Abstract
We developed a DNA microarray suitable for simultaneous detection and discrimination between multiple bacterial species based on 16S ribosomal DNA (rDNA) polymorphisms using glass slides. Microarray probes (22- to 31-mer oligonucleotides) were spotted onto Teflon-masked, epoxy-silane-derivatized glass slides using a robotic arrayer. PCR products (ca. 199 bp) were generated using biotinylated, universal primer sequences, and these products were hybridized overnight (55°C) to the microarray. Targets that annealed to microarray probes were detected using a combination of Tyramide Signal Amplification and Alexa Fluor 546. This methodology permitted 100% specificity for detection of 18 microbes, 15 of which were fish pathogens. With universal 16S rDNA PCR (limited to 28 cycles), detection sensitivity for purified control DNA was equivalent to <150 genomes (675 fg), and this sensitivity was not adversely impacted either by the presence of competing bacterial DNA (1.1 × 106 genomes; 5 ng) or by the addition of up to 500 ng of fish DNA. Consequently, coupling 16S rDNA PCR with a microarray detector appears suitable for diagnostic detection and surveillance for commercially important fish pathogens.
Bacterial pathogens that infect fish include a phylogenetically diverse collection of organisms. Consequently, detection of these pathogens requires a wide diversity of sometimes time-consuming assays. For example, detection of the obligate, intracellular pathogen Piscirickettsia salmonis requires a lengthy, antibiotic-free, cell culture assay (11), while detecting Renibacterium salmoninarum can take up to 12 weeks on specialized media (2, 22). Assays for other species, such as Flavobacterium psychrophilum, require 3 to 7 days for growth with specialized media and growth conditions. Mycobacterium also requires specialized media and growth conditions, and mycobacteria are often overgrown by competing microflora. Thus, the heterogeneity of assays combined with often fastidious growth conditions makes these organisms excellent candidates for detection by molecular methods, such as PCR.
PCR-based assays are designed to amplify specific fragments of DNA that are subsequently identified by size using gel electrophoresis (3, 9, 13). By combining primer sets for multiple species-specific markers, a single PCR can be used to simultaneously detect several microbes. There are, however, practical limits to these assays. It can be difficult to incorporate more than six primer sets because of challenges in optimizing reaction conditions and challenges inherent in size discrimination between PCR products by conventional electrophoresis. Additional methods, such as Southern blotting or sequencing, are often needed to confirm product identity. An alternative approach is to use species-specific 16S ribosomal DNA (rDNA) PCR (17, 26) or universal PCR based on the 16S rDNA gene. Products from a universal reaction can be screened to identify species-specific polymorphisms in the DNA sequence using oligonucleotide probes (15), nested PCR (16), or automated sequencing. Sequencing is practical only if one 16S rDNA sequence is present in a sample. If more than one 16S rDNA sequence is present in a sample, then DNA microarrays can be used to efficiently screen a complex mixture of different sequences (27).
DNA microarrays permit simultaneous product interrogation with a large number of probe sequences (5, 7, 8, 27). In this format, probes for specific targets are typically deposited on a glass substrate to which PCR product or genomic DNA is then hybridized and detected (4). Because product size is no longer a factor in product identification, PCR assays can be designed to amplify similar-sized products and thus reduce PCR template bias. Furthermore, PCR assays can be designed to generate small products and thus maximize PCR efficiency and maximize the probability of template recovery from degraded DNA. Differences as small as 1-nucleotide base can be distinguished with an oligonucleotide-based microarray, although this degree of specificity is dependent on the sequence context (e.g., local melting temperature), hybridization conditions, and detection chemistry. In this study, we demonstrate a microbial detection system that couples PCR amplification of the 16S rDNA gene with an oligonucleotide-based microarray to simultaneously screen for 18 bacteria, 15 of which are fish pathogens.
MATERIALS AND METHODS
Bacterial positive controls.
Positive-control isolates were type strains (except for P. salmonis) obtained from the American Type Culture Collection, (Manassas, Va.) and from the Washington Animal Disease Diagnostic Laboratory (Pullman, Wash.) (Table 1) and were grown following American Type Culture Collection guidelines. Briefly, the following organisms were cultured on Columbia blood agar (incubation temperature/time): Aeromonas hydrophila (22 to 25°C/24 h), Aeromonas salmonicida subsp. salmonicida (22 to 25°C/24 to 48 h), Edwardsiella ictaluri (22 to 25°C/36 to 48 h), Staphylococcus aureus (37°C/24 to 36 h), Streptococcus iniae (22 to 25°C/24 to 48 h), Vagococcus salmoninarum (22 to 25°C/24 to 48 h), and Yersinia ruckeri type I (22 to 25°C/24 to 48 h). Escherichia coli (37°C/18 to 24 h) and Photobacterium phosphoreum (15°C/24 to 36 h) were cultivated on Luria-Bertani agar. Flavobacterium branchiophilum (22 to 25°C/6 to 10 days), Flavobacterium columnare (22 to 25°C/72 h), and Flavobacterium psychrophilum (15 to 17°C/5 to 7 days) were cultured on tryptone yeast extract agar with 1% milk. Tenacibaculum maritimum (22 to 25°C/36 to 96 h) was cultured on tryptone yeast extract agar with 1% milk and 1% sea salt. Mycobacterium chelonae (22 to 25°C/3 to 7 days), Mycobacterium fortuitum (37°C/3 to 5 days), and Mycobacterium marinum (22 to 25°C/5 to 10 days) were cultivated on Middlebrook 7H11 (with 0.0001% malachite green) or 7H10 agar (with 0.000025% malachite green). R. salmoninarum (15 to 17°C/10 days to >3 weeks) was cultivated on SKDM agar with 0.005% cycloheximide, 0.00125% d-cycloserine, 0.0025% polymyxin B sulfate, and 0.00025% oxolinic acid (1, 20, 24, and 28).
TABLE 1.
Oligonucleotides used in this study
| Organism or primer name | ATCCa | GenBankb | Probe or primer sequence | Low clusterc | SDc | High clusterc |
|---|---|---|---|---|---|---|
| Organisms | ||||||
| A. hydrophila | ATCC 7966 | X74676 | AAGGTTGATGCCTAATACGTATCAACTG | 1,598 | 614 | 44,503 |
| A. salmonicida | ATCC 33658 | AB027005 | TTGGCGCCTAATACGTGTCAAC | 410 | 245 | 12,339 |
| E. ictaluri | ATCC 33202 | AF310622 | GTGTGAGCGTTAATAGCGTTCACAA | 894 | 290 | 53,800 |
| E. coli | ATCC 27662 | AE000452 | GGGAGTAAAGTTAATACCTTTGCTCAT | 1,104 | 251 | 50,738 |
| F. branchiophilum | ATCC 35035 | D14017 | AGAAACACTTCTACGAGTAGAAGCTTG | 411 | 167 | 14,130 |
| F. columnare | ATCC 23463 | AY577821 | CCCTCCCTTGTAAGGGAGCTTGA | 971 | 253 | 3,946 |
| F. psychrophilum | ATCC 49418 | AY577822 | GAAACACTACCTCGTGAGGTAGCT | 515 | 158 | 17,221 |
| T. maritimum | ATCC 43398 | D14023 | GAAACGTACCTACGAGTAGGTATTT | 447 | 129 | 8,547 |
| M. chelonae | ATCC 35752 | X82235 | TTCAGTAGGGACGAAGCGAAAGT | 19,386 | 2205 | 65,535 |
| M. fortuitum | ATCC 6841 | X65528 | TTCAATAGGGACGAAGCGCAAGT | 352 | 185 | 17,399 |
| M. marinum | ATCC 927 | X52920 | GACGAAGGTTCGGGTTTTCTCG | 8,342 | 2550 | 60,552 |
| P. salmonis | ATCC 1361 | U36915 | AGGTAAGCTAATTAATACTTGGCTTAAT | 403 | 173 | 14,905 |
| R. salmoninarum | ATCC 33209 | X51601 | GAACAAGACATCATTTTTGTGGTGTTGAGG | 1,138 | 525 | 62,548 |
| S. aureus | ATCC 29213 | Y15856 | AACATATGTGTAAGTAACTGTGCACATCTTG | 1,151 | 536 | 63,323 |
| S. iniae | ATCC 29178 | AY577823 | CGGTAATGGGAGTGGAAAATCCATTAC | 623 | 363 | 61,591 |
| V. salmoninarum | ATCC 51200 | AY577824 | GTGGGAGAGTAACTGTTCCCACC | 1,299 | 459 | 43,884 |
| P. phosphoreum | ATCC 35080 | AY577825 | GTTGGAGTTAATAGCTTCAGCGTTTG | 612 | 215 | 31,932 |
| Y. ruckeri | ATCC 29473 | AJ289197 | AGGGTTAAGTGTTAATAGCACTGAACAT | 413 | 119 | 39,638 |
| Primers | ||||||
| 16S_517rvs | ATTACCGCGGCTGCTGG | |||||
| 16S_336fwd | AGACTCCTACGGGAGGCAGC | |||||
| 16S_008fwd | AGAGTTTGATCMTGGCTCAG |
American Type Culture Collection (ATCC) accession number for positive-control strains.
Representative GenBank accession number from which the probe sequence was derived.
Quantified results from Fig. 1. Low cluster represents average signal intensity for nonspecific probe hybridizations, SD is the standard deviation for the average low cluster, and High cluster is the average signal intensity for positive probes (excluding the biotin controls). In all cases, the low cluster was at least 3 standard deviations less than the high cluster.
The species identification of all strains was confirmed using morphological parameters and standard biochemical tests employed at the Washington Animal Disease Diagnostic Laboratory. In addition, fluorescent antibody testing using commercially prepared antibody (Bayotek International, Inc., Saanichton, British Columbia, Canada) was performed on A. salmonicida subsp. salmonicida, Y. ruckeri type I, and E. ictaluri as well as antibody agglutination for A. salmonicida subsp. salmonicida and Y. ruckeri type I (Bayotek). Monoclonal enzyme-linked assay (DiagXotics, Inc., Wilton, Conn.) was performed on the R. salmonicida isolate. PCR assays were used to confirm species identification of Y. ruckeri type I (17) and F. psychrophilum (25) isolates.
DNA was prepared by one of three methods. Cells were washed from agar plates using 1× phosphate-buffered saline (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4 [dibasic anhydrous], 2 mM KH2PO4 [monobasic anhydrous] [pH 7.4]), and DNA was extracted using a DNeasy kit (Qiagen, Valencia, Calif.). Alternatively, a single isolate was picked, put into fresh broth, and grown to stationary phase in the appropriate medium, and DNA was extracted using a DNeasy kit. DNA template was also prepared as boiled lysate when a colony was picked, resuspended in 200 μl of TE buffer (10 mM Tris base [pH 8], 1 mM EDTA [pH 8]), and boiled for 5 min. Template was removed after centrifuging the lysed cells for 5 min at 14,000 × g.
P. salmonis is an obligate intracellular pathogen and consequently was cultivated in CHSE-214 cells without antibiotics as described in the Office International des Epizooties (22). The CHSE-214 cells were harvested after a cytopathic effect was observed, approximately 10 to 14 days postinoculation. Harvested cells were extracted using the DNeasy kit, and species identification was confirmed using a PCR assay (19). The same kit was used to extract DNA from Chinook salmon (Oncorhynchus tshawytscha) kidneys obtained from a Pacific Northwest hatchery.
Our choice of such a diverse set of organisms reflects our interest in applying this technique across a phylogenetically disparate group of organisms. This selection also reflects broad regulatory concerns and the interests of clients that regularly use the Aquatic Health Inspection Service at the Washington Animal Disease Diagnostic Laboratory. E. coli, S. aureus, and P. phosphoreum were included as control organisms for this and other ongoing research projects (D. R. Call, A. Warsen, and M. Soule, unpublished data).
16S rDNA probe sequences.
16S rDNA sequences were retrieved from GenBank (www.ncbi.nlm.nih.gov). In some cases, the GenBank data were ambiguous; therefore, we generated new sequences for this project (F. columnare, F. psychrophilum, S. iniae, V. salmoninarum, and P. phosphoreum) using 16S universal primers 16s_008fwd and 16S_517rvs (Table 1). The resulting ca. 513- to 534-bp PCR products were subsequently cloned (TA kit; Invitrogen Corp., Carlsbad, Calif.) and sequenced (GenBank accession numbers AY577821 to AY577825) (Amplicon Express, Pullman, Wash.). Sequence alignments were used to identify polymorphic regions (Vector NTI version 7.1; InforMax, Inc., Bethesda, Md.) in the vicinity of position 453 (E. coli) (GenBank accession number AE000452). Oligonucleotide probes were designed for the polymorphic region using Primer Premier software (Biosoft International, Palo Alto, Calif.) (Table 1). All oligonucleotides were normalized to have a calculated annealing temperature of 63°C ± 3°C. Unmodified, desalted oligonucleotide probes were commercially synthesized (Invitrogen Corp.).
Slide preparation.
Multiple microarrays were printed on glass slides so that independent microarrays were contained within eight individual wells defined by Teflon masking (Erie Scientific, Portsmouth, N.H.); the hydrophobic nature of the masking permitted independent samples to be hybridized within each well. Slides were derivatized with an epoxy-silane monolayer (6). Prior to printing, the slides were sonicated in 2.5% Contrad 70 detergent (Fisher Scientific, Pittsburgh, Pa.) for 2 min, rinsed three times with distilled water, and dried using compressed air. Slides were then soaked for 1 h in 3 N HCl, rinsed three times with deionized water, and dried with compressed air. Slides were derivatized with a 2% solution of 3-glycidoxypropyltrimethoxysilane (Sigma Aldrich, St. Louis, Mo.) in methanol (high-performance liquid chromatography grade) for 15 min, rinsed twice with 100% methanol, and dried with compressed air.
Microarray construction.
Probes were reconstituted in TE buffer, quantified using a biophotometer (Eppendorf Scientific, Westburg, N.Y.), diluted to 60 μM in print buffer (0.1 M Na2HPO4, 0.2 M NaCl, 0.01% sodium dodecyl sulfate) with a pH of ∼11 and transferred to 96-well plates for printing. Arbitrary biotinylated oligonucleotides (25-mer; 5 μM) were included with every array. These biotin pseudoprobes served as positive controls for the detection chemistry and to orient the array for image processing. All probes were deposited as four replicates at a fixed location within each masked well using a Microgrid II arrayer (BioRobotics, Woburn, Mass.) with humidity held at 45%. Printing parameters included washing the pins in a recirculating bath (four pins washed twice for 4 s each time), followed by 0.5 s of flushing and 6 s of drying. This washing procedure was repeated twice between probes to minimize possible probe carryover. Printed slides were baked under a vacuum (22 Hg/mm) for 1 h (130°C) and stored away from light at room temperature.
Target preparation.
Biotinylated primers 16s_336 (this study) and 16s_517 (21) were used to amplify a ca.199-bp product from the 16S rDNA gene in both gram-positive and gram-negative bacteria. Each 50-μl PCR mixture contained reaction buffer (Fisher Scientific), 0.2 mM concentration of each deoxynucleoside triphosphate, 2.0 mM MgCl2, 2 to 5 U of Taq polymerase, 0.4 μM concentration of each biotinylated primer, and ca. 20 to 50 ng of genomic DNA or 10 μl of boiled lysate. PCR cycling conditions included an initial 5-min incubation at 95°C, followed by 25 to 35 cycles of PCR, with 1 cycle consisting of denaturation (95°C for 30 s), annealing (62°C for 60 s), and extension (72°C for 60 s), with a final extension step at 72°C for 10 min. Agarose gel electrophoresis was used to confirm the presence of PCR products.
Hybridization.
Prior to hybridization, slides were preblocked at room temperature for 30 min in TNB buffer. TNB buffer consists of 100 mM Tris-HCl (pH 7.5), 150 mM NaCl, and 0.5% blocking reagent (Biotin Tyramide Signal Amplication [TSA] kit; PerkinElmer Life Sciences, Boston, Mass.). PCR product (21.4 μl) was combined with hybridization buffer (final concentration, 4× SSC [1× SSC is 15 mM NaCl plus 0.15 mM sodium citrate, pH 7.0]) and 5× Denhardt's solution [0.001% Ficoll, 0.001% polyvinylpyrrolidone, 0.001% bovine serum albumin] in a final volume of 100 μl. This mixture was denatured by heat treatment (95°C for 2 min), cooled, and held on ice until application to two independent wells on the slide. Blocking buffer was aspirated and immediately replaced with 45 μl of denatured target per well. Slides were placed in a humidified chamber (50-ml conical tube with paper moistened with hybridization buffer). Chambers were then submerged in a 55°C water bath for incubation overnight. After removal from hybridization chambers, targets were removed by aspiration. Throughout these procedures, it was critical that the surface of the slide did not dry out. Slides were washed three times for 1 min each time in TNT buffer (0.1 M Tris-HCl [pH 7.5], 0.15 M NaCl, 0.05% Tween 20). A streptavidin-horseradish peroxidase conjugate (TSA kit) diluted 1:100 in TNB buffer was then incubated in each well for 30 min, and then each well was washed with TNT buffer three times for 1 min each time. Fetal equine serum (10%) in 2× SSC was incubated in each well for 30 min to provide a protein surface for tyramide binding, and then each well was washed with TNT buffer three times for 1 min each time. Biotinyl tyramide (1:50 in amplification diluent) was incubated in each well for 10 min. Slides were washed and incubated with 2 μg of streptavidin Alexa Fluor 546 conjugate (Molecular Probes, Eugene, Oreg.) per ml prepared in a mixture consisting of 1× SSC and 5× Denhardt's solution (total volume, 45 μl) for 1 h at room temperature (shielded from ambient light). After incubation, slides were washed and dried with a Microarray high-speed centrifuge (ArrayIt, Sunnyvale, Calif.) and imaged with an arrayWoRxe scanner (Applied Precision, Issaquah, Wash.).
Assay sensitivity.
We determined the baseline sensitivity using serial dilution curves (10-fold) between 6.6 × 10−8 and 6.6 × 10−17 g. We arbitrarily selected genomic DNA from Y. ruckeri for these experiments, and each dilution series was repeated three times. Dilution series were assessed using (i) Y. ruckeri DNA only, (ii) Y. ruckeri DNA plus 5 ng of S. aureus DNA in each dilution, or (iii) Y. ruckeri DNA plus 500 ng of fish DNA (O. tshawytscha) in each dilution. NCSS 2004 (Number Cruncher Statistical Systems, Kaysville, Utah) was used for statistical analysis.
Signal detection and array analysis.
SoftWoRx Tracker software (Applied Precision) was used to quantify median spot intensity, and data were managed with a custom relational database (Access; Microsoft Corp., Redmond, Wash.). All experiments were performed in triplicate. For most purposes, we were able to identify positive hybridization simply by visual inspection. We also developed a cluster algorithm to score microarray data and detect positive hybridization. For each hybridization experiment, probe intensity data were sorted from the lowest signal intensity (ca. 200 U) to the highest signal intensity (65,535 U). The lowest and highest intensity values were identified as cluster seeds. The next lowest intensity value was compared to the cluster seeds and assigned to the nearest cluster. Cluster seeds were then recalculated as the average of all cluster members. This process continued until all probes were assigned to either the low (CL1) or high (CH1) cluster. Because this procedure will produce a higher variance for the CL1 cluster, the procedure was repeated, except that cluster membership was assigned in the opposite order, from the highest intensity to the lowest intensity to calculate CL2 and CH2. Final cluster averages were calculated as CL = (CL1 + CL2)/2 and CH = (CH1 + CH2)/2. All probe intensity values were then reassigned to the low and high clusters on the basis of the proximity to CL and CH. Standard deviations were then calculated for the low and high clusters and if CL and CH were separated by at least 3 standard deviations, then probes belonging to the high cluster were classified as positive. This procedure was programmed using Visual Basic (Microsoft Corp.) and is available upon request.
RESULTS
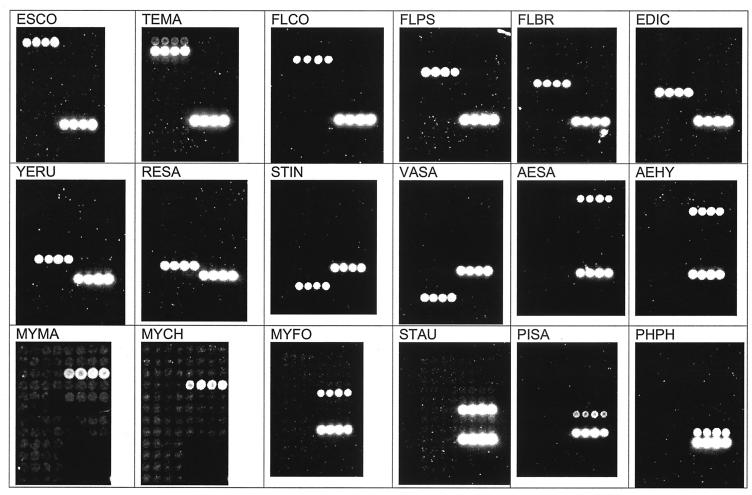
We constructed an oligonucleotide microarray for simultaneous detection of 18 microbes based on 16S rDNA polymorphisms (Table 1). In all cases, the microarray detector demonstrated a one-to-one correspondence between a positive-control bacterium and its probe (Fig. 1). These results were quite explicit by either visual inspection or our cluster analysis (Table 1).
FIG. 1.
Probe specificity. Positive-control hybridization signals for A. hydrophila (AEHY), A. salmonicida subsp. salmonicida (AESA), E. ictaluri (EDIC), E. coli (ESCO), F. branchiophilum (FLBR), F. columnare (FLCO), F. psychrophilum (FLPS), M. chelonae (MYCH), M. fortuitum (MYFO), M. marinum (MYMA), R. salmoninarum (RESA), P. salmonis (PISA), S. aureus (STAU), S. iniae (STIN), T. maritimum (TEMA), V. salmoninarum (VASA), P. phosphoreum (PHPH), and Y. ruckeri type I (YERU) are shown. The lower right row of spots in each panel (not shown for MYMA or MYCH) is a biotinylated oligonucleotide control for detection chemistry.
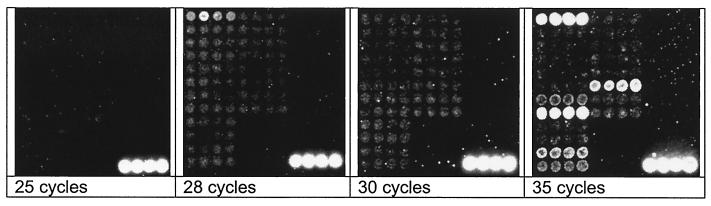
Figure 1 illustrates hybridization results when there was no competing template DNA and when there was abundant positive-control DNA in the reaction mixture. Most of these results were also produced using our original protocol that included 35 cycles of PCR. With this protocol, however, we often encountered amplification of unexpected PCR products in the negative-control PCRs (no-template controls) (Fig. 2). The E. coli probe was frequently positive when we used a recombinant source of Taq polymerase. We also encountered amplification of targets that readily hybridized to multiple probes on the microarray (Fig. 2), which is consistent with low-level contamination of our laboratory with either amplicons or template DNA. This was an unexpected problem, because all reaction components were mixed in a room separately from where samples were added to the PCR tubes and separately from where we conducted agarose electrophoresis. Additional precautions included UV treatment of reaction tubes and pipettes and storage of PCR reagents separate from template DNA. Clearly, if the level of precaution undertaken in our facility still permitted contaminating PCR template to routinely amplify in no-template control reactions, then the assay would not be very useful.
FIG. 2.
Representative examples of microarray hybridization from no-template control PCRs. Little to no background is evident with 30 cycles or less, but considerable background is evident with 35 cycles of PCR.
Our initial solution to this difficulty was to spike each PCR mixture with the equivalent of 50 genomes (225 fg) of a positive-control organism (e.g., S. aureus). This strategy was predicated on the assumption that if we provided a low level of known template DNA in the reaction mixture, this DNA would be preferentially amplified in the presence of very low concentrations of contaminating DNA. Our strategy worked (data not shown), but we found it difficult to accurately deliver the same low concentration of DNA with every experiment; therefore, this strategy was not robust. Our second strategy was to risk decreasing the assay sensitivity and reduce the number of PCR cycles to avoid amplifying contaminating DNA to the point where it was detected by the microarray. We found that 30 or fewer PCR cycles almost always eliminated background amplification and detection with the microarray (Fig. 2). Even with 28 cycles, we frequently detected E. coli DNA, and we attributed this to DNA contamination from the commercial reagents used in this study.
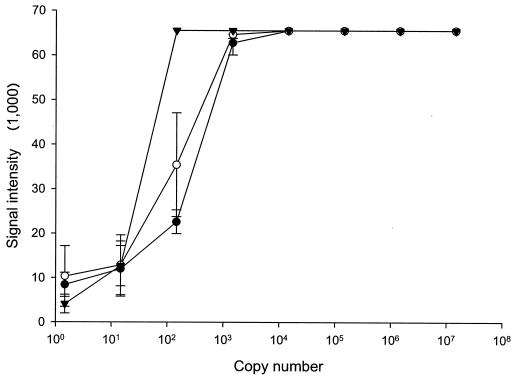
The sensitivity of the assay, when limited to 28 PCR cycles, was equivalent to <150 genomes for purified template DNA (Fig. 3). This conclusion was based on the magnitude of probe signal (>0 U; P < 0.05 by the t test) and our cluster algorithm, which indicated positive detection with this starting concentration of template DNA. Importantly, this level of sensitivity was not compromised by the addition of a competing template DNA (5 ng of S. aureus DNA added; ca. 1.1 × 106 genomes), and the addition of 500 ng of fish DNA did not affect assay sensitivity (Fig. 3). Hybridization signals from nontarget probes (excluding Y. ruckeri type I, S. aureus, and E. coli) were significantly lower for dilution curves mixed with fish DNA (3,733 U) than for dilution curves with purified DNA (9,789 U) and purified DNA mixed with competing bacterial DNA (10,022 U) (analysis of variance with Tukey-Kramer multiple-comparison test; P < 0.05).
FIG. 3.
Assay sensitivity. Data showing assay sensitivity for purified pathogen DNA alone (•) or for DNA with competing bacterial (○) or fish (▾) DNA. Competing DNA does not impact assay sensitivity (<150 copies; P < 0.05) using this protocol.
DISCUSSION
Simultaneous testing for multiple pathogens is clearly advantageous when diagnostic testing would otherwise require a heterogeneous suite of assays. This may not be as important in cases where clinical disease is evident and where the number of suspected pathogens can be reduced to a manageable number on the basis of pathological indicators. Indeed, it is probably reasonable to assume that many cases of infectious disease are caused by a single microbial species. Nevertheless, an assay such as the one we describe herein could still be used for final discrimination or confirmation. If, however, the goal is surveillance at a population level in the absence of pathological indicators, then simultaneous testing becomes potentially very valuable. This is also true for environmental sampling where the water column or other matrices are tested as potential sources of microbial pathogens (18).
Our assay clearly detects and discriminates between 18 bacterial species. It is, however, important to note a potential “polymorphism paradox” in detecting these genetic differences. That is, we are purposely selecting a region of the 16S rDNA gene that has a high degree of variation; therefore, this region may be prone to frequent mutations. A strain that should be identified using the microarray might be missed due to a random mutation within this region and thus produce a false negative. We do not know what the frequency of these events might be at a population level, but we have evidence that this might be the case for one strain of F. pyschophilum (Call et al., unpublished data). An additional challenge with this strategy is that two “distinct” species may share the same sequence identity within the probe region (10), although this was not an issue for the 18 species tested here.
Because we had to reduce the number of PCR cycles in this assay, the assay sensitivity (<150 genomes) was not as high as can be achieved with PCR (<100 genomes [5, 12]). We submit, however, that the most effective gains in assay sensitivity can be achieved “upstream” of the microarray detector itself. That is, processing larger sample volumes (water) or mass (tissue) combined with more efficient DNA extraction techniques may be the most effective means to improve overall assay sensitivity (23).
For the assay detection sensitivity to be 100%, all truly positive samples must be detected and differentiated with the array. Nevertheless, depending on the sample matrix and “upstream” activities, false-negative results can be an important problem (18). In the format described here, coextracted sample impurities can interfere with PCR amplification. Failed amplification would not be readily apparent if only the microarray were used with no additional controls. One strategy to test for failed amplification is to spike samples with a known 16S rDNA sequence concurrently with the PCR. The internal control sequence would be amplified by PCR along with templates from the sample, and all products would be differentiated with the microarray. Adding template DNA runs the risk of reduced sensitivity if the spiked template is preferentially amplified during the PCR (Call et al., unpublished data). Alternatively, when complex prokaryotic DNA mixtures are amplified (e.g., from a water sample), we routinely see an amplified product by gel electrophoresis even when no targets of interest hybridize to the microarray. In these cases, nonspecific amplification of the 16S rDNA gene serves as an internal control for false-negative results due to PCR inhibition. Amplification of an independent eukaryotic marker would be very useful if tissues are being tested, although this is not part of the current assay.
We developed a relatively simple assay for screening PCR products for species-specific sequence polymorphisms. Our multiwell format permits higher sample throughput than conventional microarrays where a separate slide is required for each test. We have also found that the multiwell format is relatively simple to use and requires less time to develop necessary technical skills compared with unmasked slides that require coverslips. Ultimately, detection methods such as the one described here will become even more useful when the probes are used in conjunction with microsphere beads (14). Suspension arrays of this nature can be processed more efficiently, and they permit beginning-to-end sample processing in a 96-well format without need for posthybridization image processing.
Acknowledgments
Excellent technical assistance was provided by Katie Snekvik and the Aquatic Health Inspection Service of the Washington Animal Disease Diagnostic Laboratory.
This work was supported in part by funding from the WSU & UI Aquaculture Initiative, WSU & UI Center for Reproductive Biology, and Agricultural Animal Health Program, Pullman, Wash.
REFERENCES
- 1.Austin, B., and D. Austin. 1999. Bacterial fish pathogens: disease of farmed and wild fish, 3rd ed. Springer-Praxis, London, United Kingdom.
- 2.Benediktsdottir, E., S. Helgason, and S. Gudmundsdottir. 1991. Incubation time for the cultivation of Renibacterium salmoninarum from Atlantic salmon, Salmon salar L., broodfish. J. Fish Dis. 14:97-102. [Google Scholar]
- 3.Brasher, C. W., A. DePaola, D. D. Jones, and A. K. Bej. 1998. Detection of microbial pathogens in shellfish with multiplex PCR. Curr. Microbiol. 37:101-107. [DOI] [PubMed] [Google Scholar]
- 4.Call, D. R., M. Borucki, and F. Loge. 2003. Detection of bacterial pathogens in environmental samples using DNA microarrays. J. Microbiol. Methods 53:235-243. [DOI] [PubMed] [Google Scholar]
- 5.Call, D. R., F. J. Brockman, and D. P. Chandler. 2001. Detecting and genotyping Escherichia coli O157:H7 using multiplexed PCR and nucleic acid microarrays. Int. J. Food Microbiol. 67:71-80. [DOI] [PubMed] [Google Scholar]
- 6.Call, D. R., D. P. Chandler, and F. Brockman. 2001. Fabrication of DNA microarrays using unmodified oligonucleotide probes. BioTechniques 30:368-376. [DOI] [PubMed] [Google Scholar]
- 7.Chizhikov, V., A. Rasooly, K. Chumakov, and D. D. Levy. 2001. Microarray analysis of microbial virulence factors. Appl. Environ. Microbiol. 67:3258-3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chizhikov, V., M. Wagner, A. Ivshina, Y. Hoshino, A. Z. Kapikian, and K. Chumakov. 2002. Detection and genotyping of human group A rotaviruses by oligonucleotide microarray hybridization. J. Clin. Microbiol. 40:2398-2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Del Cerro, A., I. Marquez, and J. A. Guijarro. 2002. Simultaneous detection of Aeromonas salmonicida, Flavobacterium psychrophilum, and Yersinia ruckeri, three major fish pathogens, by multiplex PCR. Appl. Environ. Microbiol. 68:5177-5180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fox, G. E., J. D. Wisotzkey, and P. Jurtshuk, Jr. 1992. How close is close: 16S rRNA sequence identity may not be sufficient to guarantee species identity. Int. J. Syst. Bacteriol. 42:166-170. [DOI] [PubMed] [Google Scholar]
- 11.Fryer, J., and R. Hedrick. 2003. Piscirickettsia salmonis: a Gram-negative intracellular bacterial pathogen in fish. J. Fish Dis. 26:251-262. [DOI] [PubMed] [Google Scholar]
- 12.Gibello, A., M. M. Blanco, M. A. Moreno, M. T. Cutuli, A. Domenech, L. Dominguez, and J. F. Fernandez-Garayzabal. 1999. Development of a PCR assay for detection of Yersinia ruckeri in tissues of inoculated and naturally infected trout. Appl. Environ. Microbiol. 65:346-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.González, S., C. R. Osorio, and Y. Santos. 2003. Development of a PCR-based method for the detection of Listonella anguillarum in fish tissues and blood samples. Dis. Aquat. Org. 55:109-115. [DOI] [PubMed] [Google Scholar]
- 14.Grate, J., C. Bruckner-Lea, A. Jarrell, and D. P. Chandler. 2003. Automated sample preparation method for suspension arrays using renewable surface separations with multiplexed flow cytometry fluorescence detection. Anal. Chim. Acta 47:85-98. [Google Scholar]
- 15.Greisen, K., M. Loeffelholz, A. Purohit, and D. Leong. 1994. PCR primers and probes for the 16S rRNA gene of most species of pathogenic bacteria, including bacteria found in cerebrospinal fluid. J. Clin. Microbiol. 32:335-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herman, L. M., J. H. De Block, and G. M. Waes. 1995. A direct PCR detection method for Clostridium tyrobutyricum spores in up to 100 milliliters of raw milk. Appl. Environ. Microbiol. 61:4141-4146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.LeJeune, J. T., and F. R. Rurangirwa. 2000. Polymerase chain reaction for definitive identification of Yersinia ruckeri. J. Vet. Diagn. Investig. 12:558-561. [DOI] [PubMed] [Google Scholar]
- 18.Loge, F. J., D. E. Thompson, and D. R. Call. 2002. PCR detection of specific pathogens in water: a risk-based analysis. Environ. Sci. Technol. 36:2754-2759. [DOI] [PubMed] [Google Scholar]
- 19.Marshall, S., S. Heath, V. Henriquez, and C. Orrego. 1998. Minimally invasive detection of Piscirickettsia salmonis in cultivated salmonids via the PCR. Appl. Environ. Microbiol. 64:3066-3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murray, P. R., E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.). 1999. Manual of clinical microbiology, 7th ed. American Society for Microbiology, Washington, D.C.
- 21.Muyzer, G., E. C. de Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Office International des Epizooties. 2003. Manual of diagnostic tests for aquatic animals. Office International des Epizooties, Paris, France.
- 23.Straub, T. M., and D. P. Chandler. 2003. Towards a unified system for detecting waterborne pathogens. J. Microbiol. Methods 53:185-197. [DOI] [PubMed] [Google Scholar]
- 24.Thoesen, J. (ed.). 1994. Suggested procedures for the detection and identification of certain finfish and shellfish pathogens, 4th ed. SOS Publications, Fair Haven, N.J.
- 25.Toyama, T., K. Kita-Tsukamoto, and H. Wakabayashi. 1994. Identification of Cytophaga psychrophila by PCR targeted 16S ribosomal RNA. Fish Pathol. 29:271-275. [Google Scholar]
- 26.Urdaci, M. C., C. Chakroun, D. Faure, and J. F. Bernardet. 1998. Development of a polymerase chain reaction assay for identification and detection of the fish pathogen Flavobacterium psychrophilum. Res. Microbiol. 149:519-530. [DOI] [PubMed] [Google Scholar]
- 27.Wilson, W. J., C. L. Strout, T. Z. DeSantis, J. L. Stilwell, A. V. Carrano, and G. L. Andersen. 2002. Sequence-specific identification of 18 pathogenic microorganisms using microarray technology. Mol. Cell. Probes 16:119-127. [DOI] [PubMed] [Google Scholar]
- 28.Woo, P., and D. Bruno (ed.). 1999. Fish disease and disorders, vol. 3. Bacterial and fungal infections. CABI Publishing, New York, N.Y.