Abstract
Pathogen colonization and transcript levels of three host chitinases, putatively representing classes I, II, and IV, were monitored with real-time PCR after wounding and bark infection by Heterobasidion annosum in 32-year-old trees of Norway spruce (Picea abies) with low (clone 409) or high (clone 589) resistance to this pathogen. Three days after inoculation, comparable colonization levels were observed in both clones in the area immediately adjacent to inoculation. At 14 days after infection, pathogen colonization was restricted to the area immediately adjacent to the site of inoculation for clone 589 but had progressed further into the host tissue in clone 409. Transcript levels of the class II and IV chitinases increased after wounding or inoculation, but the transcript level of the class I chitinase declined after these treatments. Transcript levels of the class II and class IV chitinases were higher in areas immediately adjacent to the inoculation site in clone 589 than in similar sites in clone 409 3 days after inoculation. This difference was even more pronounced 2 to 6 mm away from the inoculation point, where no infection was yet established, and suggests that the clones differ in the rate of chitinase-related signal perception or transduction. At 14 days after inoculation, these transcript levels were higher in clone 409 than in clone 589, suggesting that the massive upregulation of class II and IV chitinases after the establishment of infection comes too late to reduce or prevent pathogen colonization.
Plants are continuously exposed to microbial pathogens and have evolved a number of strategies to resist infection. These defense mechanisms include preformed physical barriers, e.g., plant cell walls. Active defense mechanisms induced by microbial infection include increased lignification and de novo synthesis of defense-related proteins (15). The level of resistance attained depends on the degree of coordination of the different defense strategies and the rapidity of the overall response (17).
Pathogenesis-related (PR) proteins, including chitinases are synthesized by the plant in response to pathogen attack (see, for example, reference 37). Chitinases (EC 3.2.1.14) hydrolyze the β-1,4 glycosidic bonds that link the N-acetylglucosamine residues of chitin and may play a direct role in plant defense by attacking chitin. Purified chitinases inhibit fungal growth by causing swelling and lysis of hyphal tips (21, 31), and chitinolytic breakdown products can elicit further defense reactions in plants (2, 10, 18, 28, 29). Constitutive overexpression of chitinases in transgenic plants increases resistance to pathogens in vivo (4, 13). Some chitinases are expressed in healthy plants in an organ-specific and developmentally regulated pattern, suggesting that these enzymes may have a role during plant growth and development in addition to protecting plants against pathogens (6). Individual plant species may contain chitinase isozymes that are distinct in terms of their regulation and function (see, for example, reference 5).
Chitinases are classified based on their structure, enzymatic properties, and localization (6, 11, 24). All chitinases are grouped into two families of glycosylhydrolases that are distinguished from one another on the basis of their hydrolytic mechanisms. Classes I, II, and IV form family 19 chitinases, which are found only in plants. The class I chitinases are structurally related to class IV chitinases since they both possess a chitin-binding domain, but their overall sequence homology otherwise is rather low. Class II chitinases lack the chitin-binding domain (11). Family 18 chitinases includes all bacterial, fungal, and animal chitinases, as well as class III chitinases from plants (15).
Heterobasidion annosum is a major root and butt rot pathogen of conifers causing tremendous economic losses worldwide. Suberized bark tissues form a strong barrier to penetration by this pathogen (20, 33). Bark wounds caused by wind, animals, insects, and timber extraction expose the trees to this pathogen, which is characterized by a high spore deposition rate and long spore viability in bark (27, 32). Clonal variation in disease resistance to this pathogen is reported for several conifer species (8, 35), although the resistance mechanism is not known.
Recently, we developed a multiplex real-time PCR procedure to monitor colonization of Norway spruce by H. annosum (14) and could reproducibly detect 1 to 2 fungal cells/860 host cells. To investigate the role of chitinases in host defense of Norway spruce, we sought to monitor H. annosum colonization rates and the expression of chitinases in two clones with differential disease resistance. Elucidation of the mechanisms underlying disease resistance will pave the way for establishing breeding programs targeted to disease resistance.
MATERIALS AND METHODS
Inoculation experiment.
Two 32-year-old clones growing at the Hogsmark Plantation of the Norwegian Forest Research Institute at Ås were used as host material. Clone 409 has limited resistance to H. annosum compared to clone 589 (14, 19). Two ramets were used for each clone, and stems were inoculated with the heterokaryotic strain 87-257/1 (S intersterility group) as previously described (14). Inoculations were made 1 m above ground level at four positions evenly spaced around each stem. At each inoculation point, a plug of bark down to the sapwood surface was excised by using a 5-mm-diameter cork borer. A similarly sized agar plug containing the actively growing fungus was inserted into the hole, and the bark plug was replaced. A rectangular strip (2 × 10 cm) of bark containing phloem and cambium, with the inoculation site in the middle, was removed 3, 7, 14, and 35 days after inoculation (dai). Immediately after excision, the samples were frozen in liquid N2 and stored at −150°C. Samples taken 35 dai were not included in the present study, because they had been analyzed previously (14) for fungal colonization. In addition, the ramets were wounded with a 5-mm-diameter cork borer a year after the initial inoculation at two positions located between the original inoculations, and harvested similarly 3 and 14 days after treatment. Bark samples taken at the time of inoculation and wounding were used as controls. Thus, eight total samples (one control sample for inoculations, four inoculated samples, one control sample for wounding, and two wounded samples) were taken from each ramet in a ring-like manner 1 m above ground level.
RNA isolation and cDNA synthesis.
Control samples and the upper part of inoculated or wounded samples from each ramet for each time point were processed for RNA isolation and real-time PCR. Prior to RNA extraction, the rhytidome and the periderm were removed and the tissue subdivided into sections (ca. 2 by 5 by 3 mm) that were processed separately. The sampled 2-mm-long sections were frozen immediately in liquid N2 and ground twice in liquid N2-chilled containers for 2 min each time in an MM 300 mill (Retsch Gmbh, Haan, Germany). RNA was isolated by using an RNAqueous small-scale phenol-free total RNA isolation kit and Plant RNA Isolation Aid (Ambion, Austin, Tex.) according to the manufacturer's instructions. Contaminating DNA was removed by using DNA-Free (Ambion) with 2 U of DNase I per sample and a 30-min incubation at 37°C. Total RNA was quantified with a VersaFluor fluorometer (Bio-Rad, Hercules, Calif.) and a RiboGreen RNA quantification kit (Molecular Probes, Eugene, Oreg.) according to the manufacturer's instructions. Total RNA was normalized between samples: 80 ng of total RNA was reverse transcribed with TaqMan reverse transcription reagents (Applied Biosystems, Foster City, Calif.) according to the manufacturer's instructions with 1.25 U of reverse transcriptase (RT) per sample and 2.5 μM random hexamer primers in a 50-μl reaction volume.
Isolation of chitinase cDNA from Norway spruce and design of oligonucleotide primers for quantification of gene expression.
Three partial chitinase sequences, referred to as PaChi1, PaChi2, and PaChi4 (GenBank accession numbers AY450922, AY450923, and AY450924, respectively), were obtained from a cDNA library from bark of Norway spruce saplings (clone 3369; Schongau, Staatliche Samenklenge und Pflanzarten Laufen, Germany) that had been sprayed with 100 μM methyljasmonate (38). Poly(A)+ RNA was isolated from total RNA by using Dynabeads (Dynal, Hamburg, Germany). cDNA was produced by using the SMART cDNA library construction kit (Stratagene, Heidelberg, Germany). In vitro packaging with Gigapack III Gold (Stratagene) yielded a library of 8 × 107 PFU. The lambda TriplEx library was converted to a pTriplEx library, and cDNA-insert containing plasmids were 5′ end sequenced and analyzed with the basic local alignment search tool (BLAST) program by GATC-Biotech AG (Konstanz, Germany). PaChi1, PaChi2 and PaChi4 were resequenced to confirm the initial sequence, and the entire coding regions were obtained for PaChi2 (GenBankAY544781) and PaChi4 (GenBank AY544780). SignalP (www.cbs.dtu.dk/services/SignalP) (25) was used to predict the presence of signal peptides and cellular sorting for the deduced N-terminal amino acid sequences of PaChi1, PaChi2, and PaChi4. To look for similarities to important amino acid motifs in proteins with known function, the sequences were searched with the online protein motif search programs InterPro (www.ebi.ac.uk/InterProScan) (3) and Motif (motif.genome.jp). Comparison analysis of the sequences to nonredundant protein and nucleotide databases was conducted by using the advanced BLAST (1) at the National Center for Biotechnology Information (www.ncbi.nlm.nih.gov/BLAST), and multiple sequence alignments were performed with CLUSTAL W 1.81 (clustalw.genome.ad.jp).
Based on the partial cDNA sequences, real-time PCR primers were designed with the Primer Express software 1.5a provided with Applied Biosystems real-time quantitative PCR systems (Applied Biosystems). For PaChi4, the chosen forward primer 5′-GCGAGGGCAAGGGATTCTAC-3′ and the reverse primer 5′-GGTGGTGCCAAATCCAGAAA-3′ amplify a 73-bp PCR product. For PaChi2, the forward primer 5′-CGATGCCACGGTCTCTTTTAA-3′ and the reverse primer 5′-GCAGGAAGGCTTGGGAGACT-3′ amplify a 70-bp PCR product. For PaChi1, the forward primer 5′-GCACAGTTCGAGGGCATAGG-3′ and the reverse primer 5′-AAGGCTGCAAGCTCCTTCTG-3′ amplify a 71-bp PCR product. An α-tubulin gene of Norway spruce (GenBank accession no. X57980) was used as an endogenous reference; the designed forward primer 5′-GGCATACCGGCAGCTCTTC-3′ and reverse primer 5′-AAGTTGTTGGCGGCGTCTT-3′ amplify the cDNA region from bases 187 to 252. The primers were ordered from the ABI Prism primers and TaqMan probes synthesis service (Applied Biosystems).
PCR conditions, specificity of primers, and amplification efficiency of transcripts.
Each 25-μl PCR was performed in SYBR Green PCR Mastermix (P/N 4309155; Applied Biosystems). A primer concentration of 50 nM was chosen for all genes assayed. As a template, 3 μl of the cDNA solution described above was used for each reaction. Each reaction was repeated twice. PCR cycling parameters were 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. Fluorescence emissions were detected with an ABI Prism 7700 (Applied Biosystems). The data acquisition and analysis were performed with the Sequence Detection System software package (version 1.7a; Applied Biosystems).
The target specificity of each primer set was examined in a melting-point analysis by using genomic DNA from H. annosum as a negative control and cDNA from both host clones as positive controls. The melting-point analysis was performed after the real-time RT-PCR program described above by using the thermal profile of 95°C for 15 s and 60°C for 20 s, followed by a melting step in which the temperature was raised linearly from 60°C to 95°C in a ramping time of 19 min 59 s.
To determine the amplification efficiencies for the chitinase and α-tubulin genes, cDNA containing a high amount of each monitored transcript was serially diluted over a 4-log range to cover the range of threshold cycles (Ct) observed for the experimental samples. Real-time PCR was performed for these dilutions. The Ct values were recorded at threshold and baseline parameters standardized for each transcript, and the results were plotted against the log of starting template concentration. The slope of the curves is dependent upon amplification efficiency (E) by the formula E = 10[−1/slope] (26). The expression level of each chitinase gene in relation to expression of α-tubulin in infected, wounded, and control samples was calculated from the Ct values and the PCR amplification efficiencies by using the formula of Pfaffl (26):
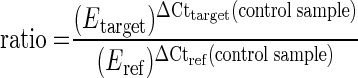 |
To compare the different treatments and time points, transcript levels observed on day 0 in the ramet 409A were treated as the basal expression level for all of the ramets, and the corresponding transcript levels were given the value 1.
Quantification of fungal colonization.
The real-time PCR detection of host and pathogen DNA was performed with TaqMan Universal PCR Master Mix (P/N 4304437; Applied Biosystems) with an ABI Prism 7700 and the laccase-polyubiquitin multiplex PCR procedure previously designed for studying the dynamics of the H. annosum/Norway spruce pathosystem (14). The PCR conditions were as described in that study. As a template, we used 2.5 μl of the total RNA solution, described above, per sample prior to DNase treatment. The amounts of pathogen and host DNA in the samples were calculated from the respective standard curves (14) by using a ΔCt-based calculation procedure, and the DNA estimates were converted to the number of host and pathogen nuclei.
Statistical analyses.
The transcript level data obtained were subjected to analysis of variance by using the GLM procedure of SAS (30). Each time point was analyzed separately. Differences between clones and treatments were considered when the P value was <0.05.
RESULTS
Characterization of the chitinases.
The 395-nucleotide (nt) partial cDNA PaChi1 gave a deduced 123-amino-acid (aa) N-terminal sequence, with a predicted 24-aa signal peptide, followed by a 20-aa chitin-binding domain and a chitinolytic domain sequence characteristic of family 19 chitinases (GenBank AY450922). PaChi1 had no significant similarity to any known Norway spruce or other gymnosperm sequence, but the deduced 123-aa N-terminal sequence was 66% similar to a 96-aa sequence from an angiosperm class I elicitor induced chitinase (GenBank BAC81645) from Pisum sativum (23).
The 895-nt cDNA PaChi2 encodes a 264-aa polypeptide with a predicted 33-aa signal peptide and a chitinolytic domain sequence characteristic of family 19 chitinases (GenBank AY544781). PaChi2 was not similar to any known Norway spruce sequence but was 80% identical to Pschi4 (GenBank U57410), an extracellullar class II chitinase gene induced by chitosan in the conifer Pinus strobus (40).
The 962-nt cDNA PaChi4 encoded a deduced 276-aa polypeptide with a predicted 26-aa signal peptide followed by a 32-aa putative chitin-binding domain and a chitinolytic domain sequence characteristic of family 19 chitinases (GenBank AY544780). PaChi4 cDNA was 96 to 97% identical to the class IV chitinase PgChi-1 from Picea glauca (GenBank L42467) (9) and to the class IV chitinases Chia4-Pa1.3 (GenBank AY270016) and Chia4-Pa1.1 (GenBank AY270017), which may promote programmed cell death in embryogenic tissue cultures of Norway spruce (39). The deduced amino acid sequence of PaChi4 differed at five or more amino acid positions from the Chia4-Pa (39) chitinases.
Specificity of primer sets and real-time PCR amplification efficiencies.
The primer pairs designed for the host α-tubulin and the three chitinases did not amplify the genomic DNA of H. annosum but did amplify cDNA from the host clones. There was a single peak with a melting temperature specific for each monitored transcript in the melting-point analysis of RT-PCR products (data not shown). The real-time PCR efficiencies (E) for the monitored transcripts were as follows: α-tubulin, 1.80; PaChi1, 1.77; PaChi2, 1.87; and PaChi4, 1.93. The Pearson correlation coefficient values in these datasets were between −0.996 and −0.999, indicating a strong linear correlation between transcript level and Ct value.
Temporal and spatial colonization profiles of H. annosum.
The colonization profiles of H. annosum differed in the two clones. By day 3, the pathogen was detected in the 2-mm-long section next to the inoculation point for both 409 ramets and for ramet 589B (Fig. 1A and B). H. annosum colonization levels were similar among these three, but the pathogen could not be detected in ramet 589A at this time. By day 7, the pathogen was still restricted to the 2-mm-long section next to the inoculation point in all four ramets, but the colonization levels were considerably higher in the 409 ramets than in the 589 ramets (Fig. 1C and D). By day 14, the pathogen had progressed 6 mm away from the inoculation point in 409A, 12 mm in 409B, 4 mm in 589A, and 2 mm in 589B (Fig. 1E and F). In all four ramets, there was a clear gradient in the pathogen/host nuclear ratio, with the highest levels immediately adjacent to the inoculation point. When we examined the host/pathogen nuclear ratios in the 2-mm-long section next to the inoculation point over the 14-day incubation time, the clones differed: the 409 ramets had a rapid and continuous increase in the pathogen/host nuclear ratio, whereas this increase was very limited for the 589 ramets. The pathogen/host nuclear ratio at 35 dai, obtained previously (14), was consistent with those obtained now, i.e., the pathogen/host nuclear ratios at 14 and 35 dai in the first section next to the inoculation point were 8.6 and 7.5 for 409A, 18 and 25 for 409B, 0.5 and 0.3 for 589A, and 0.1 and 0.02 for 589B, respectively.
FIG. 1.
Relative transcript levels of PaChi4 and pathogen/host nuclear ratios in the sampled sections of inoculated or wounded 409 ramets (left panels) and 589 ramets (right panels) on days 3 (A and B), 7 (C and D), and 14 (E and F) after treatment. The transcript levels in inoculated (P) and wounded (W) samples are indicated on the left y axis, and the pathogen/host nuclear ratios (+) are indicated on the right y axis. The basal transcript levels in the control tissues at the times of inoculation (— — —) and wounding (- - -) are included. The basal transcript level in the 409A ramet at the time of inoculation was used as a reference for all calculations and is referred to with the value 1. Each datum point represents the mean of two ramets. The error bars (for transcript levels [—] and pathogen/host nuclear ratios [+]) indicate ± 1 standard error derived from the error mean square in an analysis of variance.
Regulation of gene transcripts.
α-Tubulin expression was stable and independent of treatment; the Ct values varied between samples by ±0.5 cycles (data not shown). This transcript was used as an endogenous reference for chitinase expression. Three replicates, i.e., three 2-mm-long sections, were analyzed for each ramet on day 0. The basal transcript levels of PaChi2 (data not shown) and PaChi4 (Fig. 1) were slightly higher in clone 589 than in clone 409. In contrast, the basal transcript levels of PaChi1 were slightly higher in clone 409 than in clone 589 (Fig. 2). At the start of the experiment, the Ct values of α-tubulin, PaChi4, PaChi2, and PaChi1 for ramet 409A were 23, 27, 29, and 23, respectively. The same rank order was observed for all four ramets.
FIG. 2.
Relative transcript levels of PaChi1 and pathogen/host nuclear ratios in the sampled sections of inoculated or wounded 409 (A) and 589 (B) ramets on day 3 after treatment. For figure explanation, see the legend for Fig. 1.
In inoculated tissue, the chitinase transcripts and the α-tubulin control were differentially regulated. There was a clear gradient in the expression of PaChi4 and PaChi2 at all time points, with the highest levels adjacent to the inoculation point. On day 3, the transcript levels of PaChi4 in the first three 2-mm-long sections adjacent to the inoculation point were significantly higher in the 589 ramets than in the 409 ramets (Fig. 1A and B). The difference between the clones was greatest in the second and third sections next to the inoculation point: the transcript level ratios between 589 and 409 in the first, second, and third sections were 179/105, 100/19 and 105/23, respectively. The corresponding P values were 0.01, 0.007, and 0.006, respectively. On day 3, transcript levels of PaChi2 in clone 589 also were higher than in clone 409 in the first three 2-mm-long sections adjacent to the inoculation point, but the differences between the clones were not statistically significant.
In contrast to day 3, by day 7 the 409 ramets had significantly higher transcript levels of PaChi4 (Fig. 1C and D) and PaChi2 (data not shown) than clone 589 in the first section next to the inoculation point. On day 14, the 409 ramets had significantly higher transcript levels of PaChi4 than the 589 ramets in the first four sections next to the inoculation point (P ≤ 0.0002) (Fig. 1E and F). On day 14, the 409 ramets had significantly higher transcript levels of PaChi2 than the 589 ramets in the first two sections next to the inoculation point (P ≤ 0.01) (data not shown). The maximum transcript levels of PaChi2 were observed on day 14 for both clones in the first section next to the inoculation point: 380- and 240-fold inductions were observed in clones 409 and 589, respectively (data not shown).
No significant differences between the clones were observed in the level of PaChi1 detected in response to infection. On day 3, the level of this transcript declined two- to sevenfold in response to infection in all ramets (Fig. 2). Compared to day 3, no further changes in the transcript level of PaChi1 were observed 7 and 14 dai in the clones (data not shown).
At the time of wounding, 1 year after the original inoculation, the transcript levels of PaChi4 (Fig. 1) and PaChi2 (data not shown) were elevated in all ramets relative to the time of inoculation, but the transcript level of PaChi1 had declined (Fig. 2). The transcript levels of PaChi4 for 589 and 409 were 12 and 3.0, respectively, and the difference was statistically significant (P = 0.01). Wounding alone resulted in a clear gradient in the expression levels of PaChi4 and PaChi2, with the highest levels immediately adjacent to the inoculation point, a result similar to the one observed after inoculation. In contrast to inoculation, the maximum induction levels of the two genes were observed on 3 days after wounding: 120- and 170-fold inductions of PaChi2 (data not shown) and 90- and 110-fold inductions of PaChi4 (Fig. 1A and B) were observed for clones 409 and 589, respectively. On day 14 the transcript levels of PaChi2 (data not shown) and PaChi4 (Fig. 1E and F) for both clones had returned to the basal levels observed at the time of wounding. No significant differences in the transcript levels of PaChi4 and PaChi2 were observed between the clones 3 and 14 days after wounding. Thus, the initial inoculation resulted in a significantly higher transcript levels of PaChi4 in the first three sections next to the inoculation point in clone 589 on day 3 (P ≤ 0.02) than did the subsequent wounding. Wounding and inoculation resulted in the same PaChi4 transcription levels in clone 409. The transcript level of PaChi1 3 days after wounding declined significantly more in the section next to the wound in clone 589 than in clone 409 (P = 0.003) (Fig. 2). For both clones, the transcript level of PaChi1 14 days after wounding did not change significantly from the levels observed 3 days after wounding.
DISCUSSION
In clone 409, with low disease resistance, H. annosum grew up to 12 mm from the inoculation point during a 14-day incubation period but was restricted to the first 2 to 4 mm in resistant clone 589. A comparison of the pathogen/host nuclear ratio profiles obtained now and in the previous study (14) indicates that the infection was established in the bark of clone 409 but not in that of clone 589. Vegetative cells of H. annosum contain 10 to 30 nuclei (34), so the pathogen/host nuclear ratios recorded on day 14 for the 409 ramets implies an approximately 1:1 ratio of pathogen and host cells. In the wounded tissues, the transcript levels of PaChi2 and PaChi4 returned to close to the basal levels after the initial rise on day 3, whereas in the inoculated tissues they increased continuously and were higher on day 14 in clone 409 than in clone 589. When coupled with the differential pathogen colonization profiles of the clones, these data suggest that the high transcript levels of PaChi2 and PaChi4 observed 14 dai are related to H. annosum colonization. The third monitored chitinase, PaChi1, was downregulated in inoculated or wounded tissue from both host clones, suggesting an endogenous role for this enzyme.
PaChi4 is similar to the basic class IV chitinases PgChi-1, Chia4-Pa1.1, and Chia4-Pa1.3, which are developmentally regulated during somatic embryogenesis in white spruce and Norway spruce, respectively (9, 39). Based on the expression patterns, Wiweger et al. (39) proposed that Chia4-Pa1.1 and Chia4-Pa1.3 promote somatic embryogenesis in Norway spruce, possibly by stimulating programmed cell death. PaChi4 differs from Chia4-Pa1.1 and Chia4.Pa1.3 at five or more amino acids. These differences probably are too numerous for alleles at the same locus and may mean that the three sequences are paralogs that represent different members of the Chia4-Pa gene (39). The genome of Norway spruce is poorly characterized, however, and the number and function of different chitinase genes and/or classes remains to be definitively determined. We think that the Northern assay used by Wiweger et al. (39) would detect transcripts of all three members of the gene family, if present. Similarly, the primer set we developed to detect PaChi4 would amplify all three sequences equally since the region recognized by the primers is identical in all three sequences. Thus, the high induction levels of PaChi4 we describe may represent the cumulative changes in all three members of this class IV gene family.
PaChi2, the other upregulated chitinase, has high similarity to an extracellular class II chitinase Pschi4 that is induced by chitosan and wounding in eastern white pine (Pinus strobus) (40, 41). Recently, Davis et al. (7) described the regulation of Pschi4 in slash pine (Pinus elliottii var. elliottii) seedlings challenged with the fungal pathogen Fusarium subglutinans f.sp. pini. In a Northern analysis, resistant seedlings accumulated Pschi4 transcripts at low levels 3 and 7 dai, but no transcript was detected 14 days after challenge. Susceptible seedlings lacked detectable Pschi4 transcripts on day 3, but accumulated high transcript levels by 7 and 14 dai. This pattern resembles that observed for PaChi2 and PaChi4 in the present study.
The present data and the studies on the PaChi2 ortholog Pschi4 (7, 40, 41) provide only circumstantial evidence for the role of chitinases in host defense. PaChi4 and PaChi2 probably are extracellular enzymes, since the predicted N-terminal signal peptides would allow secretion. Extracellular chitinases probably are part of an early defense response that could reduce or prevent hyphal growth in intercellular spaces and release other plant defense response elicitors from the pathogen cell wall (22). The chitinases induced by infection probably are not redundant defense enzymes but instead are complementary hydrolases with synergistic action on N-acetylglucosamine-containing substrates (5). PaChi2 and PaChi4 differ in their domain structure since the former lacks the chitin-binding domain. Kinetic studies of chitin hydrolysis by purified proteins would help elucidate the role(s) of spruce chitinases. Purified proteins also could be applied to healthy host tissue to evaluate microscopically the proposed connection to programmed cell death (39).
Maximum transcript levels for PaChi2 and PaChi4 were observed in clone 409 on day 14. This observation is consistent with other studies, in which a strong upregulation of chitinases occurs in susceptible, but not resistant, plants during the later stages of infection (see, for example, references 7 and 12). Despite the high transcript levels in clone 409 at 14 dai, the amount of pathogen DNA did not show any clear decline 35 dai. This result suggests that once the infection is established in clone 409, these chitinases have little impact on the pathogen. On the other hand, on day 3 clone 589 had higher transcript levels of PaChi2 and PaChi4 than did clone 409 in areas adjacent to the inoculation site. This observation suggests that the time from signal perception and transduction to the induction of these genes was shorter in the more resistant clone. Chitinase enzyme activity and protein and transcript levels often are higher in resistant cultivars than in susceptible ones shortly after inoculation (6), when a lower level of chitinases may suffice to prevent or reduce hyphal penetration.
The higher PaChi4 and PaChi2 transcript levels in clone 589 during the early stages of infection also could result in earlier production of exogenous elicitors from the fungal cell wall and an earlier triggering of other host defense reactions, e.g., increased lignification (2). To test the hypothesis that the rapidity of the overall response and the degree of coordination of the different defense strategies contribute to the level of resistance (17), studies of transcriptional activation of phenylalanine lyase and genes related to lignification at an early stage of H. annosum infection could be helpful. To allow an efficient screening of a larger amount of clones, a sampling of bark inoculations could be restricted to the first 6 mm away from the inoculation point, an area where the clones now studied showed pronounced differences in chitinase expression.
The low Ct values of PaChi1 in healthy control tissues indicate that the constitutive expression level of this gene is severalfold higher than that of PaChi2 and PaChi4. The high constitutive expression level and the downregulation after wounding and inoculation are expected if PaChi1 does not have a defense role. Constitutively expressed chitinases may regulate plant development by generating signal molecules from endogenous substrates such as arabinogalactan proteins containing N-acetylglucosamine (6, 36). The observed downregulation of PaChi1 could result from transcriptional resources being directed toward expression of defense-related chitinases such as PaChi4 and PaChi2. The differential expression of the studied chitinases makes their promoters interesting candidates for future studies of gene regulation in conifers.
Real-time RT-PCR is a sensitive and robust tool for monitoring gene expression and determining whether further, more time-consuming analyses, e.g., purification and activity analysis of the corresponding protein, are justified. The sampling scheme we used could not have been combined with a traditional Northern analysis due to the small sample size and low RNA yield (<3 μg of total RNA per sampled section). In addition, real-time RT-PCR can be used to monitor gene expression in the pathogen. The genome of H. annosum currently is poorly characterized, but recent work (16) and ongoing projects are likely to provide interesting candidate genes for monitoring pathogen gene expression at different stages of infection in order to identify, e.g., genes related to parasitic and saprophytic modes of H. annosum. In summary, spruce clones 589 and 409 accumulate PaChi4 and PaChi2 in patterns that are consistent with their resistance classifications and that suggest that more detailed work on these genes in relation to the resistance of Norway spruce toward H. annosum is warranted.
Acknowledgments
This study was financially supported by the Academy of Finland (grant to Ari Hietala), by the Norwegian Research Council (NFR-140682/130), and by the EU (QLK5-CT-2000-00241).
We thank Inger Heldal for assistance in processing the samples and John F. Leslie for comments improving the manuscript.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Barber, M. S., R. E. Bertram, and J. P. Ride. 1989. Chitin oligosaccharides elicit lignification in wounded wheat leaves. Physiol. Mol. Plant Pathol. 34:3-12. [Google Scholar]
- 3.Biswas, M., J. F. O'Rourke, E. Camon, G. Fraser, A. Kanapin, Y. Karavidopoulou, P. Kersey, E. Kriventseva, V. Mittard, N. Mulder, I. Phan, F. Servant, and R. Apweiler. 2002. Applications of InterPro in protein annotation and genome analysis. Brief Bioinform. 3:285-295. [DOI] [PubMed] [Google Scholar]
- 4.Broglie, K., I. Chet, M. Holliday, R. Cressman, P. Biddle, S. Knowlton, C. J. Mauvais, and R. Broglie. 1991. Transgenic plants with enhanced resistance to the fungal pathogen Rhizoctonia solani. Science 254:1194-1197. [DOI] [PubMed] [Google Scholar]
- 5.Brunner, F., A. Stintzi, B. Fritig, and M. Legrand. 1998. Substrate specificities of tobacco chitinases. Plant J. 14:225-234. [DOI] [PubMed] [Google Scholar]
- 6.Collinge, D. B., K. M. Kragh, J. D. Mikkelsen, K. K. Nielsen, U. Rasmussen, and K. Vad. 1993. Plant chitinases. Plant J. 3:31-40. [DOI] [PubMed] [Google Scholar]
- 7.Davis, J. M., H. G. Wu, J. E. K. Cooke, J. M. Reed, K. S. Luce, and C. H. Michler. 2002. Pathogen challenge, salicylic acid, and jasmonic acid regulate expression of chitinase gene homologs in pine. Mol. Plant-Microbe Interact. 15:380-387. [DOI] [PubMed] [Google Scholar]
- 8.Delatour, C., K. Von Weissenberg, and L. Dimitri. 1998. Host resistance, p. 143-166. In S. Woodward, J. Stenlid, R. Karjalainen, and A. Hüttermann (ed.), Heterobasidion annosum: biology, ecology, impact, and control. CAB International, Oxford, United Kingdom.
- 9.Dong, J., and D. I. Dunstan. 1997. Endochitinase and β-1,3-glucanase genes are developmentally regulated during somatic embryogenesis in Picea glauca. Planta 201:189-194. [DOI] [PubMed] [Google Scholar]
- 10.Felix, G., M. Regenass, and T. Boller. 1993. Specific perception of subnanomolar concentrations of chitin fragments by tomato cells: induction of extracellular alkalinization, changes in protein phosphorylation, and establishment of a refractory state. Plant J. 4:307-316. [Google Scholar]
- 11.Graham, L. S., and M. B. Sticklen. 1994. Plant chitinases. Can. J. Bot. 72:1057-1083. [Google Scholar]
- 12.Gregersen, P. L., H. Thordal-Christensen, H. Förster, and D. B. Collinge. 1997. Differential gene transcript accumulation in barley leaf epidermis and mesophyll in response to attack by Blumeria graminis f.sp. hordei (syn. Erysiphe graminis f.sp. hordei). Physiol. Mol. Plant Pathol. 51:85-97. [Google Scholar]
- 13.Grison, R., B. Grezes-Besset, M. Schneider, N. Lucante, L. Olsen, J. Leguay, and A. Toppan. 1996. Field tolerance to fungal pathogens of Brassica napus constitutively expressing a chimeric chitinase gene. Nat. Biotechnol. 14:643-646. [DOI] [PubMed] [Google Scholar]
- 14.Hietala, A. M., M. Eikenes, H. Kvaalen, H. Solheim, and C. G. Fossdal. 2003. Multiplex real-time PCR for monitoring Heterobasidion annosum colonization in Norway spruce clones that differ in disease resistance. Appl. Environ. Microbiol. 69:4413-4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iseli, B., S. Armand, T. Boller, J.-M. Neuhaus, and B. Henrissat. 1996. Plant chitinases use two different hydrolytic mechanisms. FEBS Lett. 382:186-188. [DOI] [PubMed] [Google Scholar]
- 16.Karlsson, M., Å. Olson, and J. Stenlid. 2003. Expressed sequences from the basidiomycetous tree pathogen Heterobasidion annosum during early infection of Scots pine. Fungal Genet. Biol. 39:51-59. [DOI] [PubMed] [Google Scholar]
- 17.Kombrink, E., and I. E. Somssich. 1995. Defense responses of plants to pathogens. Adv. Bot. Res. 21:1-34. [Google Scholar]
- 18.Kurosaki, F., N. Tashiro, and A. Nishi. 1988. Role of chitinase and chitin oligosaccharides in lignification response of cultured carrot cells treated with mycelial walls. Plant Cell Physiol. 29:527-531. [Google Scholar]
- 19.Kvaalen, H., and H. Solheim. 2000. Coinoculation of Ceratocystis polonica and Heterobasidion annosum with callus of two Norway spruce clones with different in vivo susceptibility. Plant Cell Tiss. Org. Cult. 60:221-228. [Google Scholar]
- 20.Lindberg, M., and M. Johansson. 1991. Growth of Heterobasidion annosum through bark of Picea abies. Eur. J. For. Pathol. 21:377-388. [Google Scholar]
- 21.Mauch, F., B. Mauch-Mani, and T. Boller. 1988. Antifungal hydrolases in pea tissue. Plant Physiol. 88:936-942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mauch, F., and L. A. Stahelin. 1989. Functional implications of the subcellular localization of ethylene-induced chitinase and β-1,3-glucanase in bean leaves. Plant Cell 1:447-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakamura, N., M. Marutani, S. Sanematsu, K. Toyoda, and Y. S. Inagaki. 2003. Phylogenetic classification of Dof-type transcription factors in pea (Pisum sativum). Plant Biotechnol. 20:247-253. [Google Scholar]
- 24.Neuhaus, J.-M., B. Fritig, H. J. M. Linthorst, F. Meins, J. D. Mikkelsen, and J. Ryals. 1996. A revised nomenclature for chitinase genes. Plant Mol. Biol. Rep. 14:102-104. [Google Scholar]
- 25.Nielsen, H., J. Engelbrecht, S. Brunak, and G. Von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 26.Pfaffl, M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:2002-2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Redfern, D. B., and J. Stenlid. 1998. Spore dispersal and infection, p. 105-124. In S. Woodward, J. Stenlid, R. Karjalainen, and A. Hüttermann (ed.), Heterobasidion annosum: biology, ecology, impact, and control. CAB International, Oxford, United Kingdom.
- 28.Ren, Y. Y., and C. A. West. 1992. Elicitation of diterpene biosynthesis in rice (Oryza sativa L.) by chitin. Plant Physiol. 99:1169-1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roby, D., A. Gadelle, and A. Toppan. 1987. Chitin oligosaccharides as elicitors of chitinase activity in melon plants. Biochem. Biophys. Res. Commun. 143:885-892. [DOI] [PubMed] [Google Scholar]
- 30.SAS. 1990. SAS user's guide: statistics, vol. 1, version 6. SAS Institute, Inc., Cary, N.C.
- 31.Schlumbaum, A., F. Mauch, U. Vögeli, and T. Boller. 1986. Plant chitinases are potent inhibitors of fungal growth. Nature 324:365-367. [Google Scholar]
- 32.Shaw, C. G. 1981. Basidiospores of Armillaria mellea survive an Alaskan winter on tree bark. Plant Dis. 65:972-974. [Google Scholar]
- 33.Solla, A., F. Tomlinson, and S. Woodward. 2002. Penetration of Picea sitchensis root bark by Armillaria mellea and Heterobasidion annosum. For. Pathol. 32:55-70. [Google Scholar]
- 34.Stenlid, J., and A. D. M. Rayner. 1991. Patterns of nuclear migration and heterokaryosis in pairings between sibling homokaryons of Heterobasidion annosum. Mycol. Res. 95:1275-1283. [Google Scholar]
- 35.Swedjemark, G., J. Stenlid, and B. Karlsson. 1998. Genetic variation among clones of Picea abies in resistance to growth of Heterobasidion annosum. Silvae Genet. 46:369-374.
- 36.van Hengel, A. J., Z. Tadesse, P. Immerzeel, H. Schols, A. van Kammen, and S. C. de Vries. 2001. N-Acetylglucosamine and glucosamine-containing arabionogalactan proteins control somatic embryogenesis. Plant Physiol. 125:1880-1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Loon, L. C., and E. A. van Strien. 1999. The families of pathogenesis-related proteins, their activities, and comparative analysis of PR-1 type proteins. Physiol. Mol. Plant Pathol. 55:85-97. [Google Scholar]
- 38.Wang, S. X., W. Hunter, and A. Plant. 2000. Isolation and purification of functional total RNA from woody branches and needles of sitka and white spruce. BioTechniques 28:292-296. [DOI] [PubMed] [Google Scholar]
- 39.Wiweger, M., I. Farbos, M. Ingouff, U. Lagercrantz, and S. von Arnold. 2003. Expression of Chia4-Pa chitinase genes during somatic and zygotic embryo development in Norway spruce (Picea abies): similarities and differences between gymnosperm and angiosperm class IV chitinases. J. Exp. Bot. 54:2691-2699. [DOI] [PubMed] [Google Scholar]
- 40.Wu, H., C. S. Echt, M. P. Popp, and J. M. Davis. 1997. Molecular cloning, structure and expression of an elicitor-inducible chitinase gene from pine trees. Plant Mol. Biol. 33:979-987. [DOI] [PubMed] [Google Scholar]
- 41.Wu, H., C. H. Michler, L. LaRussa, and J. M. Davis. 1999. The pine Pschi4 promoter directs wound-induced transcription. Plant Sci. 142:199-207. [Google Scholar]




