Abstract
We designed a real-time PCR assay able to recognize dioxygenase large-subunit gene sequences with more than 90% similarity to the Ralstonia sp. strain U2 nagAc gene (nagAc-like gene sequences) in order to study the importance of organisms carrying these genes in the biodegradation of naphthalene. Sequencing of PCR products indicated that this real-time PCR assay was specific and able to detect a variety of nagAc-like gene sequences. One to 100 ng of contaminated-sediment total DNA in 25-μl reaction mixtures produced an amplification efficiency of 0.97 without evident PCR inhibition. The assay was applied to surficial freshwater sediment samples obtained in or in close proximity to a coal tar-contaminated Superfund site. Naphthalene concentrations in the analyzed samples varied between 0.18 and 106 mg/kg of dry weight sediment. The assay for nagAc-like sequences indicated the presence of (4.1 ± 0.7) × 103 to (2.9 ± 0.3) × 105 copies of nagAc-like dioxygenase genes per μg of DNA extracted from sediment samples. These values corresponded to (1.2 ± 0.6) × 105 to (5.4 ± 0.4) × 107 copies of this target per g of dry weight sediment when losses of DNA during extraction were taken into account. There was a positive correlation between naphthalene concentrations and nagAc-like gene copies per microgram of DNA (r = 0.89) and per gram of dry weight sediment (r = 0.77). These results provide evidence of the ecological significance of organisms carrying nagAc-like genes in the biodegradation of naphthalene.
Natural environments harbor diverse and often uncharacterized microbial populations able to degrade a wide variety of pollutants (36). Difficulties studying these microbial consortia arise because they are highly complex and because conventional cultivation methods can isolate only a small fraction of the microorganisms present in the environment (37). Real-time PCR provides the opportunity of monitoring and enumerating bacterial populations in contaminated environments without cultivation biases (4). This technique detects the accumulation of PCR products in each cycle of the process, which can be used to estimate the amount of target present prior to the start of the amplification (10).
Polycyclic aromatic hydrocarbons (PAHs), which are common constituents of petroleum, coal tar, and shale oil and which are formed in the combustion of fossil fuels can be toxic, mutagenic, and/or carcinogenic (30). Naphthalene is often used as a model compound to study PAH biodegradation (11), and the majority of the isolated naphthalene-degrading bacteria are species of Pseudomonas (41). In these organisms, naphthalene biodegradation genes (called nah genes) are located in two operons: the upper pathway encodes the conversion of naphthalene to salicylate, and the lower pathway encodes the conversion of salicylate to central metabolites via catechol (18). The initial reaction in the degradation of naphthalene is catalyzed by a multicomponent enzyme system that adds both atoms of dioxygen to the aromatic ring to form cis-1,2-dihydroxy-1,2-dihydronaphthalene (6). This enzyme system consists of an iron-sulfur flavoprotein reductase, an iron-sulfur ferredoxin, and a catalytic oxygenase component (3). The catalytic component, called naphthalene 1,2-dioxygenase (NDO), is an α3β3 hexamer (17). The α subunit (NDOα), containing the active site and frequently called nahAc when identified in archetypical Pseudomonas-type genes, is usually used as a target for molecular studies (22, 40) because it is believed to confer substrate specificity (28).
A more recently studied naphthalene biodegradation pathway is the one from Ralstonia sp. strain U2, isolated from oil-contaminated soil in Venezuela (8). Ralstonia sp. strain U2 converts naphthalene to central metabolites via gentisate, and the genes for the complete pathway (called nag genes) appear to be on a single operon carried on a large plasmid (9, 43). Genes encoding the conversion of naphthalene to salicylate in this strain are homologous to those of the archetypical nah upper pathway, suggesting a common ancestry, with the NDOα gene being called nagAc (9). Two additional open reading frames (nagGH), encoding the salicylate 5-hydroxylase enzyme, which converts salicylate to gentisate, are located between the nagAa and nagAb genes (42).
Comamonas testosteroni GZ42, with the ability to grow on naphthalene as well as phenanthrene, also presents a nag-like arrangement of genes (44), with high sequence similarity with the nag operon (9). Genes dntAc, ntdAc, and nbzAc, involved in 2,4-dinitrotoluene (16, 34), 2-nitrotoluene (27), and nitrobenzene (23) degradation, respectively, also present high sequence similarities with the NDOα gene from Ralstonia sp. strain U2 (9). In addition, sequences with more than 90% similarity to the nagAc gene of Ralstonia sp. strain U2 were detected in mRNA extracted from groundwater samples (40). Zhou and coworkers suggested that the nag-like route of naphthalene biodegradation might be a relative rarity (42). However, the abundance of nagAc-like NDO gene sequences in libraries from environmental samples and the isolation of naphthalene-degrading strains with nag-type NDOα sequences in several continents (8, 15, 38, 44) suggest that these organisms may be broadly distributed.
The objective of this study was to investigate the abundance of organisms carrying dioxygenase gene sequences with more than 90% similarity to that of the Ralstonia sp. strain U2 nagAc gene (nagAc-like genes) in the environment. To accomplish this goal, we designed a real-time PCR assay, which was applied to DNA extracted from surficial freshwater sediment samples obtained in or in close proximity to a coal tar-contaminated Superfund site. A positive correlation between the abundance of nagAc-like gene sequences and naphthalene concentrations measured in these sediments was found. This work represents a significant step in the elucidation of the ecological significance of indigenous bacteria carrying nagAc-like NDOα gene sequences in aerobic naphthalene biodegradation.
MATERIALS AND METHODS
Bacterial strains.
Ralstonia sp. strain U2 (ATCC BAA-423 [8]) liquid cultures were grown in BBL Trypticase soy broth (Becton Dickinson, Sparks, Md.), and Pseudomonas putida G7 was grown in the phosphate-buffered mineral salts medium PAS broth (2), with naphthalene added as solids to the liquid medium. All strains were cultivated at room temperature with constant shaking (150 rpm).
Sediment samples.
Nine surficial sediment samples were collected in January 2002 from Chattanooga Creek, located in southeast Tennessee (Chattanooga, Tenn.). Samples were immediately frozen in dry ice and stored at −80°C for nucleic acid extraction.
Chemical analysis.
PAHs were extracted from sediment samples with an accelerated solvent extractor (ASE-300; Dionex Corp., Salt Lake City, Utah). Sample extracts were analyzed with a Hewlett-Packard (HP) gas chromatograph (model 6890) equipped with mass-selective detector (model 5973) and an HP autosampler complying with Environmental Protection Agency (EPA) standard method 8270D.
DNA extractions. (i) Pure cultures.
Total DNA was extracted from cells recovered from 10 ml of pure cultures of P. putida G7 and Ralstonia sp. strain U2 in stationary phase with the FastDNA kit (Q-BIOgene, Carlsbad, Calif.) by following the manufacturer's instructions.
(ii) Sediment samples.
Total DNA was purified in triplicate from sediment samples with the FastDNA SPIN kit for soil (Q-BIOgene) with minor modifications as follows. In each DNA extraction 109 copies of plasmid pUTK215 carrying the Vibrio fischeri luxCDABE cassette (13) were added to 0.5 g of wet sediment, in order to assess DNA recovery. Samples were homogenized twice for 30 s each at a speed of 5.5 m/s with a FastPrep FP120 instrument (Q-BIOgene). This treatment produced adequate cell lysis without producing excessive shearing of the DNA (data not shown). The washing of the binding matrix-DNA complex and elution were performed as described previously (5), except that DNA was eluted in a total volume of 100 μl of 10 mM Tris-HCl, pH 8.0. DNA recovery was calculated for each individual extraction from the recovery of the luxE gene measured by real-time PCR as described below. A recovery of 109 copies of luxE gene per extraction corresponded to 100% DNA recovery. DNA recovery values were used to compensate for DNA losses during extraction only when copies of the target were expressed per gram of dry weight sediment (dw). DNA was quantified with a PicoGreen double-stranded DNA quantitation kit (Molecular Probes, Eugene, Oreg.), with lambda DNA as the standard, in accordance with the manufacturer's instructions.
Real-time PCR assays.
Real-time PCR assays were carried out using 1× QuantiTect probe supermix (QIAGEN, Valencia, Calif.) in a total volume of 25 μl. Primers and probe were synthesized by Biosearch Technologies, Inc. (Novato, Calif.). Amplification and detection were performed with the DNA Engine Opticon continuous fluorescence detection system (MJ Research, Waltham, Mass.). All PCR runs included control reactions without template and standard DNA with concentrations between 10 and 108 copies/reaction for nagAc-like gene and luxE assays or 103 to 108 copies for the 16S rRNA gene assay. The numbers of copies of each target per PCR was calculated by comparison of threshold cycles obtained in each PCR run from standard DNA.
(i) nagAc-like gene real-time PCR assay.
Sequences and concentrations of forward primer (nagAc-like-F), reverse primer (nagAc-like-R), and probe (nagAc-like-P) are shown in Table 1. The PCR program included 1 cycle at 50.0°C for 2 min, 1 cycle of 95.0°C for 15 min, and then 40 cycles of 95.0°C for 30 s and 59°C for 30 s. Standard DNA consisted of plasmid pCR4-TOPO (Invitrogen, Carlsbad, Calif.) carrying a PCR product of an NDOα gene amplified from sediment sample DNA and sequenced to confirm its identity. The closest match of standard DNA was the Polaromonas naphthalenivorans strain CJ2 naphthalene dioxygenase gene (15) (GenBank accession number AY194932; 100% similarity). In addition, eight NDOα clones from uncultured bacteria have 100% similarity with standard DNA (GenBank accession numbers AF099763, AF099764, AF099767, AF099770, AF099772, AF099773, AY194929, and AY250113). For the analysis of sediment DNA, triplicate real-time PCR measurements were performed at two different DNA concentrations (between 5 and 50 ng of sediment DNA/PCR). Specificity of the assay was analyzed by cloning real-time PCR products from three different sediment samples into pCR4-TOPO (Invitrogen) and randomly selecting and sequencing 10 clones from each library.
TABLE 1.
Primers and probes used for the quantification of nagAc-like, bacterial 16S rRNA, and luxE genes
| Target | Namea | Sequence (5′-3′) | Concn (nM) | Amplicon size (bp) | Reference or source |
|---|---|---|---|---|---|
| nagAc-like genes | nagAc-like-F | GGCTGTTTTGATGCAGA | 600 | 107 | This study |
| nagAc-like-R | GGGCCTACAAGTTCCA | 600 | |||
| nagAc-like-P | FAM-CCAGGCTGCATCACCCAGATA-BHQ-1 | 300 | |||
| luxE (Vibrio fischeri) | luxE-F | TATTGCACGTGATCGTCTTA | 500 | 84 | This study |
| luxE-R | TGCTCAAACCAATCTCCA | 500 | |||
| luxE-P | FAM-ACGCTTGCTTGGTTCTGTCAACTTC-BHQ-1 | 100 | |||
| 16S rRNA genes (bacteria) | 1055f | ATGGCTGTCGTCAGCT | 600 | 352 | 12 |
| 1392r | ACGGGCGGTGTGTAC | 600 | |||
| 16STaq1115-BHQ | FAM-CAACGAGCGCAACCC-BHQ-1 | 125 |
F, forward primer; R, reverse primer; P, TaqMan probe; FAM, 6-carboxyfluorescein; BHQ-1, Black Hole Quencher.
(ii) luxE real-time PCR.
Forward primer (luxE-F), reverse primer (luxE-R), and probe (luxE-P) sequences and concentrations are shown in Table 1. The PCR program used for the luxE assay was as follows: 1 cycle at 50.0°C for 2 min, 1 cycle of 95.0°C for 10 min, and then 50 cycles of 95.0°C for 15 s and 61°C for 30 s. Standard DNA consisted of the pUTK215 plasmid (13), used as an internal spike. To calculate DNA recovery, template was added as 5 μl of a 1:100 dilution of each sediment DNA (up to 3.7 ng of DNA/reaction) and measured in triplicate reactions.
(iii) Bacterial 16S rRNA gene real-time PCR.
Bacterial 16S rRNA gene quantifications were performed by a previously described real-time PCR assay (12) with minor modifications, as follows. The concentrations of 1055f and 1392r primers and 16STaq1115-BHQ (carrying a Black Hole Quencher instead of carboxytetramethylrhodamine) are shown in Table 1. The PCR program used for the bacterial 16S rRNA gene assay included 1 cycle at 50.0°C for 2 min, 1 cycle of 95.0°C for 15 min, and then 45 cycles of 95.0°C for 30 s and 55°C for 45 s. Standard DNA consisted of dilutions of plasmid pCR2.1 carrying a bacterial 16S rRNA gene (GenBank accession number AF420301). The bacterial 16S rRNA gene target was quantified in sediment samples in triplicate with 1 ng of sediment DNA/PCR.
Multiple sequence alignments.
Sequence alignments were performed with the ClustalX (version 1.64b) program, with all parameters set at their default values (35).
Statistical analysis.
One-way analysis of variance was performed using SPSS, version 11.5 (SPSS Inc., Chicago, Ill.), with mean differences significant at the 0.05 level. The Tukey (b) post hoc test was used for multiple comparisons between groups. Data were normally distributed, and a log10 transformation was used when the assumption of equality of variance was not met with untransformed data (copies of nagAC-like gene per gram dw and copies of 16S rRNA gene per microgram of DNA and per gram dw). Correlations between variables were analyzed with the bivariate two-tailed Pearson correlation in SPSS version 11.5.
RESULTS
Design of a real-time PCR assay for the quantification of nagAc-like genes.
We designed a TaqMan real-time PCR assay to study the abundance and distribution of organisms carrying nagAc-like genes in environmental samples (Table 1). Target sequences of the assay were Ralstonia sp. strain U2 nagAc (GenBank accession number AF036940) (9), as well as 32 dioxygenase sequences publicly available by March 2002 and having similarities between 91.7 and 97.8% to nagAc. The majority of the target sequences represented NDOα genes from uncultured bacteria cloned from groundwater by reverse transcription PCR (GenBank accession numbers AF099748, AF099749, AF099752, AF099753, AF099755 to AF099759, AF099761 to AF099767, AF099769, AF099770, and AF099772 to AF099774) (40). Target NDOα genes from cultured representatives besides nagAc were pahAc, cloned from C. testosteroni (AF252550) (26), nahAc from Burkholderia sp. strain S1-17 (AF448048) and SOD-5b (AF448053), and nahAc from the PAH-degrading bacteria NJ2, NK2, NK3, and Ralstonia sp. strain NI1 (AB066442 to AB066445, respectively) (38). In addition, we targeted other closely related dioxygenase genes involved in 2,4-dinitrotoluene (U62430 and AF169302), 2-nitrotoluene (U49504), and nitrobenzene degradation (AF379638), with 93.1 to 94.6% similarities at the nucleotide level to the nagAc gene (16, 23, 27, 34). The most related nontarget sequences of the designed assay were Pseudomonas-type NDOα genes, with similarities to the nagAc gene between 76.3 and 80.7%.
Specificity of the nagAc-like gene real-time PCR assay.
The designed nagAc-like gene real-time PCR assay was optimized and applied to freshwater sediment samples. Cloning and sequencing of PCR products was used to analyze the specificity of the amplification. Three gene libraries were constructed from real-time PCRs of sediment samples with different degrees of naphthalene contamination (0.32, 52.8, and 106 mg/kg dw). Ten clones from each library were sequenced, and the closest match of each clone with BLAST (www.ncbi.nlm.nih.gov/BLAST) corresponded to a target nagAc-like sequence. In addition, sequences showed some degree of diversity in the 74 bp located between primers, suggesting that the assay is capable of detecting a variety of target sequences. For 20 clones, the closest cultured match corresponded to sequence AF252550 from C. testosteroni (97.3 to 100% similarity). In 10 clones the closest cultured match corresponded to the nahAc gene from the recently isolated P. naphthalenivorans strain CJ2 (15) (91.9 to 100% similarity). No sequence in the libraries was identical to the nagAc gene from Ralstonia sp. strain U2. In addition, 29 out of 30 clones had no mismatches with the probe sequence, and one clone carried one mismatch approximately in the center of the probe.
In addition to sequence analysis, Ralstonia sp. strain U2 and P. putida G7 total DNA was used to test the specificity of the nagAc-like gene real-time PCR assay. Ralstonia sp. strain U2 total DNA generated a strong amplification signal with the assay, with detection of 1.3 ± 0.1 × 108 nagAc copies per μg of total DNA. In contrast, total DNA from P. putida G7 produced in a very low and variable amplification signal and target quantifications of (8.2 ± 7.1) × 102 copies of the nagAc gene per μg of DNA. Overall, these results suggest that the designed real-time PCR assay is specific for nagAc-like sequences in sediment samples showing low levels of background signal from nontarget organisms, at least 5 orders of magnitude lower than specific amplifications.
Real-time PCR amplification efficiencies.
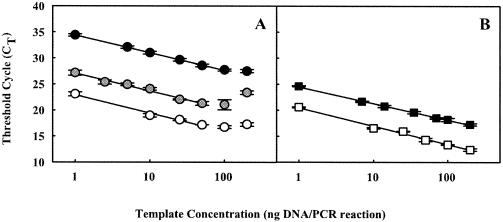
Optimum template concentrations and amplification efficiencies for the nagAc-like gene assay were determined by using total DNA extracted from Ralstonia sp. strain U2 or a PAH-contaminated sediment sample. Linear relationships between the logarithm of the template concentration and threshold cycle values indicated that 1 to 100 ng of DNA per 25-μl reaction mixture resulted in accurate target quantification in both cases (Fig. 1). However, at least 25 ng per reaction was needed to surpass the detection limit of 10 copies/reaction when using DNA extracted from sediments with low levels of naphthalene contamination (data not shown). Amplification efficiencies for this assay calculated based on the slope of the regressions shown in Fig. 1 were 0.97 and 1.04 for sediment and Ralstonia sp. strain U2 DNA, respectively. These amplification efficiencies suggest the duplication of almost all target molecules in each cycle, in the conditions tested for the assay.
FIG. 1.
Effect of template concentration in real-time PCR assays. (A) Total DNA extracted from contaminated sediments; (B) total DNA extracted from Ralstonia sp. strain U2. Black symbols, nagAc-like gene assay; gray symbols, luxE assay; white symbols, bacterial 16S rRNA gene assay. Equations of the regressions are as follows: y = 34.40 − 3.38x, r2 = 0.999 (nagAc assay plus sediment DNA); y = 24.51 − 3.22x, r2 = 0.998 (nagAc assay plus Ralstonia sp. strain U2 DNA); y = 27.11 − 3.42x, r2 = 0.982 (luxE assay plus sediment DNA); y = 22.92 − 3.49x, r2 = 0.986 (16S rRNA gene assay plus sediment DNA); y = 20.48 − 3.53x, r2 = 0.992 (16S rRNA gene assay plus Ralstonia sp. strain U2 DNA). Error bars indicate the standard deviations of triplicate real-time PCRs. CT, threshold cycle.
The V. fischeri luxE gene (Table 1) was used for spiking sediment samples in order to calculate DNA recovery. The amplification efficiency of the luxE assay was 0.96 for contaminated-sediment DNA at template concentrations between 1 and 50 ng per reaction (Fig. 1A). Amplification efficiencies obtained for 16S rRNA gene assay with pure culture and environmental sample DNA were 0.92 and 0.93, respectively. It has been previously reported that the 16S rRNA gene assay shows low amplification efficiency (12). However, because of high target concentration in most environmental samples, this assay was proven to be reliable as a measure of total bacterial biomass (5, 12). An inhibitory effect of real-time PCRs at high DNA concentrations was observed when using environmental samples, but not Ralstonia sp. strain U2 total DNA (Fig. 1). This effect may be associated with a low level of inhibitory-substance carryover from the sediment sample during the DNA extraction.
Analysis of freshwater sediment samples. (i) Sediment samples and DNA extractions.
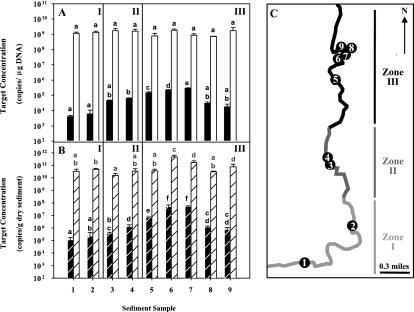
Nine sediment samples from three different sections of Chattanooga Creek were selected for molecular analysis (Fig. 2C). Zone I is located upstream of the PAH source, and chemical analysis of these samples indicated the presence of 0.18 to 0.32 mg of naphthalene/kg dw. The EPA recently remediated zone II by removing coal tar deposits and dredging contaminated sediments along a 1-mile section of the creek (phase I remediation plan, completed in November 1998). Naphthalene concentrations in analyzed samples of this section were 1.13 to 44.8 mg/kg dw. EPA is currently considering remediation of zone III. Samples of this section contained 44.4 to 106 mg of naphthalene/kg dw in samples 5 to 7, but much lower naphthalene concentrations downstream, in samples 8 and 9 (7.8 and 6.9 mg/kg dw, respectively). All samples showed, in addition, various levels of higher-molecular-weight PAHs (data not shown).
FIG. 2.
Molecular analysis of sediment samples. (A) Copies of target per microgram of DNA of the nagAc-like gene (black bars) and bacterial 16S rRNA gene (white bars) in Chattanooga Creek sediment samples. (B) Copies of target per gram dw of sediment of the nagAc gene (black bars with white stripes) and bacterial 16S rRNA gene (white bars with black stripes). Values of copies of the target per gram dw were corrected by DNA recovery calculated based on real-time PCR quantification of the spiked luxE gene, as indicated in Materials and Methods. Letters above bars indicate homogeneous subsets by the multiple-comparison Tukey (b) post hoc tests of untransformed data (copies of the nagAc-like gene per microgram of DNA) or log10-transformed data (copies of the nagAc-like gene per gram dw and copies of the 16S rRNA gene per microgram of DNA or per gram dw). Error bars indicate the standard deviations of values measured in triplicate DNA extractions. (C) Map of sampling area. I to III represent different segments of the creek. Zone I, background section upstream of the contaminated area of the creek (sampling sites 1 and 2); zone II, remediated section of Chattanooga Creek (sampling sites 3 and 4); zone III, section of the creek considered for remediation (sampling sites 5 to 9). Arrow, direction of the creek, as well as north (N).
Total DNA was extracted in triplicate from each sediment sample. Observed differences in DNA yields (Table 2) may be due to different amounts of biomass in the samples or, alternatively, to uneven losses of DNA during the extraction. We estimated the percentage of DNA recovery in each extraction by adding 109 copies of plasmid pUTK215, which contains the luxCDABE operon from V. fischeri (13), and analyzing its recovery by real-time PCR of the luxE gene. Total DNA recoveries varied significantly among samples (P < 0.001), although they were fairly consistent in triplicate extractions from the same sample (Table 2). Control DNA extractions without spiking with pUTK215 had very low luxE background signals (Table 2). Percentages of luxE recovery obtained in each DNA extraction were used to compensate for differences in nucleic acid recoveries when concentrations of the target were calculated as copies per gram dw.
TABLE 2.
DNA yields and luxE recovery in surficial sediment samples of Chattanooga Creek
| Zonea | Sample | DNA yieldb (μg of DNA/g dw) | luxE recoveryb (%) |
|---|---|---|---|
| I | 1 | 4.5 ± 0.8 (4.3) | 18.4 ± 8.0 (0.001) |
| 2 | 28.4 ± 1.5 | 82 ± 13.7 | |
| II | 3 | 9.6 ± 0.2 | 107.9 ± 3.3 |
| 4 | 10.3 ± 0.2 | 50.8 ± 10.8 | |
| III | 5 | 26.4 ± 4.1 | 55.2 ± 14.9 |
| 6 | 30.8 ± 1.3 (29) | 65.4 ± 16.5 (0.0004) | |
| 7 | 20 ± 0.9 | 54 ± 9.0 | |
| 8 | 15.7 ± 0.6 | 37.9 ± 3.4 | |
| 9 | 17.4 ± 0.2 | 37.8 ± 6.7 |
Zones I, II, and III correspond to background upstream from the source of contamination, remediated, and contaminated nonremediated sections of the creek, respectively (Fig. 2C).
Averages ± standard deviations for triplicate extractions are indicated. Values in parentheses for samples 1 and 6 indicate DNA yields and luxE recoveries for DNA extractions without luxE addition.
(ii) Target quantification.
Concentrations of nagAc-like gene targets calculated as copies per microgram of DNA are shown in Fig. 2A. Multiple comparisons of these data indicated that the concentration of the nagAc-like gene was significantly higher in samples 5 to 7 (P ≤ 0.008). After compensating for differences in DNA recovery among extractions, it was found that the concentration of the nagAc-like gene target varied between (1.2 ± 0.6) × 105 and (5.4 ± 0.4) × 107 copies per g dw (Fig. 2B). Multiple comparisons of log10-transformed data indicated that copies of nagAc-like gene per gram dw were also significantly higher (P ≤ 0.002) in samples 5 to 7 than in the rest of the analyzed samples.
As a way of estimating total bacterial biomass in sediment samples, a real-time PCR assay targeting most of the bacterial 16S rRNA gene (12) was applied to Chattanooga Creek total-DNA sediment samples. Copies of 16S rRNA gene per microgram of DNA were not significantly different among samples (log10-transformed data, P > 0.05; Fig. 2A). This was not surprising, as we expected to find approximately the same average chromosome size and average 16S rRNA gene copy number per cell in different sediment samples, assuming similar amounts of eukaryotic DNA. Similar amounts of 16S rRNA gene target per microgram of DNA were also observed in activated sludge samples from bench scale reactors operating at different retention times, with different amounts of suspended biosolids (5). In contrast, significant differences among samples in 16S rRNA gene copies were found when amounts were calculated as copies of this target per gram dw (log10-transformed data, P < 0.001; Fig. 2B). This result suggests that differences in DNA yields were due not only to uneven DNA recoveries but also to different amounts in initial biomass.
(iii) Correlation with naphthalene concentrations.
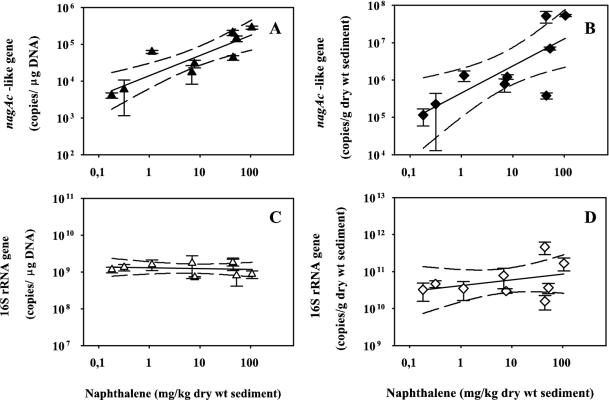
A positive correlation between naphthalene concentrations and the number of copies of the nagAc-like gene target measured in sediment samples was found (Fig. 3A and B). The correlation coefficient for these two variables was higher when the target was calculated as copies per microgram of DNA (r = 0.89) than as copies per gram dw (log10-transformed data, r = 0.77). Both correlations were significant at the 0.01 level. This difference may be due to inaccuracies of the measurement of DNA recovery introduced to the values of the nagAc-like target during normalization. Alternatively, copies per microgram of DNA may be a better indicator, indirectly evaluating the proportion of the population carrying the measured gene, since no significant differences among samples in 16S rRNA gene target per microgram of DNA were found (Fig. 2A). In contrast, there was no significant correlation between naphthalene concentrations and copies of 16S rRNA gene target per microgram of DNA (log10-transformed data, r = −0.213, P = 0.285; Fig. 3C). A low but significant correlation (log10-transformed data, r = 0.409, P = 0.034; Fig. 3D) between the number of copies of 16S rRNA gene per gram dw and naphthalene concentrations was found. These results suggest that naphthalene concentration may influence the sizes of populations carrying the nagAc-like gene in surficial freshwater sediments but has a small effect on the general bacterial population.
FIG. 3.
Correlations between target and naphthalene concentrations in Chattanooga Creek sediment samples. (A) Copies of the nagAc-like gene target per microgram of DNA (y = 4.14 + 0.55x, r2 = 0.747); (B) copies of the nagAc-like gene target per gram dw (y = 5.65 + 0.72x, r2 = 0.586); (C) copies of the bacterial 16S rRNA gene per microgram of DNA (y = 9.11 − 0.023x, r2 = 0.021); (D) copies of the bacterial 16S rRNA gene per gram dw (y = 10.62 + 0.16x, r2 = 0.128). Values of copies of the target per gram dw were corrected by DNA recovery calculated based on real-time PCR quantification of the spiked luxE gene, as indicated in Materials and Methods. Dashed lines, 95% confidence intervals; error bars, standard deviations of triplicate DNA extractions.
DISCUSSION
Biodegradation usually modifies the microbial community, in particular if targets promote cell growth, and these secondary effects (e.g., population expansion or shifts in the relative proportion of cell types) can be used to evaluate this process (32). We were interested in studying the importance of organisms carrying nagAc-like genes in the biodegradation of naphthalene. We designed a real-time PCR assay able to recognize NDOα gene sequences more than 90% similar to that of the Ralstonia sp. strain U2 nagAc gene. Sequences cloned from both uncultured organisms and pure cultures were used for the design of this assay, as a way of obtaining a more realistic representation of this group's diversity. The assay was optimized and applied to PAH-contaminated freshwater sediment samples.
Since the design of the assay, new nagAc-like sequences have been incorporated into the databases. An analysis of NDOα genes from uncultured bacteria as well as pure cultures publicly available to date indicated 0 mismatches of the designed nagAc-like real-time PCR primers and probe with 37 sequences with similarities of between 91.7 and 97.8% to the Ralstonia sp. strain U2 nagAc gene. In addition, six sequences of the naphthalene dioxygenase gene with more than 91.7% similarity to the nagAc gene had one mismatch in a primer or the probe, but none of them in a critical position, such as in the 3′ end of a primer. The designed forward primer has one mismatch with the Ralstonia sp. strain U2 nagAc gene in the third base close to its 5′ end, where this gene carries a T instead of a C, as do the rest of the available nagAc-like sequences. No sequences with similarities higher than 91.7% to the nagAc gene in the databases has more than one mismatch with the designed real-time PCR system.
Due to their high similarity to nag-type NDOα gene sequences, dntAc, ntdAc, and nbzAc genes, involved in the degradation of nitroaromatic compounds, can also be recognized by the nagAc-like gene real-time PCR assay. It has been suggested that the gene encoding the initial dioxygenase involved in the biodegradation of these compounds recently diverged from a naphthalene degradation pathway similar to that of nag of Ralstonia sp. strain U2 (16). Although strains carrying these genes are not able to grow with naphthalene or salicylate as a carbon source, their initial dioxygenases present relaxed substrate specificities and are able to transform naphthalene (23, 24, 28).
All organisms isolated so far carrying nag-like genes for the degradation of naphthalene or nitroarenes belong to the beta subdivision of Proteobacteria, although some of them were formerly classified as Pseudomonas spp. It has been suggested that these distinctive nag-type NDOα genes may be related to the use of a gentisate pathway such as that for the well-characterized strain Ralstonia sp. strain U2 (26), although this important aspect remains to be determined. C. testosteroni CZ42 presents genes homologous to nagG and nagH encoding salicylate 5-hydroxylase, the enzyme that converts salicylate to gentisate (44). In addition, in C. testosteroni strain H part of the gene encoding salicylate 5-hydroxylase has been sequenced (26) and catechol 2,3-dioxygenase activity could not be measured (25). It will be of interest to know if the recently isolated P. naphthalenivorans strain CJ2 (14), a beta proteobacterium with an NDOα gene that belongs to a subcluster separate from nagAc (15) and the only cultured representative of this group of sequences (called clade I in reference 15), follows this alternative biodegradation pathway.
The closest nontarget sequences of the nagAc-like gene assay are NDOα genes cloned from Pseudomonas spp. and uncultured bacteria closely related to them, with similarities to the nagAc gene between 76.3 and 80.7%. The total number of mismatches of the nagAc-like gene real-time PCR assay with archetypical nah-type sequences (such as M83949) is 13 to 16 out of a total of 54 nucleotides present in the primers and probe set. Low unspecific amplification from pure cultures of P. putida G7 indicated that these sequences will not affect the quantification if both populations are present at approximately equal concentrations. In addition, no Pseudomonas-type sequences were found in 30 analyzed clones from libraries constructed from real-time PCRs of sediment samples. The designed primer and probe set has 12 or 13 mismatches with phnAc-type dioxygenase sequences, such as the one cloned from Burkholderia sp. strain RP007 (21) and related sequences from uncultured bacteria (GenBank accession numbers AY032935, AY032936, and AY032937) (22). On the other hand, the narAa gene cloned from the gram-positive bacterium Rhodococcus sp. strain NCIMB12038 (20) has 26 mismatches with the nagAc-like primers and probe.
The area chosen for testing the nagAc-like gene real-time PCR assay is located in an EPA Superfund site in Tennessee. Chattanooga Creek has historically been subjected to gross pollution by industrial waste discharges mainly, but not only, from a former coal carbonization facility. The main contaminant compounds found in the stream are PAHs, chlorinated solvents, organic solvents, pesticides, polychlorinated biphenyls, metals, and phenols (www.epa.gov/region4/waste/npl/npltn/tennprtn.htm). Naphthalene concentrations in Chattanooga Creek surficial sediment samples retrieved in January 2002 varied between 0.18 and 106 mg/kg dw. The lowest naphthalene concentration was found in sample 1, the furthermost sampling point upstream of the main source of contamination (zone I; Fig. 2B). The highest naphthalene concentration was found in sample 7, belonging to the nonremediated zone III. This sample also showed the highest phenanthrene, fluoranthene, fluorene, anthracene, pyrene, benzo[a]anthracene, chrysene, and benzo[b]fluoranthene concentrations of all analyzed samples (11,453, 8,426, 4,651, 2,658, 5,287, 2,144, 1,277, and 1,042 mg/kg dw, respectively) and highest nagAc-like gene target concentrations (Fig. 2). PAH concentrations in the remediated section of the creek (zone II) were in general lower than in section III, indicating that the excavation of sediments up to the bedrock reduced the level of contamination present in this Superfund site. However, fresh sediments covering section II still show considerable PAH contamination, and it is therefore of interest to study the intrinsic bioremediation capacity at this site.
A strong positive correlation between naphthalene and nagAc-like gene target concentrations was found (Fig. 3A and B). A lower but significant correlation (0.01 level) between phenanthrene and nagAc-like target concentrations was also found (r = 0.74 and 0.57 for copies per microgram of DNA and log10-transformed copies per gram dw, respectively; data not shown). These results suggest a strong influence of the levels of these contaminants on population size for organisms carrying nagAc-like genes. Such a high correlation was surprising, considering other factors that can affect population size, for instance, toxicity of naphthalene or other contaminants present in the sediment, oxygen and nutrient concentrations, and substrate bioavailability (30). In addition, populations of bacteria carrying nagAc-like NDOα genes should compete for the same growth substrate with other populations of this guild with different aerobic as well as anaerobic naphthalene biodegradation pathways.
Results from Laurie and Lloyd-Jones's work (22) suggest that the Pseudomonas-type genotype may not always be ecologically dominant. Quantitative studies using competitive PCR showed that phnAc genes similar to the one cloned from Burkholderia sp. strain RP007 were present at equivalent or greater concentrations than nahAc-like genes in contaminated soils (22). In the same study, neither target was detected in pristine soils, while the addition of naphthalene enriched phnAc but not nahAc genes (22). In contrast, other studies indicated an effect of naphthalene concentrations on nah-type populations. In one of these studies, nahAc transcript levels correlated positively with [14C]naphthalene mineralization rates, soil naphthalene concentrations, and nahAc gene frequency in soil samples (7). Furthermore, Sanseverino et al. (31) established a threshold level of naphthalene of approximately 100 mg/kg of soil, below which the nahAc-like naphthalene-degrading population is relatively inactive.
As found in this study with nagAc-like populations, higher concentrations of the nahAc gene were found in PAH-contaminated freshwater sediments than in sediments with only trace amounts of these compounds (19). In addition, transient responses of indigenous microorganisms to hydrocarbon exposure were detected in groundwater at a field scale study, where an enrichment of nah-like genes was followed by a population crash (33). In bioslurry treatments of dredged harbor sediments contaminated with PAHs the microbial community structure has been correlated with the presence of catabolic genes (such as the naphthalene dioxygenase gene) and PAH loss (29). Recently, a primer set for the detection and quantification of a larger group of NDOα sequences belonging to both nah- and nag-type organisms using SYBR green I has been published (1), and its application to environmental samples can provide important information of the distribution of these gene types in the future.
The quantification of distinct naphthalene dioxygenase genes in environmental samples is starting to provide important information concerning the ecological significance and the distribution of different naphthalene-degrading populations. The quantification of nagAc-like genes in sediment samples of Chattanooga Creek suggests an important role of bacteria carrying these genes in naphthalene biodegradation. Efforts are under way to quantify other important naphthalene-degrading populations and also to implement the use of the technique reverse transcription real-time PCR to allow the analysis of the expression of different types of NDOα genes in environmental samples.
Acknowledgments
This work was supported by the Center for Environmental Biotechnology, Research Center of Excellence, University of Tennessee, and by the Waste Management Research and Education Institute at the University of Tennessee. H.M.D. is a recipient of a postdoctoral fellowship from CONICET (National Research Council, Argentina).
Special thanks are extended to D. Gibson for providing P. putida strain G7. We thank W. Housain for technical assistance, K. Cook for the design of luxE real-time PCR assay, V. Vulava for his help during the sampling process, and M. Newman from the statistical consulting center, University of Tennessee, for his advice in the statistical analysis of the data. We also thank A. Layton, J. Sanseverino, and D. Castle for helpful suggestions to improve the manuscript.
REFERENCES
- 1.Baldwin, B. R., C. H. Nakatsu, and L. Nies. 2003. Detection and enumeration of aromatic oxygenase genes by multiplex and real-time PCR. Appl. Environ. Microbiol. 69:3350-3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bopp, L. H. 1986. Degradation of highly chlorinated PCBs by Pseudomonas strain LB400. J. Ind. Microbiol. 1:23-29. [Google Scholar]
- 3.Boyd, D. R., N. D. Sharma, and C. C. R. Allen. 2001. Aromatic dioxygenases: molecular biocatalysis and applications. Curr. Opin. Biotechnol. 12:564-573. [DOI] [PubMed] [Google Scholar]
- 4.Brunk, C., J. Li, and E. Avaniss-Aghajani. 2002. Analysis of specific bacteria from environmental samples using a quantitative polymerase chain reaction. Curr. Issues Mol. Biol. 4:13-18. [PubMed] [Google Scholar]
- 5.Dionisi, H. M., G. Harms, A. C. Layton, I. R. Gregory, J. Parker, S. A. Hawkins, K. G. Robinson, and G. S. Sayler. 2003. Power analysis for real-time PCR quantification of genes in activated sludge and analysis of the variability introduced by DNA extraction. Appl. Environ. Microbiol. 69:6597-6604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ensley, B., and D. Gibson. 1983. Naphthalene dioxygenase: purification and properties of a terminal oxygenase component. J. Bacteriol. 155:505-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fleming, J. T., J. Sanseverino, and G. S. Sayler. 1993. Quantitative relationship between naphthalene catabolic gene frequency and expression in predicting PAH degradation in soils at town gas manufacturing sites. Environ. Sci. Technol. 27:1068-1074. [Google Scholar]
- 8.Fuenmayor, S. L., and V. Rodriguez-Lemoine. 1992. Characterization of polycyclic aromatic hydrocarbons degradative soil Pseudomonas. Acta Cient. Venez. 43:349-354. [PubMed] [Google Scholar]
- 9.Fuenmayor, S. L., M. Wild, A. L. Boyes, and P. A. Williams. 1998. A gene cluster encoding steps in conversion of naphthalene to gentisate in Pseudomonas sp. strain U2. J. Bacteriol. 180:2522-2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ginzinger, D. G. 2002. Gene quantification using real-time quantitative PCR: a new technology hits the mainstream. Exp. Hematol. 30:503-512. [DOI] [PubMed] [Google Scholar]
- 11.Goyal, A. K., and G. J. Zylstra. 1997. Genetics of naphthalene and phenanthrene degradation by Comamonas testosteroni. J. Ind. Microbiol. Biotechnol. 19:401-407. [DOI] [PubMed] [Google Scholar]
- 12.Harms, G., A. Layton, H. Dionisi, I. Gregory, V. Garrett, S. Hawkins, K. Robinson, and G. Sayler. 2003. Real-time PCR quantification of nitrifying bacteria in a municipal wastewater treatment plant. Environ. Sci. Technol. 37:343-351. [DOI] [PubMed] [Google Scholar]
- 13.Hay, A. G., J. F. Rice, B. M. Applegate, N. G. Bright, and G. S. Sayler. 2000. A bioluminescent whole-cell reporter for detection of 2,4-dichlorophenoxyacetic acid and 2,4-dichlorophenol in soil. Appl. Environ. Microbiol. 66:4589-4594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jeon, C. O., W. Park, W. C. Ghiorse, and E. L. Madsen. 2004. Polaromonas naphthalenivorans sp. nov., a naphthalene-degrading bacterium from naphthalene-contaminated sediment. Int. J. Syst. Evol. Microbiol. 54:93-97. [DOI] [PubMed] [Google Scholar]
- 15.Jeon, C. O., W. Park, P. Padmanabhan, C. DeRito, J. R. Snape, and E. L. Madsen. 2003. Discovery of a bacterium, with distinctive dioxygenase, that is responsible for in situ biodegradation in contaminated sediment. Proc. Natl. Acad. Sci. USA 100:13591-13596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson, G. R., R. K. Jain, and J. C. Spain. 2002. Origins of the 2,4-dinitrotoluene pathway. J. Bacteriol. 184:4219-4232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kauppi, B., K. Lee, E. Carredano, R. Parales, D. Gibson, H. Eklund, and S. Ramaswamy. 1998. Structure of an aromatic-ring-hydroxylating dioxygenase-naphthalene 1,2-dioxygenase. Structure 6:571-586. [DOI] [PubMed] [Google Scholar]
- 18.Kurkela, S., H. Lehväslaiho, E. T. Palva, and T. H. Teeri. 1988. Cloning, nucleotide sequence and characterization of genes encoding naphthalene dioxygenase of Pseudomonas putida strain NCIB 9816. Gene 73:355-362. [DOI] [PubMed] [Google Scholar]
- 19.Langworthy, D. E., R. D. Stapleton, G. S. Sayler, and R. H. Findlay. 1998. Genotypic and phenotypic responses of a riverine microbial community to polycyclic aromatic hydrocarbon contamination. Appl. Environ. Microbiol. 64:3422-3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Larkin, M. J., C. C. Allen, L. A. Kulakov, and D. A. Lipscomb. 1999. Purification and characterization of a novel naphthalene dioxygenase from Rhodococcus sp. strain NCIMB12038. J. Bacteriol. 181:6200-6204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laurie, A. D., and G. Lloyd-Jones. 1999. The phn genes of Burkholderia sp. strain RP007 constitute a divergent gene cluster for polycyclic aromatic hydrocarbon catabolism. J. Bacteriol. 181:531-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laurie, A. D., and G. Lloyd-Jones. 2000. Quantification of phnAc and nahAc in contaminated New Zealand soils by competitive PCR. Appl. Environ. Microbiol. 66:1814-1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lessner, D. J., G. R. Johnson, R. E. Parales, J. C. Spain, and D. T. Gibson. 2002. Molecular characterization and substrate specificity of nitrobenzene dioxygenase from Comamonas sp. strain JS765. Appl. Environ. Microbiol. 68:634-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lessner, D. J., R. E. Parales, S. Narayan, and D. T. Gibson. 2003. Expression of the nitroarene dioxygenase genes in Comamonas sp. strain JS765 and Acidovorax sp. strain JS42 is induced by multiple aromatic compounds. J. Bacteriol. 185:3895-3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meyer, S., R. Moser, A. Neef, U. Stahl, and P. Kampfer. 1999. Differential detection of key enzymes of polyaromatic-hydrocarbon-degrading bacteria using PCR and gene probes. Microbiology 145:1731-1741. [DOI] [PubMed] [Google Scholar]
- 26.Moser, R., and U. Stahl. 2001. Insights into the genetic diversity of initial dioxygenases from PAH-degrading bacteria. Appl. Microbiol. Biotechnol. 55:609-618. [DOI] [PubMed] [Google Scholar]
- 27.Parales, J. V., A. Kumar, R. E. Parales, and D. T. Gibson. 1996. Cloning and sequencing of the genes encoding 2-nitrotoluene dioxygenase from Pseudomonas sp. JS42. Gene 181:57-61. [DOI] [PubMed] [Google Scholar]
- 28.Parales, R. E., M. D. Emig, N. A. Lynch, and D. T. Gibson. 1998. Substrate specificities of hybrid naphthalene and 2,4-dinitrotoluene dioxygenase enzyme systems. J. Bacteriol. 180:2337-2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ringelberg, D. B., J. W. Talley, E. J. Perkins, S. G. Tucker, R. G. Luthy, E. J. Bouwer, and H. L. Fredrickson. 2001. Succession of phenotypic, genotypic, and metabolic community characteristics during in vitro bioslurry treatment of polycyclic aromatic hydrocarbon-contaminated sediments. Appl. Environ. Microbiol. 67:1542-1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Samanta, K. S., O. V. Singh, and R. K. Jain. 2002. Polycyclic aromatic hydrocarbons: environmental pollution and bioremediation. Trends Biotechnol. 20:243-248. [DOI] [PubMed] [Google Scholar]
- 31.Sanseverino, J., C. Werner, J. Fleming, B. Applegate, J. M. King, and G. S. Sayler. 1993. Molecular diagnostics of polycyclic aromatic hydrocarbon biodegradation in manufactured gas plant soils. Biodegradation 4:303-321. [DOI] [PubMed] [Google Scholar]
- 32.Shannon, M. J. R., and R. Unterman. 1993. Evaluating bioremediation: distinguishing fact from fiction. Annu. Rev. Microbiol. 47:715-736. [DOI] [PubMed] [Google Scholar]
- 33.Stapleton, R. D., and G. S. Sayler. 2000. Changes in subsurface catabolic gene frequencies during natural attenuation of petroleum hydrocarbons. Environ. Sci. Technol. 34:1991-1999. [Google Scholar]
- 34.Suen, W., B. Haigler, and J. Spain. 1996. 2,4-Dinitrotoluene dioxygenase from Burkholderia sp. strain DNT: similarity to naphthalene dioxygenase. J. Bacteriol. 178:4926-4934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The ClustalX Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Watanabe, K. 2001. Microorganisms relevant to bioremediation. Curr. Opin. Biotechnol. 12:237-241. [DOI] [PubMed] [Google Scholar]
- 37.Watanabe, K., and P. W. Baker. 2000. Environmentally relevant microorganisms. J. Biosci. Bioeng. 89:1-11. [DOI] [PubMed] [Google Scholar]
- 38.Widada, J., H. Nojiri, K. Kasuga, T. Yoshida, H. Habe, and T. Omori. 2002. Molecular detection and diversity of polycyclic aromatic hydrocarbon-degrading bacteria isolated from geographically diverse sites. Appl. Microbiol. Biotechnol. 58:202-209. [DOI] [PubMed] [Google Scholar]
- 39.Widada, J., H. Nojiri, and T. Omori. 2002. Recent developments in molecular techniques for identification and monitoring of xenobiotic-degrading bacteria and their catabolic genes in bioremediation. Appl. Microbiol. Biotechnol. 60:45-59. [DOI] [PubMed] [Google Scholar]
- 40.Wilson, M. S., C. Bakermans, and E. L. Madsen. 1999. In situ, real-time catabolic gene expression: extraction and characterization of naphthalene dioxygenase mRNA transcripts from groundwater. Appl. Environ. Microbiol. 65:80-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yen, K.-M., and C. M. Serdar. 1988. Genetics of naphthalene catabolism in Pseudomonas. CRC Crit. Rev. Microbiol. 15:247-268. [DOI] [PubMed] [Google Scholar]
- 42.Zhou, N.-Y., J. Al-Dulayymi, M. S. Baird, and P. A. Williams. 2002. Salicylate 5-hydroxylase from Ralstonia sp. strain U2: a monooxygenase with close relationships to and shared electron transport proteins with naphthalene dioxygenase. J. Bacteriol. 184:1547-1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou, N.-Y., S. L. Fuenmayor, and P. A. Williams. 2001. nag genes of Ralstonia (formerly Pseudomonas) sp. strain U2 encoding enzymes for gentisate catabolism. J. Bacteriol. 183:700-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zylstra, G. J., E. Kim, and A. K. Goyal. 1997. Comparative molecular analysis of genes for polyaromatic hydrocarbon degradation. Genet. Eng. 19:257-269. [DOI] [PubMed] [Google Scholar]