Abstract
Households provide environments that encourage the formation of microbial communities, often as biofilms. Such biofilms constitute potential reservoirs for pathogens, particularly for immune-compromised individuals. One household environment that potentially accumulates microbial biofilms is that provided by vinyl shower curtains. Over time, vinyl shower curtains accumulate films, commonly referred to as “soap scum,” which microscopy reveals are constituted of lush microbial biofilms. To determine the kinds of microbes that constitute shower curtain biofilms and thereby to identify potential opportunistic pathogens, we conducted an analysis of rRNA genes obtained by PCR from four vinyl shower curtains from different households. Each of the shower curtain communities was highly complex. No sequence was identical to one in the databases, and no identical sequences were encountered in the different communities. However, the sequences generally represented similar phylogenetic kinds of organisms. Particularly abundant sequences represented members of the α-group of proteobacteria, mainly Sphingomonas spp. and Methylobacterium spp. Both of these genera are known to include opportunistic pathogens, and several of the sequences obtained from the environmental DNA samples were closely related to known pathogens. Such organisms have also been linked to biofilm formation associated with water reservoirs and conduits. In addition, the study detected many other kinds of organisms at lower abundances. These results show that shower curtains are a potential source of opportunistic pathogens associated with biofilms. Frequent cleaning or disposal of shower curtains is indicated, particularly in households with immune-compromised individuals.
Cases of opportunistic infections in humans have increased steadily over the past decade, and often the source of infection remains unidentified (6, 13, 20, 25, 27). The expanding caseloads of opportunistic infections correspond to a rising number of immune-compromised patients, many of whom self-medicate (20, 25, 37, 43). Potential or adventitious pathogens in households pose a particular threat to such patients (24). Households provide many environments in which microorganisms can thrive, often with the formation of biofilms. Bacteria have been cultured from many environments in and around homes, particularly in moist settings such as those involving water pipes, toothbrushes, and spas (12, 14, 29, 30, 38). Although evidence of microbial growth and biofilm formation is ubiquitous in households, little is known about the diversity and complexity of the organisms that make up household microbial communities.
Several studies have shown that domestic water supplies can be a source of opportunistic infectious agents, and household plumbing accumulates numerous microorganisms (5, 55). Potentially infectious agents such as Mycobacterium spp. and Legionella spp. have been detected in water systems and may serve as reservoirs for infection (10, 24, 32, 33). One water-related setting that may provide a persistent reservoir for pathogenic microorganisms is shower curtain biofilms, although there is little information on the makeup of the microbial communities that compose such biofilms.
In order to identify the kinds of organisms that colonize shower curtain biofilms, we undertook a molecular survey of several such communities. In our study, we isolated microbial-community DNA from shower curtains and used a PCR-based molecular survey to determine the phylogenetic diversity of 16S rRNA gene sequences from these communities. Numerous studies have utilized 16S rRNA sequences to assess the nature of microbial organisms in the environment without the requirement for culture (1, 2, 22, 39). The rRNA sequence collection constitutes a rough census of this particular community. The phylogenetic identification of the constituent microbes can provide some insight into their natures by comparison with available cultured organisms.
MATERIALS AND METHODS
Samples.
DNA was extracted from five different samples taken from four different vinyl shower curtains, all in use for more than 6 months in Boulder, Colo. These samples consisted of (i) two samples from the same shower curtain, one from the bottom section (whitish pink flakes) of a dry shower curtain that had been stored at 25°C for about 1 week subsequent to wetting (SC1A) and one sample from a pink film on a corner of the curtain that was folded over and constantly wet (SC1B); (ii) one sample of white flakes from a dry curtain that had been stored at 25°C for about 1 week subsequent to a previous wetting (SC2); (iii) one sample of pinkish flakes from the bottom section of a dry curtain that had been stored at 25°C for about 1 week subsequent to wetting (SC3); and (iv) one sample of a pinkish orange biofilm from a wet shower curtain that was often used and essentially continuously moist (SC4). Dry biofilm flakes or moist biofilms were scraped from shower curtains and stored at −80°C pending extraction of DNA.
Epifluorescence microscopy.
Biofilm (“shower scum”) was hydrated with sterile phosphate-buffered saline (0.01 M) dispersed by vortex mixing, and a smear was dried on a microscope slide, stained with 10 μg of 4′6-diamidino-2-phenylindole (DAPI; Sigma, St. Louis, Mo.)/ml, and mounted with antifadent (CitiFluor Ltd., Leicester, England). Slides were examined with an epifluorescence microscope (Eclipse E400; Nikon Instruments Inc., Melville, N.Y.) with a SPOT camera and software (Diagnostic Instruments Inc., Sterling Heights, Mich.).
DNA extraction.
Samples from shower curtains were dispersed in sterile purified water (Fluka Chemical Corp., Milwaukee, Wis.) prior to extraction. All chemical buffers were made by using purified water to minimize contamination with external DNA sources. After mixing, buffers were filtered sterilized, exposed to a UV germicidal lamp for 20 min, and stored frozen. Baked (300°C) 0.1-mm-diameter zirconium-silica beads (0.4 g) were added to 2-ml screw-cap microfuge tubes with 500 μl of biofilm suspension containing about 20 mg of dry biofilm. A negative extraction control (pure water only) was processed in parallel with the samples to test for reagent contamination. The following solutions were then added: 500 μl of TEN buffer (200 mM Tris HCl [pH 8.0], 20 mM EDTA, 200 mM NaCl), 200 μl of 20% sodium dodecyl sulfate, and 500 μl of phenol/chloroform/isoamyl alcohol (24:24:1). Suspensions were reciprocated with a Mini-Beadbeater (Biospec Products) at high speed for 2 min. The aqueous phase was collected following centrifugation, and the DNA was precipitated by the addition of 90 μl of 3M NaAc and 900 μl of isopropanol, rinsed in 70% ethanol, air dried, resuspended in 50 μl of 10 mM Tris-HCl (pH 7.5)-1 mM EDTA buffer, and stored at −20°C.
PCR.
The bacterium-specific primers used to amplify 16S rRNA gene fragments were 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 805R (5′-GACTACCAGGGTATCTAATCC-3′). The ∼800-bp fragment amplified with this primer pair includes a region of 16S rRNA that is useful for database identification and comparisons with published sequences (52). PCR was carried out with a total reaction volume of 50 μl including 1 μl of sample DNA as template, each deoxynucleoside triphosphate at 200 μM, 1.5 mM MgCl2, each primer at 0.4 μM, 4 μl of a 10-mg/ml concentration of bovine serum albumin, and 5 U of AmpliTaq Gold (Applied-Biosystems, Foster City, Calif.). To minimize outside contamination, reaction mixtures were assembled in a biological hood after all pipettes, pipette tips, and Eppendorf tubes and the PCR master mix (prior to the addition of the Taq DNA polymerase) had been irradiated with UV light for 20 min. Thirty cycles of PCR amplification were conducted. Each cycle included an initial denaturing step at 94°C for 1 min followed by a 45-s annealing step at 55°C and a 1.5-min extension step at 72°C. The amplification cycles were preceded by a one-time denaturing step at 94°C for 2 min prior to the first cycle and included a final 72°C extension step for 20 min to ensure complete extension for efficient cloning.
Cloning and RFLP analysis.
Samples were cloned by using a pGEM T-easy vector system cloning kit (Promega Corp., Madison, Wis.) according to the manufacturer's instructions. After cloning, colonies with inserts (96 per sample) were randomly selected and grown overnight in 1.5 ml of 2XYT broth containing 1 μM ampicillin. Restriction fragment length polymorphism (RFLP) analyses were used to identify unique clones for sequencing and analysis. In preparation for RFLP analysis, 25 μl of the cultures were heated at 95°C for 10 min, the cell debris was pelleted, and 1 μl of the supernatant was used in PCR with primers that flank the insertion site. The PCR products were digested simultaneously with HinP1I and Msp1 in NEBuffer 2 (New England Biolabs, Beverly, Mass.). The digested DNA was separated on a 3.5% 1× Tris-borate-EDTA low-melt agarose gel including ethidium bromide at 80 V for approximately 2 h (46). Fragment banding patters were visualized under UV light with a NucleoVision digital imaging system (NucleoTech Corp., San Carlos, Calif.).
Sequence and phylogenetic analysis.
Clones with unique RFLP band patterns were sequenced on a sequencer (Licor Corp., Lincoln, Nebr.) according to the manufacturer's instructions. The sequencing reaction mixtures were prepared with the primers Sp6 (5′-ATTTAGGTGACACTATAG-3′) and T7 (5′-TAATACGACTCACTATA-3′). Sequences were screened for chimeras (none were detected in the sequences analyzed) and compared to GenBank sequences by using a standard nucleotide basic local alignment search tool (BLAST) search. BLAST results for all sequences analyzed are tabulated at http://pacelab.colorado.edu/Publications/publications.html. Sequences were aligned and manually refined by using the ARB program (http://www.arb-home.de) and considering secondary structure.
All phylogenetic analyses were performed by using PAUP* (51). Phylogenetic trees were estimated by using maximum likelihood (ML). Ten heuristic random-addition sequence searches were performed to find the highest-likelihood tree. Maximum-parsimony (MP) and neighbor-joining (NJ) analyses were performed in addition to the ML search. MP analysis included 100 heuristic random-addition sequence searches to find the most parsimonious tree or set of trees. NJ analysis used the uncorrected distance measure to find the best tree. Bootstrap analyses were performed with the MP and NJ criteria. (ML bootstrap analyses were not performed because of the extensive computational time needed for such analyses.) MP analyses were performed with 100 resampling replicates with 10 random-addition sequence searches per replicate. The NJ bootstrap analysis included 5,000 resampling replicates. We do not report bootstrap values lower than 50% for either NJ or MP analyses.
Nucleotide sequence accession numbers.
Sequences obtained in this study have been deposited in GenBank under accession numbers AY268226 to AY268349.
RESULTS
Vinyl shower curtains over time accumulate a film, flakey when dry, that is popularly referred to as “soap scum.” Examination of this material from shower curtains by epifluorescence microscopy, as shown in Fig. 1, revealed that the “soap scum” is in fact a lush bed of microbes, generally imbedded in a biofilm matrix. In order to survey rRNA gene sequences, DNA was purified from biofilms scraped from four different shower curtains, including two different patches from one curtain, one dry and one continuously wet. All of the DNAs extracted from shower curtain samples produced PCR products with bacteria-specific 16S rRNA gene (rDNA) primers. None of the extraction controls showed detectable amounts of amplified rDNA. PCR products were cloned, and unique sequences were identified by RFLP analysis and sequenced (22). In the course of the study, 337 clones were screened and 117 unique rRNA sequences were determined.
FIG. 1.
Epifluoresence microscopy of biofilm samples SC4 (A and B) and SC2 (C) stained with DAPI (Materials and Methods).
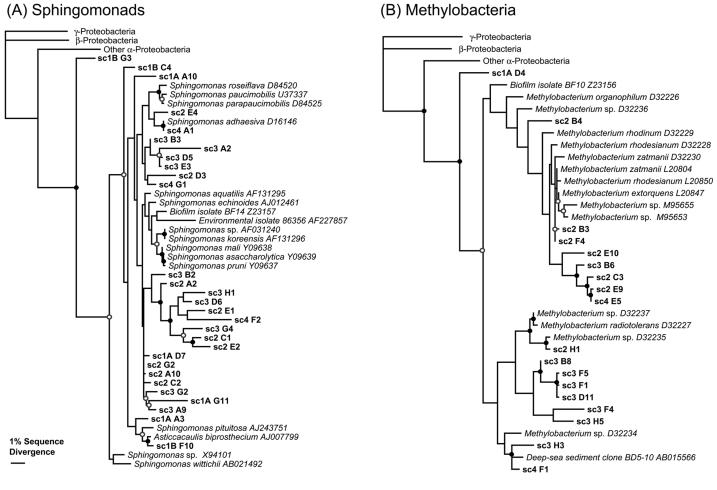
The sequences obtained from the shower curtain microbes were compared to each other and to sequences in GenBank. Results of phylogenetic analyses for the Sphingomonas- and Methylobacterium-related sequences are presented in Fig. 2. There were no identical sequences between different shower curtain samples. Instead, as generally occurs with environmental samples, members of phylogenetic clusters of closely related sequences were seen. Several of the clone DNA sequences were closely related to cultured organisms. The resolution of the trees was fairly low in several places, likely because the data set was limited (only 670 positions). Despite the limitations of the data set, the relevant relationships were supported with high MP and NJ bootstrap values (>70%). In relating the new sequences to known ones, we took relatedness clusters with 97% or higher sequence identity to correspond to a species-level relationship and clusters with 95% or higher sequence identity to correspond to a genus-level relationship (50).
FIG. 2.
Results of phylogenetic analyses with Sphingomonas spp.-related (A) and Methylobacterium spp.-related (B) 16S rRNA sequences obtained in this study. The analyses performed were based on alignments of approximately 670 nucleotide positions. The alignments included cultured Sphingomonas and Methylobacterium spp. and outgroup γ-proteobacteria (Escherichia coli L10328), β-proteobacteria (Nitrosospira mutiformis L35509), and α-proteobacteria (Roseobacter denitrificans M59063). The phylogenetic trees shown were estimated by using ML. MP and NJ analyses converged on very similar tree topologies. Filled circles indicate both MP and NJ bootstrap support exceeding 70%, and open circles indicate bootstrap support exceeding 50%. (See Materials and Methods for details on the phylogenetic and bootstrap analyses.).
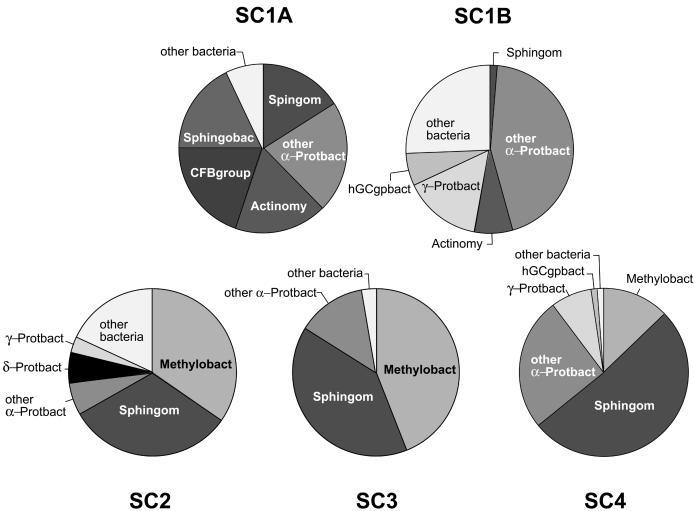
The pie chart diagrams in Fig. 3 illustrate the diversity and abundance of organisms found in the various shower curtain samples. The sequences represented are only the most abundant of the sequences encountered. Approximately 15% of the clones analyzed on the basis of RFLP or sequence were observed only once, so we did not nearly exhaust the diversity of organisms that comprise the communities. Most of the clones we sequenced were >95% identical to sequences already in GenBank. Only 38 out of 337 clones screened (11.2%) were less than 95% identical (genus-level relatedness) to known sequences in GenBank. Few clones had less than 90% identities to known sequences. Although the specific sequences associated with the different shower curtains differed in detail, the phylogenetic comparisons revealed common themes. The most prevalent types of organisms found in all of the samples belonged to the phylogenetic group α-proteobacteria. Sphingomonas spp. and Methylobacterium spp. prevailed in three of the four shower curtains in dry (SC2 and SC3) and moist (SC4) samples. Methylobacterium and Sphingomonas related clones found in the shower curtains spanned the diversity of these genera, and several of the clones were closely related to cultured species (Fig. 2). Although there were types of organisms consistently associated with all of the curtains, each of the samples contained significantly different proportions of specific organismal types. Indeed, one of the shower curtain communities (SC1) (Fig. 3) did not contain Methylobacterium spp. rRNA genes. Instead, other α-proteobacterial rRNA genes, perhaps with the same ecological roles as the Methylobacteria, were encountered. A χ2 test comparing the abundances of different clonal types (four categories: Sphingomonas spp., Methylobacterium spp., other α-proteobacteria, and γ-proteobacteria) found significant differences between the shower curtain samples (χ2 = 58.4, 9 df; P < 0.0001), including dry (SC1A) and moist (SC1B) patches from one sampled curtain. Thus, each shower curtain is itself an anecdote with respect to the specific organisms that make up these complex communities. In general, however, the different shower curtain communities tend to be composed of similar suites of genera.
FIG. 3.
Compositions of rRNA gene libraries obtained from shower curtain samples. Numbers of clones of each rRNA gene sequence from libraries are grouped as identified on the pie charts as follows: Methylobact, Methylobacterium spp.; Sphingom, Sphingomonas spp.; α-Protbact, α-proteobacteria; δ-Protbact, δ-proteobacteria; γ-Protbact, γ-proteobacteria; Actinomy, Actinomycetales; hGCgpbact, high-G+C gram-positive bacteria; CFBgroup, Cytophaga-Flavobacteria-Bacteroides group; Sphingobac, sphingobacteria.
DISCUSSION
The biofilms from the various shower curtains analyzed in this study contained a substantial diversity of microorganisms. DAPI staining of films on shower curtains revealed high concentrations of microorganisms (Fig. 1), and amplifications of DNA extracted from the shower curtains produced PCR products with bacterium-specific primers. Members of two genera of α-proteobacteria, Sphingomonas spp. and Methylobacterium spp., composed the largest proportion of organisms generally encountered in the shower curtain biofilms (Fig. 3). Sphingomonas spp. were found in the greatest abundance on all of the curtains analyzed, followed by Methylobacterium spp. (Fig. 3). Several of the Sphingomonas spp. clones appeared to be closely related to the rRNA sequences of cultured representatives, such as Sphingomonas adhaesiva and Sphingomonas pituitosa (Fig. 2A). However, many of the sequences clustered together phylogenetically and apart from those of cultured representatives, suggesting that the corresponding organisms consist of novel groups within the genus. Based on their phylogenetic distinctness, the novel organisms may have unique and interesting properties not present in known examples of the genus (Fig. 2A). Similar patterns were found in the phylogenetic analysis of the sequences related to Methylobacterium spp. (Fig. 2B).
Sphingomonads, generally the most abundant of the biofilm organisms detected, are ubiquitous in the environment and are frequently isolated from soil, water, and sediments (55). Methylobacterium spp., especially Methylobacterium mesophilicum, have been cultured from pink-colored biofilms of wet environments such as automobile air-conditioning systems, printing paper machines, and dental unit waterlines (3, 28, 44, 53). Considering the abundance of Methylobacterium spp. rRNA gene clones detected in this study, such organisms likely are responsible for the pink color of some shower curtain biofilms. We acknowledge, however, that the abundance of rRNA gene clones does not directly indicate the relative abundances of the corresponding cells. Different species of organisms contain different numbers of rRNA genes per cell, and potential biases in the recovery of different rRNA genes by PCR techniques must be recognized. Nevertheless, we believe that the relative frequencies of the rRNA genes detected represent some rough reflection of the environmental abundance of the corresponding organisms.
In addition to Sphingomonas spp. and Methylobacterium spp., the shower curtains also contained other representatives of the Proteobacteria phylogenetic division, particularly α-proteobacteria. Proteobacteria are diverse in physiology and ubiquitous in water environments, ranging from deep seawater to waste and drinking water (8, 48, 49). The sources of carbon and energy for these biofilm communities are unclear. Potential food resources include soap products, sloughed-off human debris, and bath area volatiles. Biofilms provide a supportive and protective environment in which many kinds of metabolism can thrive. Sphingomonads, for instance, use a broad range of carbon compounds, including complex organics such as dibenzofuran (18) and hexachlorocyclohexane (23). Some isolates from the deep subsurface have been shown to metabolize aromatic compounds such as toluene, naphthalene, and others, although laboratory strains have not shown these properties (17). Sphingomonads colonize new environments readily and adhere to surfaces through the production of exopolysaccharides, such as gellan, and welan (41). Methylobacterium spp. also thrive on numerous different kinds of carbon sources, such as succinate, ethanol, ethanolamine, methanol, and methylamine (26, 54).
Potential pathogens.
In this limited survey, we did not encounter any known specific pathogens. However, several sphingomonads are known to be opportunistic pathogens. For instance, Sphingomonas paucimobilis has a history of infecting immune-compromised patients or persons with predisposing conditions (21). Infection with S. paucimobilis can lead to intravascular catheter-related bacteremia, urinary tract infections, pneumonia, cutaneous infections, and visceral abscesses (7, 21, 45). In hospitals, sources of S. paucimobilis infections have been traced to fluid in humidifiers and tap water (21, 31, 40).
Methylobacterium spp. also are known to cause infections in immune-compromised patients or patients with other diseases that render them prone to infection (15, 16, 20, 25, 35, 47). We found several clones representing organisms closely related to Methylobacterium extorquens and Methylobacterium zatmanii (Fig. 2B), both known to cause illness in immune-compromised patients (20, 25). We also encountered abundant M. mesophilicum in a 16S rDNA clone library prepared from humidifier filter samples (data not shown). In patients, M. mesophilicum has been detected in blood, peritoneal fluid, and ascitic fluid and has been known to cause pneumonia, skin ulcers, empyema, keratitis, and bacteremia (25, 35, 47). Based on the rising number of cases, it has been proposed that Methylobacterium spp. infections could increase dramatically in the future (16, 19, 20, 42, 47).
In addition to the consistent presence of Sphingomonas spp. and Methylobacterium spp., we also uncovered a number of other bacterial species at low frequencies. Some of these species are closely related to known opportunistic pathogens, including Nocardia spp. and Gordonia spp. (high-G+C gram-positive bacteria). Infections with Gordonia terrae can invade wounds and result in bacteremia and brain abscess (34). Patients with such infections usually have underlying diseases predisposing them to opportunistic infections, but infections of healthy patients have been reported (9). There have been reports of severe infections in immune-compromised patients by several Nocardia species, which often are misidentified by standard culture methods (11). Other microorganisms detected in samples include close relatives of the known opportunistic pathogens Afipia felis (36) and Moraxella osloensis (4).
Our results suggest that shower curtains harbor potential opportunistic pathogens that can threaten immune-compromised or otherwise ill patients. For immune-compromised people, consistent exposure to sources of infection, such as shower curtains, is a public health problem. Exposure can be minimized by regular cleaning or by changing shower curtains.
Acknowledgments
We thank Mark Hernandez (Department of Civil, Environmental and Architectural Engineering, University of Colorado at Boulder) for contributions to this study. We thank Ruth Ley and Amy Buck for contributing their shower curtains.
This work was supported in part by a grant from the National Institutes of Health to N.R.P.
REFERENCES
- 1.Acinas, S. G., J. Anton, and F. Rodriguez-Valera. 1999. Diversity of free-living and attached bacteria in offshore Western Mediterranean waters as depicted by analysis of genes encoding 16S rRNA. Appl. Environ. Microbiol. 65:514-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann, R. I., W. Ludwig, and K. H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barbeau, J., R. Tanguay, E. Faucher, C. Avezard, L. Trudel, L. Cote, and A. P. Prevost. 1996. Multiparametric analysis of waterline contamination in dental units. Appl. Environ. Microbiol. 62:3954-3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berrocal, A. M., I. U. Scott, D. Miller, and H. W. Flynn, Jr. 2002. Endophthalmitis caused by Moraxella osloensis. Graefe's Arch. Clin. Exp. Ophthalmol. 240:329-330. [DOI] [PubMed] [Google Scholar]
- 5.Boe-Hansen, R., H. J. Albrechtsen, E. Arvin, and C. Jorgensen. 2002. Bulk water phase and biofilm growth in drinking water at low nutrient conditions. Water Res. 36:4477-4486. [DOI] [PubMed] [Google Scholar]
- 6.Casadevall, A. 1996. Crisis in infectious diseases: time for a new paradigm? Clin. Infect. Dis. 23:790-794. [DOI] [PubMed] [Google Scholar]
- 7.Casadevall, A., L. F. Freundlich, and L. Pirofski. 1992. Septic shock caused by Pseudomonas paucimobilis. Clin. Infect. Dis. 14:784. [DOI] [PubMed] [Google Scholar]
- 8.Cottrell, M. T., and D. L. Kirchman. 2000. Community composition of marine bacterioplankton determined by 16S rRNA gene clone libraries and fluorescence in situ hybridization. Appl. Environ. Microbiol. 66:5116-5122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drancourt, M., J. Pelletier, A. A. Cherif, and D. Raoult. 1997. Gordona terrae central nervous system infection in an immunocompetent patient. J. Clin. Microbiol. 35:379-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.du Moulin, G. C., K. D. Stottmeier, P. A. Pelletier, A. Y. Tsang, and J. Hedley-Whyte. 1988. Concentration of Mycobacterium avium by hospital hot water systems. JAMA 260:1599-1601. [DOI] [PubMed] [Google Scholar]
- 11.Eggink, C. A., P. Wesseling, P. Boiron, and J. F. Meis. 1997. Severe keratitis due to Nocardia farcinica. J. Clin. Microbiol. 35:999-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Embil, J., P. Warren, M. Yakrus, R. Stark, S. Corne, D. Forrest, and E. Hershfield. 1997. Pulmonary illness associated with exposure to Mycobacterium avium-complex in hot tub water. Hypersensitivity pneumonitis or infection? Chest 111:813-816. [DOI] [PubMed] [Google Scholar]
- 13.Excoffier, L., P. E. Smouse, and J. M. Quattro. 1992. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Falkinham, J. O., III, C. D. Norton, and M. W. LeChevallier. 2001. Factors influencing numbers of Mycobacterium avium, Mycobacterium intracellulare, and other mycobacteria in drinking water distribution systems. Appl. Environ. Microbiol. 67:1225-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fernandez, M., Z. Dreyer, M. Hockenberry-Eaton, and C. J. Baker. 1997. Methylobacterium mesophilica as a cause of persistent bacteremia in a child with lymphoma. Pediatr. Infect. Dis. J. 16:1007-1008. [DOI] [PubMed] [Google Scholar]
- 16.Flournoy, D. J., R. L. Petrone, and D. W. Voth. 1992. A pseudo-outbreak of Methylobacterium mesophilica isolated from patients undergoing bronchoscopy. Eur. J. Clin. Microbiol. Infect. Dis. 11:240-243. [DOI] [PubMed] [Google Scholar]
- 17.Fredrickson, J. K., D. L. Balkwill, G. R. Drake, M. F. Romine, D. B. Ringelberg, and D. C. White. 1995. Aromatic-degrading Sphingomonas isolates from the deep subsurface. Appl. Environ. Microbiol. 61:1917-1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fukuda, K., S. Nagata, and H. Taniguchi. 2002. Isolation and characterization of dibenzofuran-degrading bacteria. FEMS Microbiol. Lett. 208:179-185. [DOI] [PubMed] [Google Scholar]
- 19.Hiraishi, A., K. Furuhata, A. Matsumoto, K. A. Koike, M. Fukuyama, and K. Tabuchi. 1995. Phenotypic and genetic diversity of chlorine-resistant Methylobacterium strains isolated from various environments. Appl. Environ. Microbiol. 61:2099-2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hornei, B., E. Luneberg, H. Schmidt-Rotte, M. Maass, K. Weber, F. Heits, M. Frosch, and W. Solbach. 1999. Systemic infection of an immunocompromised patient with Methylobacterium zatmanii. J. Clin. Microbiol. 37:248-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hsueh, P. R., L. J. Teng, P. C. Yang, Y. C. Chen, H. J. Pan, S. W. Ho, and K. T. Luh. 1998. Nosocomial infections caused by Sphingomonas paucimobilis: clinical features and microbiological characteristics. Clin. Infect. Dis. 26:676-681. [DOI] [PubMed] [Google Scholar]
- 22.Hugenholtz, P., C. Pitulle, K. L. Hershberger, and N. R. Pace. 1998. Novel division level bacterial diversity in a Yellowstone hot spring. J. Bacteriol. 180:366-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Imai, R., Y. Nagata, M. Fukuda, M. Takagi, and K. Yano. 1991. Molecular cloning of a Pseudomonas paucimobilis gene encoding a 17-kilodalton polypeptide that eliminates HCl molecules from gamma-hexachlorocyclohexane. J. Bacteriol. 173:6811-6819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kahana, L. M., J. M. Kay, M. A. Yakrus, and S. Waserman. 1997. Mycobacterium avium complex infection in an immunocompetent young adult related to hot tub exposure. Chest 111:242-245. [DOI] [PubMed] [Google Scholar]
- 25.Kaye, K. M., A. Macone, and P. H. Kazanjian. 1992. Catheter infection caused by Methylobacterium in immunocompromised hosts: report of three cases and review of the literature. Clin. Infect. Dis. 14:1010-1014. [DOI] [PubMed] [Google Scholar]
- 26.Korotkova, N., and M. E. Lidstrom. 2001. Connection between poly-beta-hydroxybutyrate biosynthesis and growth on C(1) and C(2) compounds in the methylotroph Methylobacterium extorquens AM1. J. Bacteriol. 183:1038-1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Korvick, J. A., J. D. Rihs, G. L. Gilardi, and V. L. Yu. 1989. A pink-pigmented, oxidative, nonmotile bacterium as a cause of opportunistic infections. Arch. Intern. Med. 149:1449-1451. [PubMed] [Google Scholar]
- 28.Kressel, A. B., and F. Kidd. 2001. Pseudo-outbreak of Mycobacterium chelonae and Methylobacterium mesophilicum caused by contamination of an automated endoscopy washer. Infect. Control Hosp. Epidemiol. 22:414-418. [DOI] [PubMed] [Google Scholar]
- 29.Le Dantec, C., J. P. Duguet, A. Montiel, N. Dumoutier, S. Dubrou, and V. Vincent. 2002. Occurrence of mycobacteria in water treatment lines and in water distribution systems. Appl. Environ. Microbiol. 68:5318-5325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee, T. C., J. E. Stout, and V. L. Yu. 1988. Factors predisposing to Legionella pneumophila colonization in residential water systems. Arch. Environ. Health 43:59-62. [DOI] [PubMed] [Google Scholar]
- 31.Lemaitre, D., A. Elaichouni, M. Hundhausen, G. Claeys, P. Vanhaesebrouck, M. Vaneechoutte, and G. Verschraegen. 1996. Tracheal colonization with Sphingomonas paucimobilis in mechanically ventilated neonates due to contaminated ventilator temperature probes. J. Hosp. Infect. 32:199-206. [DOI] [PubMed] [Google Scholar]
- 32.Leoni, E., P. Legnani, M. T. Mucci, and R. Pirani. 1999. Prevalence of mycobacteria in a swimming pool environment. J. Appl. Microbiol. 87:683-688. [DOI] [PubMed] [Google Scholar]
- 33.Leoni, E., P. P. Legnani, M. A. Bucci Sabattini, and F. Righi. 2001. Prevalence of Legionella spp. in swimming pool environment. Water Res. 35:3749-3753. [DOI] [PubMed] [Google Scholar]
- 34.Lesens, O., Y. Hansmann, P. Riegel, R. Heller, M. Benaissa-Djellouli, M. Martinot, H. Petit, and D. Christmann. 2000. Bacteremia and endocarditis caused by a Gordonia species in a patient with a central venous catheter. Emerg. Infect. Dis. 6:382-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu, J. W., J. J. Wu, H. M. Chen, A. H. Huang, W. C. Ko, and Y. C. Chuang. 1997. Methylobacterium mesophilicum synovitis in an alcoholic. Clin. Infect. Dis. 24:1008-1009. [DOI] [PubMed] [Google Scholar]
- 36.Luhrmann, A., K. Streker, A. Schuttfort, J. J. Daniels, and A. Haas. 2001. Afipia felis induces uptake by macrophages directly into a nonendocytic compartment. Proc. Natl. Acad. Sci. USA 98:7271-7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martin, D. S., P. Oray-Schrom, and Y. Amoateng-Adjepong. 2002. Emerging significance of Mycobacterium avium-complex infection in an inner-city hospital. Conn. Med. 66:323-330. [PubMed] [Google Scholar]
- 38.Nelson, F. P., S. Macari, G. Faria, S. Assed, and I. Y. Ito. 2000. Microbial contamination of toothbrushes and their decontamination. Pediatr. Dent. 22:381-384. [PubMed] [Google Scholar]
- 39.Pace, N. R. 1997. A molecular view of microbial diversity and the biosphere. Science 276:734-740. [DOI] [PubMed] [Google Scholar]
- 40.Perola, O., T. Nousiainen, S. Suomalainen, S. Aukee, U. M. Karkkainen, J. Kauppinen, T. Ojanen, and M. L. Katila. 2002. Recurrent Sphingomonas paucimobilis -bacteraemia associated with a multi-bacterial water-borne epidemic among neutropenic patients. J. Hosp. Infect. 50:196-201. [DOI] [PubMed] [Google Scholar]
- 41.Pollock, T. J., W. A. van Workum, L. Thorne, M. J. Mikolajczak, M. Yamazaki, J. W. Kijne, and R. W. Armentrout. 1998. Assignment of biochemical functions to glycosyl transferase genes which are essential for biosynthesis of exopolysaccharides in Sphingomonas strain S88 and Rhizobium leguminosarum. J. Bacteriol. 180:586-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rice, E. W., D. J. Reasoner, C. H. Johnson, and L. A. DeMaria. 2000. Monitoring for methylobacteria in water systems. J. Clin. Microbiol. 38:4296-4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rose, A. M., K. Sinka, J. M. Watson, J. Y. Mortimer, and A. Charlett. 2002. An estimate of the contribution of HIV infection to the recent rise in tuberculosis in England and Wales. Thorax 57:442-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rose, L. J., R. B. Simmons, S. A. Crow, and D. G. Ahearn. 2000. Volatile organic compounds associated with microbial growth in automobile air conditioning systems. Curr. Microbiol. 41:206-209. [DOI] [PubMed] [Google Scholar]
- 45.Salazar, R., R. Martino, A. Sureda, S. Brunet, M. Subira, and A. Domingo-Albos. 1995. Catheter-related bacteremia due to Pseudomonas paucimobilis in neutropenic cancer patients: report of two cases. Clin. Infect. Dis. 20:1573-1574. [DOI] [PubMed] [Google Scholar]
- 46.Sambrook, J., and D. W. Russel (ed.). 2001. Molecular cloning, 3rd ed., vol. 1. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 47.Sanders, J. W., J. W. Martin, M. Hooke, and J. Hooke. 2000. Methylobacterium mesophilicum infection: case report and literature review of an unusual opportunistic pathogen. Clin. Infect. Dis. 30:936-938. [DOI] [PubMed] [Google Scholar]
- 48.Schwartz, T., S. Hoffmann, and U. Obst. 1998. Formation and bacterial composition of young, natural biofilms obtained from public bank-filtered drinking water systems. Water Res. 32:2787-2797. [Google Scholar]
- 49.Seviour, R. J., A. M. Maszenan, J. A. Soddell, V. Tandoi, B. K. Patel, Y. Kong, and P. Schumann. 2000. Microbiology of the ′G-bacteria' in activated sludge. Environ. Microbiol. 2:581-593. [DOI] [PubMed] [Google Scholar]
- 50.Stackebrandt, E., and B. M. Goebel. 1994. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Bacteriol. 44:846-849. [Google Scholar]
- 51.Swofford, D. 1998. PAUP*: phylogenetic analysis using parsimony (*and other methods), 4th ed. Sinauer Associates, Sunderland, Mass.
- 52.Tanner, M. A., D. Shoskes, A. Shahed, and N. R. Pace. 1999. Prevalence of corynebacterial 16S rRNA sequences in patients with bacterial and “nonbacterial” prostatitis. J. Clin. Microbiol. 37:1863-1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vaisanen, O. M., A. Weber, A. Bennasar, F. A. Rainey, H. J. Busse, and M. S. Salkinoja-Salonen. 1998. Microbial communities of printing paper machines. J. Appl. Microbiol. 84:1069-1084. [DOI] [PubMed] [Google Scholar]
- 54.Vorholt, J. A., C. J. Marx, M. E. Lidstrom, and R. K. Thauer. 2000. Novel formaldehyde-activating enzyme in Methylobacterium extorquens AM1 required for growth on methanol. J. Bacteriol. 182:6645-6650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.White, D. C., S. D. Sutton, and D. B. Ringelberg. 1996. The genus Sphingomonas: physiology and ecology. Curr. Opin. Biotechnol. 7:301-306. [DOI] [PubMed] [Google Scholar]