors interpreted their data to strongly suggest that most of the increase in bacterial numbers was a result of bacterial growth and not coadhesion (10). The presence of glucose and sucrose to mFUM and saliva gives the five-membered consortium significant added nutrient; is known to support the growth of streptococci, actinomyces, and fusobacteria; and is in clear contrast to unamended saliva used in our experiments. The hydroxyapatite model is an excellent model system that offers an opportunity to study the roles of coadhesion and growth of oral bacteria.
In our study, examination of the growth of each bacterial species as monocultures as well as in sequentially and coaggregate-inoculated biofilms also revealed differences in overall biofilm development. Each species bound to the saliva-conditioned flow cell substratum. As monocultures, only S. gordonii significantly grew within the first 14 h: A. naeslundii, V. atypica, and F. nucleatum were unable to grow independently. Additional time beyond 14 h may have been required for monocultures of A. naeslundii, V. atypica, and F. nucleatum to demonstrate significant growth.
In mixed-species communities, however, differences in the growth of all the organisms were detected by 14 h. Examination with FISH revealed that S. gordonii in sequentially inoculated biofilms grew as well as S. gordonii monocultures. In coaggregate-inoculated flow cells the biovolumes of S. gordonii were significantly lower than with mixed-species biofilms inoculated sequentially or with monocultures of S. gordonii. Nevertheless, significant growth of S. gordonii in coaggregate-inoculated biofilms did occur between 1 and 14 h. Thus, S. gordonii is well adapted to growth independently or with coaggregation partners in flowing saliva.
In addition, these results suggest that the ability of S. gordonii to bind to the flow cell surface is lower when added to the flow cell as a coaggregate. The streptococci may be sequestered within the multigeneric coaggregates and, thus, less available for binding to the saliva-conditioned substratum. Previous studies examining the coaggregating partners Streptococcus oralis C104 (formerly named S. sanguis) and Prevotella loescheii PK1295 (formerly named Bacteroides loescheii) demonstrated that by forming coaggregates of S. oralis in the presence of a 10-fold excess of P. loescheii, dual-species rosettes were produced (14). S. oralis was effectively sequestered within the rosettes and unable to attach to its coaggregation partners; however, the rosettes formed large coaggregates with partners of P. loescheii (14). In the present study a coaggregate-inoculated biofilm may contain S. gordonii cells that are partially sequestered by surrounding coaggregating partners. Given that S. gordonii is the major binding species of the four examined here (57 to 73% of the total biovolume bound [Fig. 4]), sequestering this species, even partially within coaggregates, would have the result of reduced binding to the conditioning film of the flow cell.
These comparisons confirmed that S. gordonii accounted for the majority of the cells in the multispecies biofilm at both 1 and 14 h. In previous studies similar results were obtained using culture-dependent methods (19, 27, 28). In those studies, the authors utilized tooth scraping to remove dental plaque and plated the sonically dispersed plaque to estimate that 60 to 90% of the total cell counts were streptococci. In the present study using FISH S. gordonii, regardless of the order of inoculation or mode of inoculation, constituted between 57 and 73% of the total biovolume, indicating that S. gordonii is especially adapted to growth in saliva with or without other members of the oral bacterial community.
The growth of S. gordonii in mixed-species biofilms contrasted the growth of A. naeslundii and V. atypica. While neither of the latter two species grew significantly in monocultures or in sequentially inoculated biofilms even after 14 h of incubation, growth of A. naeslundii and V. atypica was detected in coaggregate-inoculated biofilms. These two species form mixed-species clusters, which may have contributed to their growth when inoculated as coaggregates. Several studies have demonstrated that symbiotic associations among oral bacteria have fostered the growth of one or both of the bacterial partners. For example, A. naeslundii has also been shown to form a mutualistic association with S. oralis 34 in biofilms, but A. naeslundii does not significantly grow in the absence of the streptococcus (29). Further, in vitro studies of veillonellae have demonstrated that veillonellae ferment short-chain organic acids, in particular lactate, excreted by streptococci and actinomyces (25). It has also been previously shown that in vivo, larger populations of veillonellae develop in cocultures with those streptococci that are coaggregating partners (23). These results suggest that for some oral bacteria, coaggregation may be an important strategy for retention and increased species diversity in the flow cell despite the lower overall growth of the total biofilm.
Although evidence for growth was detected in all three of the earlier colonizers, no growth was detected for the late colonizer, F. nucleatum. In fact a significant decrease in the biovolumes of F. nucleatum was observed in both sequentially and coaggregate-inoculated biofilms. The results of the LIVE/DEAD vitality staining indicated that even after 14 h in the flow cell more than 90% of the F. nucleatum cells were viable. Previous in vivo studies have shown that F. nucleatum was not prominent in dental plaque until early colonizers such as Streptococcus, Actinomyces, and Veillonella had colonized the tooth surface (34). The higher biovolumes of F. nucleatum at 1 h could be the result of the ability of F. nucleatum to coaggregate with all three of the other species. However, as the total biovolume of early colonizers increased during the 14 h incubation period, F. nucleatum failed to grow. In two other in vitro multispecies non-flow cell model systems, F. nucleatum, as part of consortia with different species composition than used here and different media supplemented with carbohydrate, became a dominant member of the multispecies communities after several days incubation (4, 10). Perhaps an anaerobic atmosphere or other environmental conditions, longer incubation times, or additional species within the flow cell used here would be needed to detect significant growth of the obligate anaerobe F. nucleatum in the mixed-species community with saliva as the sole source of nutrient.
Together these results indicate that although larger biofilm biomass can be obtained through the sequential addition of oral bacteria to the biofilm, the relative amounts of certain species can increase through the formation of planktonically preformed coaggregates. Coaggregates may enable the proper spatial location of different species and facilitate the opportunity to form essential partnerships within the growing biofilms, thus influencing the overall development of the complex microbial community.
Acknowledgments
We thank D. Blehert, P. Diaz, B. Duval, P. Egland, and R. Palmer for their useful comments on the manuscript and S. Taylor and B. Swaim for their technical support.
REFERENCES
- 1.Blehert, D. S., R. J. Palmer, Jr., J. B. Xavier, J. S. Almeida, and P. E. Kolenbrander. 2003. Autoinducer 2 production by Streptococcus gordonii DL1 and the biofilm phenotype of a luxS mutant are influenced by nutritional conditions. J. Bacteriol. 185:4851-4860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bos, R., H. C. van der Mei, and H. J. Busscher. 1996. Co-adhesion of oral microbial pairs under flow in the presence of saliva and lactose. J. Dent. Res. 75:809-815. [DOI] [PubMed] [Google Scholar]
- 3.Bos, R., H. C. van der Mei, J. M. Meinders, and H. J. Busscher. 1994. A quantitative method to study co-adhesion of microorganisms in a parellel plate flow chamber: basic principles of the analysis. J. Microbiol. Meth. 20:289-305. [Google Scholar]
- 4.Bradshaw, D. J., P. D. Marsh, C. Allison, and K. M. Schilling. 1996. Effect of oxygen, inoculum composition and flow rate on development of mixed-culture oral biofilms. Microbiology 142:623-629. [DOI] [PubMed] [Google Scholar]
- 5.Ciardi, J. E., G. F. McCray, P. E. Kolenbrander, and A. Lau. 1987. Cell-to-cell interaction of Streptococcus sanguis and Propionibacterium acnes on saliva-coated hydroxyapatite. Infect. Immun. 55:1441-1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cisar, J. O., P. E. Kolenbrander, and F. C. McIntire. 1979. Specificity of coaggregation reactions between human oral streptococci and strains of Actinomyces viscosus or Actinomyces naeslundii. Infect. Immun. 24:742-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dawes, C., S. Watanabe, P. Biglow-Lecomte, and G. H. Dibdin. 1989. Estimation of the velocity of the salivary film at some different locations in the mouth. J. Dent. Res. 68:1479-1482. [DOI] [PubMed] [Google Scholar]
- 8.de Jong, M. H., and J. S. van der Hoeven. 1987. The growth of oral bacteria on saliva. J. Dent. Res. 66:498-505. [DOI] [PubMed] [Google Scholar]
- 9.Gibbons, R. J., and M. Nygaard. 1970. Interbacterial aggregation of plaque bacteria. Arch. Oral Biol. 15:1397-1400. [DOI] [PubMed] [Google Scholar]
- 10.Guggenheim, M., S. Shapiro, R. Gmur, and B. Guggenheim. 2001. Spatial arrangements and associative behavior of species in an in vitro oral biofilm model. Appl. Environ. Microbiol. 67:1343-1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holmes, A. R., R. McNab, and H. F. Jenkinson. 1996. Candida albicans binding to the oral bacterium Streptococcus gordonii involves multiple adhesin-receptor interactions. Infect. Immun. 64:4680-4685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kolenbrander, P. E. 1988. Intergeneric coaggregation among human oral bacteria and ecology of dental plaque. Annu. Rev. Microbiol. 42:627-656. [DOI] [PubMed] [Google Scholar]
- 13.Kolenbrander, P. E. 2000. Oral microbial communities: biofilms, interactions, and genetic systems. Annu. Rev. Microbiol. 54:413-437. [DOI] [PubMed] [Google Scholar]
- 14.Kolenbrander, P. E., and R. N. Andersen. 1988. Intergeneric rosettes: sequestered surface recognition among human periodontal bacteria. Appl. Environ. Microbiol. 54:1046-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kolenbrander, P. E., R. N. Andersen, D. S. Blehert, P. G. Egland, J. S. Foster, and R. J. Palmer, Jr. 2002. Communication among oral bacteria. Microbiol. Mol. Biol. Rev. 66:486-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kolenbrander, P. E., R. N. Andersen, and L. V. Moore. 1989. Coaggregation of Fusobacterium nucleatum, Selenomonas flueggei, Selenomonas infelix, Selenomonas noxia, and Selenomonas sputigena with strains from 11 genera of oral bacteria. Infect. Immun. 57:3194-3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kroes, I., P. W. Lepp, and D. A. Relman. 1999. Bacterial diversity within the human subgingival crevice. Proc. Natl. Acad. Sci. USA 96:14547-14552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li, Y. H., N. Tang, M. B. Aspiras, P. C. Lau, J. H. Lee, R. P. Ellen, and D. G. Cvitkovitch. 2002. A quorum-sensing signaling system essential for genetic competence in Streptococcus mutans is involved in biofilm formation. J. Bacteriol. 184:2699-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liljemark, W. F., C. G. Bloomquist, C. L. Bandt, B. L. Pihlstrom, J. E. Hinrichs, and L. F. Wolff. 1993. Comparison of the distribution of Actinomyces in dental plaque on inserted enamel and natural tooth surfaces in periodontal health and disease. Oral Microbiol. Immunol. 8:5-15. [DOI] [PubMed] [Google Scholar]
- 20.Maidak, B. L., J. R. Cole, T. G. Lilburn, C. T. Parker, Jr., P. R. Saxman, R. J. Farris, G. M. Garrity, G. J. Olsen, T. M. Schmidt, and J. M. Tiedje. 2001. The RDP-II (Ribosomal Database Project). Nucleic Acids Res. 29:173-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Malik, A., and K. Kakii. 2003. Intergeneric coaggregations among Oligotropha carboxidovorans and Acinetobacter species present in activated sludge. FEMS Microbiol. Lett. 224:23-28. [DOI] [PubMed] [Google Scholar]
- 22.Malik, A., M. Sakamoto, S. Hanazaki, M. Osawa, T. Suzuki, M. Tochigi, and K. Kakii. 2003. Coaggregation among nonflocculating bacteria isolated from activated sludge. Appl. Environ. Microbiol. 69:6056-60643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McBride, B. C., and J. S. van der Hoeven. 1981. Role of interbacterial adherence in colonization of the oral cavities of gnotobiotic rats infected with Streptococcus mutans and Veillonella alcalescens. Infect. Immun. 33:467-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McNab, R., S. K. Ford, A. El-Sabaeny, B. Barbieri, G. S. Cook, and R. J. Lamont. 2003. LuxS-based signaling in Streptococcus gordonii: autoinducer 2 controls carbohydrate metabolism and biofilm formation with Porphyromonas gingivalis. J. Bacteriol. 185:274-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mikx, F. H., and J. S. van der Hoeven. 1975. Symbiosis of Streptococcus mutans and Veillonella alcalescens in mixed continuous cultures. Arch. Oral Biol. 20:407-410. [DOI] [PubMed] [Google Scholar]
- 26.Moore, W. E. C., and L. V. H. Moore. 1994. The bacteria of periodontal diseases. Periodontol. 2000 5:66-77. [DOI] [PubMed] [Google Scholar]
- 27.Nyvad, B., and M. Kilian. 1990. Comparison of the initial streptococcal microflora on dental enamel in caries-active and in caries-inactive Individuals. Caries Res. 24:267-272. [DOI] [PubMed] [Google Scholar]
- 28.Nyvad, B., and M. Kilian. 1987. Microbiology of the early colonization of human enamel and root surfaces in vivo. Scand. J. Dent. Res. 95:369-380. [DOI] [PubMed] [Google Scholar]
- 29.Palmer, R. J., Jr., K. Kazmerzak, M. C. Hansen, and P. E. Kolenbrander. 2001. Mutualism versus independence: strategies of mixed-species oral biofilms in vitro using saliva as the sole nutrient source. Infect. Immun. 69:5794-5804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paster, B. J., I. M. Bartoszyk, and F. E. Dewhirst. 1998. Identification of oral streptococci using PCR-based, reverse-capture, checkerboard hybridization. Methods Cell Sci. 20:223-231. [Google Scholar]
- 31.Paster, B. J., S. K. Boches, J. L. Galvin, R. E. Ericson, C. N. Lau, V. A. Levanos, A. Sahasrabudhe, and F. E. Dewhirst. 2001. Bacterial diversity in human subgingival plaque. J. Bacteriol. 183:3770-3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rickard, A. H., P. Gilbert, N. J. High, P. E. Kolenbrander, and P. S. Handley. 2003. Bacterial coaggregation: an integral process in the development of multi-species biofilms. Trends Microbiol. 11:94-100. [DOI] [PubMed] [Google Scholar]
- 33.Rickard, A. H., A. J. McBain, R. G. Ledder, P. S. Handley, and P. Gilbert. 2003. Coaggregation between freshwater bacteria within biofilm and planktonic communities. FEMS Microbiol. Lett. 220:133-140. [DOI] [PubMed] [Google Scholar]
- 34.Socransky, S. S., A. D. Haffajee, M. A. Cugini, C. Smith, and R. L. Kent, Jr. 1998. Microbial complexes in subgingival plaque. J. Clin. Periodontol. 25:134-144. [DOI] [PubMed] [Google Scholar]
- 35.Whittaker, C. J., C. M. Klier, and P. E. Kolenbrander. 1996. Mechanisms of adhesion by oral bacteria. Annu. Rev. Microbiol. 50:513-552. [DOI] [PubMed] [Google Scholar]