Abstract
The prevalence and distribution of Cryptosporidium spp. in the fecal droppings of the free-living waterfowl Canada geese were examined at 13 sites in Ohio and Illinois. On the basis of the analysis of the small-subunit rRNA gene by PCR, followed by restriction fragment length polymorphism analysis and DNA sequencing, 49 (23.4%) of 209 fecal specimens collected from 10 sites (76.9%) were positive for Cryptosporidium spp. The following five Cryptosporidium species and genotypes were identified: Cryptosporidium goose genotype I (in 36 specimens), Cryptosporidium goose genotype II (in 9 specimens), Cryptosporidium duck genotype (in 1 specimen), Cryptosporidium parvum (in 4 specimens), and C. hominis (in 2 specimens). Cryptosporidium goose genotype I was the most prevalent parasite and was found at all five Cryptosporidium-positive sites in Ohio and at four of five positive sites in Illinois, followed by Cryptosporidium goose genotype II, which was found at two of five positive sites in Ohio and at four of five positive sites in Illinois. Cryptosporidium goose genotype II was detected for the first time, and it is phylogenetically related to goose genotype I and the duck genotype. All three genotypes have not so far been reported in humans, and their pathogenicity in geese has not been determined. Only 10.2% of the Cryptosporidium-positive specimens had C. parvum and C. hominis. The results of this study indicate that Canada geese might only serve as accidental carriers of cryptosporidia infectious to humans and probably play a minor role in the animal-to-human transmission cycle of the pathogen.
The Canada goose population in the United States has been increasing in the last decade. Most Canada geese are migratory, wintering in the United States and migrating to their summer breeding grounds in Canada (21). However, the availability of suitable habitats, such as grassy areas with bodies of water in urban and suburban areas, has increased the numbers of these geese that have become year-round residents in the United States. Their feces litter the ground in many public areas such as parks, golf courses, cemeteries, and residential areas. That these Canada geese might carry and distribute human pathogens is a major public health concern (2, 4, 6, 10, 11).
Cryptosporidia are among the pathogens potentially disseminated by Canada geese (6, 9, 10). Cryptosporidium parvum oocysts have been shown to retain viability and infectivity following passage through Canada geese (5). It has been suggested that migratory Canada geese might disseminate infectious oocysts of C. parvum through their feces into public water sources (6). This is of particular significance because Canada geese prefer aquatic habitats and contaminated water is a major source of human Cryptosporidium infection in the United States (3). In recent years, laboratory-confirmed cases of waterborne cryptosporidiosis have increased significantly in the United States (3).
Many Cryptosporidium species are present in animals, of which C. parvum and C. hominis are the most important causes of cryptosporidiosis in humans (12, 23). C. hominis was previously known as C. parvum human genotype or genotype 1, infects mostly humans, and is presumably transmitted from humans to humans. C. parvum infects both ruminants and humans and can be acquired by both human-to-human and zoonotic transmission pathways (14, 18). Lately, some other Cryptosporidium species and genotypes have also been found in both immunocompetent and immunocompromised persons, including C. meleagridis, C. felis, C. canis, C. muris, and Cryptosporidium pig and cervine genotypes (1, 7, 15-20, 22, 23). Many other host-adapted Cryptosporidium spp. have also been found in a variety of animal species, including farm animals, pets, and wildlife (24).
The number of Cryptosporidium species in Canada geese is not known. Only one study has indicated that Canada geese might play a role as mechanical carriers of infectious C. parvum (6). Another study reported a host-adapted Cryptosporidium sp. (goose genotype) in the feces of migratory Canada geese (24) that so far has not been found in humans. Cryptosporidiosis is apparently very common in Canada geese because cryptosporidia (species and genotypes undetermined) were found in a recent study in feces of Canada geese at 9 of 10 sites in the Toledo, Ohio, area (10).
The purpose of this study was to determine if Canada geese are sources of C. parvum and C. hominis, which are the dominant species infecting humans. Feces of Canada geese were collected from 13 sites in Ohio and Illinois and examined for the prevalence and distribution of Cryptosporidium species by PCR analysis of the small-subunit (SSU) rRNA gene. Results of this study have shown the presence of two host-adapted Cryptosporidium genotypes in Canada geese, with only occasional detection of human-pathogenic Cryptosporidium spp. in the feces of these animals.
MATERIALS AND METHODS
Specimen collection and DNA extraction.
Canada goose fecal specimens were collected from seven sites in Toledo, Ohio, and six sites in DuPage County, Ill. To reduce the chance of one animal contributing to more than one specimen, only recently deposited feces, appearing wet and loose and warm to the touch, were collected from each sampling site. In some cases, specimens were collected immediately after Canada geese were seen defecating. Each fecal specimen contained about 1 g of feces and was placed in a vial containing 2.5% potassium dichromate solution. A total of 209 individual specimens, of which 148 (70.8%) were from Ohio and 61 (29.2%) were from Illinois, were collected and analyzed for Cryptosporidium species and genotypes as described below. The majority (>90%) of the birds were Canada geese. However, a small number of ducks and other waterfowl were seen in some of the sites. Most of the Canada geese were adults, with only a few juveniles. The specimens from Illinois were collected in July 1999, whereas the specimens from Ohio were collected between July and October 2002. Most of the birds were migratory, because by the end of October or the beginning of November there were only few Canada geese at most of the study sites.
A portion of about 200 mg from each specimen was washed twice in a Microfuge tube with 1 ml of distilled water and centrifugation at 12,000 × g for 15 min. No oocyst concentration or purification and no microcopy were done before DNA extraction. The pellet was treated initially with 66.7 μl of 1 M KOH and 18.6 μl of 1 M dithiothreitol, followed by neutralization with 8.6 μl of 25% (vol/vol) hydrochloric acid. The DNA lysate was then extracted once with phenol-chloroform-isoamyl alcohol (25:24:1) solution, and genomic DNA was extracted with the QIAamp DNA Stool Mini Kit (QIAGEN Inc., Valencia, Calif.) as previously described (24). Extracted DNA was stored at −70°C prior to being analyzed.
Differentiation of Cryptosporidium spp. by PCR-RFLP.
Cryptosporidium species and genotypes were determined by a previously described technique based on PCR-restriction fragment length polymorphism (RFLP) analysis of the SSU rRNA gene (22, 23). In this method, a fragment of 826 to 864 bp of the SSU rRNA gene was amplified by nested PCR. For the detection and differentiation of Cryptosporidium species and genotypes, 10 μl of the secondary PCR product was subjected to restriction digestions with SspI (New England BioLabs, Beverly, Mass.) and VspI (GIBCO BRL, Grand Island, N.Y.). Differences in SspI and VspI banding patterns in 2% agarose electrophoresis were used in the determination of Cryptosporidium species or genotypes (24, 25). Each DNA specimen was analyzed at least three times by PCR-RFLP with 0.5, 1.0, or 2.0 μl of DNA as the template. Positive (C. serpentis DNA) and negative (no template DNA) controls were included in each PCR run.
DNA sequencing and phylogenetic analysis.
At least two independent PCR products from each positive specimen were sequenced in both directions with an ABI 3100 Genetic Analyzer (Applied Biosystems, Foster City, Calif.). The sequences obtained were aligned with each other and those of known Cryptosporidium spp. with the ClustalX software (http://inn-prot.weizmann.ac.il/software/ClustalX.html). A neighbor-joining tree was generated with the TREECON software (http://www.psb.rug.ac.be/bioinformatics/psb/Userman/treeconw.html) on the basis of genetic distances calculated by the Kimura two-parameter model. The reliability of branches was assessed by bootstrap analysis with 1,000 replicates.
Nucleotide sequence accession numbers.
The unique partial SSU rRNA sequences generated in this study have been deposited in the GenBank database under accession no. AY504512 to AY504517.
RESULTS
Cryptosporidium spp. in feces of Canada geese.
Of the 209 fecal specimens examined in this study, 49 (23.4%) were positive for cryptosporidia by the SSU rRNA-based PCR assay used. The real Cryptosporidium prevalence rate in Canada geese could have been underestimated, because only 200 mg of fecal specimen was analyzed for each bird and no oocyst concentration was determined prior to DNA extraction. RFLP analysis of PCR products with SspI and VspI showed three banding patterns. Most of the specimens had only one RFLP banding pattern. Three samples, however, had mixed RFLP patterns, indicating the presence of more than one Cryptosporidium genotype. Altogether, the banding pattern of the previously identified Cryptosporidium goose genotype (24) was seen in 45 specimens, the pattern of C. parvum was seen in 5 specimens, and the pattern of C. hominis was seen in 2 specimens.
Because some Cryptosporidium spp. are known to have similar RFLP banding patterns, all PCR products were sequenced to verify the species or genotype identity. Two types of DNA sequences were obtained from the 45 specimens that showed the SspI and VspI banding pattern for the Cryptosporidium goose genotype; 36 (80%) specimens produced sequences similar or identical to the previously identified Cryptosporidium goose genotype (24), which is now renamed Cryptosporidium goose genotype I, and 9 (20%) specimens produced sequences for a new Cryptosporidium genotype, which is now named Cryptosporidium goose genotype II. Of the five specimens with the C. parvum banding pattern, four produced sequences identical or similar (with a 1- or 2-bp difference) to C. parvum, and one produced sequences identical to the previously identified Cryptosporidium duck genotype (13). All sequences obtained from the two specimens with the C. hominis banding pattern were identical to those of C. hominis.
Of these positive specimens, most (46 of 49 or 93.9%) had only one Cryptosporidium genotype. Three specimens, however, had two Cryptosporidium genotypes each: one specimen (no. 6846) had both goose genotype I and the duck genotype, one specimen (no. 6869) had both goose genotype I and C. parvum, and one specimen (no. 6888) had both C. parvum and C. hominis (Table 1 and Fig. 1).
TABLE 1.
Detection of cryptosporidia in fecal specimens from Canada geese
| State and site no. | No. of positive specimens/total (% positive) | No. of infections | Cryptosporidium genotype and/or spp. (no. positive) |
|---|---|---|---|
| Ohio | |||
| 1 | 2/12 (17) | 2 single | Goose genotype I (1) |
| Goose genotype II (1) | |||
| 2 | 3/17 (18) | 3 single | Goose genotype I (3) |
| 3 | 0/21 (0) | 0 | None |
| 4 | 0/18 (0) | 0 | None |
| 5 | 1/40 (3) | 1 single | Goose genotype I (1) |
| 6 | 15/25 (60) | 13 single | Goose genotype I (14) |
| 2 double | Duck genotype (1) | ||
| C. parvum (2) | |||
| 7 | 12/15 (80) | 11 single | Goose genotype I (7) |
| 1 double | Goose genotype II (2) | ||
| C. hominis (2) | |||
| C. parvum (2) | |||
| Illinois | |||
| 1 | 3/10 (30) | 3 single | Goose genotype I (2) |
| Goose genotype II (1) | |||
| 2 | 0/10 (0) | 0 | None |
| 3 | 3/7 (43) | 3 single | Goose genotype II (3) |
| 4 | 1/10 (10) | 1 single | Goose genotype I (1) |
| 5 | 2/10 (20) | 2 single | Goose genotype I (1) |
| Goose genotype II (1) | |||
| 6 | 7/14 (50) | 7 single | Goose genotype I (6) |
| Goose genotype II (1) | |||
| Total (13 sites) | 49/209 (24)a | 46 single 3 double | 5 Cryptosporidium species and genotypes |
Ten of 13 sites.
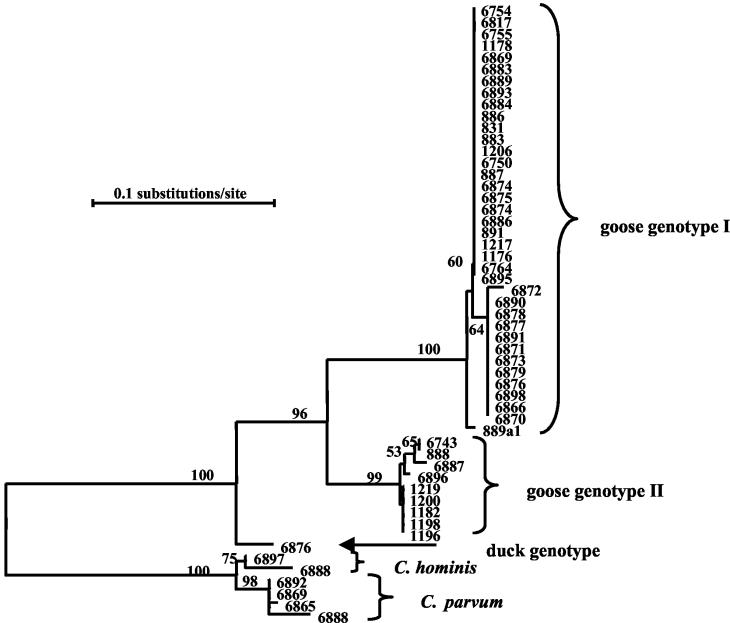
FIG. 1.
Phylogenetic relationships among Cryptosporidium species and genotypes from Canada geese based on SSU rRNA sequences. The Kimura two-parameter model was used for distance calculation. Numbers on branches are percent bootstrap values from 1,000 resamplings.
Distribution of Cryptosporidium spp.
Of the seven sites in Ohio investigated, five (71.4%) were positive for the Cryptosporidium goose genotypes and two of the five sites were also positive for C. parvum and C. hominis. One goose at a site in Ohio was also positive for the Cryptosporidium duck genotype. Of the six sites in Illinois, five (83.3%) were positive for cryptosporidia, all of which belonged to Cryptosporidium goose genotype I or II. The percentages of specimens positive for cryptosporidia ranged from 0 to 80% in Ohio and from 0 to 50% in Illinois (Table 1). Cryptosporidium goose genotype I was the most widely distributed, being detected at all five of the positive sites in Ohio and at four of five positive sites in Illinois, followed by Cryptosporidium goose genotype II, which was detected at four of five positive sites in Illinois and at two of five positive sites in Ohio.
Genetic relationships among Cryptosporidium spp. in Canada geese.
Among the five Cryptosporidium spp. and genotypes identified in Canada geese, the SSU rRNA gene of Cryptosporidium goose genotypes I and II and the duck genotype showed high sequence homology to each other, as reflected by direct sequence alignment, as well as genetic distance calculation. Nucleotide differences of the SSU rRNA gene among the three genotypes were 2.4 to 3.1%. In contrast, the differences between these genotypes and C. parvum or C. hominis were 4.5 to 7.1% (Table 2). As expected, the differences between C. parvum and C. hominis were very small (<1.0%). There were minor sequence variations (1 bp each) within both Cryptosporidium goose genotypes I and II.
TABLE 2.
Genetic distances among Cryptosporidium species and genotypes in Canada geese
| Genotype or species (specimen no.) | No. of nucleotide differences/100 bases from specimen no.:
|
||||
|---|---|---|---|---|---|
| 6754 | 888 | 6876 | 6888 | 6869 | |
| Goose genotype I (6754) | 0.00 | 2.94 | 3.13 | 7.05 | 6.86 |
| Goose genotype II (888) | 0.00 | 2.39 | 6.07 | 6.26 | |
| Duck genotype (6876) | 0.00 | 5.10 | 4.52 | ||
| C. hominis (6888) | 0.00 | 0.90 | |||
| C. parvum (6869) | 0.00 | ||||
Phylogenetic analysis confirmed the close relatedness among Cryptosporidium goose genotypes I and II and the duck genotype. These three parasites clustered together in a neighbor-joining tree constructed with sequences from all known Cryptosporidium spp. (24), with a sequence of Eimeria tenella (accession no. AF026388) as the outgroup, with 89% bootstrap support (data not shown). As expected, C. parvum and C. hominis were related and formed a cluster together. Among Cryptosporidium goose genotypes I and II and the duck genotype, the two goose genotypes were closely related, forming a group within the cluster, with high bootstrap support. There were two subdivisions in both the goose genotype I and II groups (Fig. 1).
DISCUSSION
The results of this study indicate that multiple Cryptosporidium species and genotypes are found in Canada geese. In addition to goose genotypes, other Cryptosporidium spp., such as the duck genotype, C. hominis, and C. parvum, were detected in the feces of Canada geese. Cryptosporidium goose genotype I had been previously detected (24), but Cryptosporidium goose genotype II had not been seen before and was detected in the feces of Canada geese for the first time. No other published study has identified the presence of more than one Cryptosporidium species or genotype in Canada geese. This study demonstrates that Canada geese can carry cryptosporidia of goose, duck, human, and ruminant origins.
Cryptosporidium goose genotypes I and II were the most common Cryptosporidium spp. in the fecal specimens studied; they had prevalence rates of 17.2 and 4.3%, were detected at 9 and 6 of the 13 study sites, respectively, and constituted 91.8% of the positive specimens. The high occurrence of these two Cryptosporidium spp. indicates that they are likely true parasites of Canada geese. Another, related Cryptosporidium sp., the duck genotype (13), was found in one goose together with goose genotype I. It remains to be determined whether this parasite is infectious to Canada geese. The two other species, C. parvum and C. hominis, were only found in five geese, reaffirming the previous conclusion that oocysts of these two species were merely passing through the digestive tracts of foraging Canada geese without establishing infection (5). When this paper was under revision, an SSU rRNA sequence characterization of eight Cryptosporidium-positive specimens in a recent study showed the presence of five Cryptosporidium genotypes in Canada geese in the United States (8). Three of the Cryptosporidium genotypes belonged to goose genotypes I (geese 1, 2, 3a, 6, and 8) and II (goose 9) and the duck genotype (goose 5) in the present study, whereas the remaining two genotypes (geese 3b and 7) represented new Cryptosporidium genotypes.
For both Ohio and Illinois, all of the sites within each state are within a 20-mile radius of one another, but three sites were negative for cryptosporidia, while two sites in Ohio were positive for C. hominis and C. parvum in addition to the two goose genotypes. All of the study sites are either used for recreational activities or close to institutional buildings. However, the two sites in Ohio positive for C. hominis and C. parvum were much more intensely used than the other sites and often were littered with garbage. It is plausible that C. hominis and C. parvum oocysts in a few geese were acquired locally from the environment contaminated by human activities, rather than mechanically carried oocysts picked up by Canada geese at distant locations.
C. hominis and C. parvum are the important pathogenic species in humans and are responsible for most of the cryptosporidiosis outbreaks worldwide (12, 18, 20). Humans and ruminants are the principal sources of these pathogens. Since sites used by Canada geese also serve as residential and/or recreational facilities, there is potential for human contact with the feces of this waterfowl. However, in this study few goose fecal specimens (5 of 209 or 2.4%) were positive for Cryptosporidium spp. that are pathogenic to humans, indicating that Canada geese might serve only as a minor source of infection in humans. Most of the specimens were positive for Cryptosporidium goose genotype I, which so far has not been seen in humans. Further research is required to determine whether similar distributions of Cryptosporidium species and genotypes also occur in Canada geese in other areas and settings.
Acknowledgments
This study was supported in part by an interagency agreement between the U.S. Environmental Protection Agency and the Centers for Disease Control and Prevention.
REFERENCES
- 1.Chalmers, R. M., K. Elwin, A. L. Thomas, and D. H. Joynson. 2002. Infection with unusual types of Cryptosporidium is not restricted to immunocompromised patients. J. Infect. Dis. 185:270-271. [DOI] [PubMed] [Google Scholar]
- 2.Dieter, R. A., Jr., R. S. Dieter, R. A. Dieter III, and G. Gulliver. 2001. Zoonotic diseases: health aspects of Canadian geese. Int. J. Circumpolar Health 60:676-684. [PubMed] [Google Scholar]
- 3.Dietz, V. J., and J. M. Roberts. 2000. National surveillance for infection with Cryptosporidium parvum, 1995-1998: what have we learned? Public Health Rep. 115:358-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Feare, C. J., M. F. Sanders, R. Blasco, and J. D. Bishop. 1999. Canada goose (Branta canadensis) droppings as a potential source of pathogenic bacteria. J. R. Soc. Health 119:146-155. [DOI] [PubMed] [Google Scholar]
- 5.Graczyk, T. K., M. R. Cranfield, R. Fayer, J. Trout, and H. J. Goodale. 1997. Infectivity of Cryptosporidium parvum oocysts is retained upon intestinal passage through a migratory water-fowl species (Canada goose, Branta canadensis). Trop. Med. Int. Health 2:341-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Graczyk, T. K., R. Fayer, J. M. Trout, E. J. Lewis, C. A. Farley, I. Sulaiman, and A. A. Lal. 1998. Giardia sp. cysts and infectious Cryptosporidium parvum oocysts in the feces of migratory Canada geese (Branta canadensis). Appl. Environ. Microbiol. 64:2736-2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guyot, K., A. Follet-Dumoulin, E. Lelievre, C. Sarfati, M. Rabodonirina, G. Nevez, J. C. Cailliez, D. Camus, and E. Dei-Cas. 2001. Molecular characterization of Cryptosporidium isolates obtained from humans in France. J. Clin. Microbiol. 39:3472-3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jellison, K. L., D. L. Distel, H. F. Hemond, and D. B. Schauer. 2004. Phylogenetic analysis of the hypervariable region of the 18S rRNA gene of Cryptosporidium oocysts in feces of Canada geese (Branta canadensis): evidence for five novel genotypes. Appl. Environ. Microbiol. 70:452-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kassa, H., B. J. Harrington, and M. S. Bisesi. 2004. Cryptosporidiosis: a brief literature review and update regarding Cryptosporidium in feces of Canada geese (Branta canadensis). J. Environ. Health 66:34-39. [PubMed] [Google Scholar]
- 10.Kassa, H., B. J. Harrington, and M. S. Bisesi. 2001. Risk of occupational exposure to Cryptosporidium, Giardia, and Campylobacter associated with the feces of giant Canada geese. Appl. Occup. Environ. Hyg. 16:905-909. [Google Scholar]
- 11.Kullas, H., M. Coles, J. Rhyan, and L. Clark. 2002. Prevalence of Escherichia coli serogroups and human virulence factors in faeces of urban Canada geese (Branta canadensis). Int. J. Environ. Health Res. 12:153-162. [DOI] [PubMed] [Google Scholar]
- 12.McLauchlin, J., C. Amar, S. Pedraza-Diaz, and G. L. Nichols. 2000. Molecular epidemiological analysis of Cryptosporidium spp. in the United Kingdom: results of genotyping Cryptosporidium spp. in 1,705 fecal samples from humans and 105 fecal samples from livestock animals. J. Clin. Microbiol. 38:3984-3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morgan, U. M., P. T. Monis, L. Xiao, J. Limor, I. Sulaiman, S. Raidal, P. O'Donoghue, R. Gasser, A. Murray, R. Fayer, B. L. Blagburn, A. A. Lal, and R. C. Thompson. 2001. Molecular and phylogenetic characterisation of Cryptosporidium from birds. Int. J. Parasitol. 31:289-296. [DOI] [PubMed] [Google Scholar]
- 14.Morgan-Ryan, U. M., A. Fall, L. A. Ward, N. Hijjawi, I. Sulaiman, R. Fayer, R. C. Thompson, M. Olson, A. Lal, and L. Xiao. 2002. Cryptosporidium hominis n. sp. (Apicomplexa: Cryptosporidiidae) from Homo sapiens. J. Eukaryot. Microbiol. 49:433-440. [DOI] [PubMed] [Google Scholar]
- 15.Ong, C. S., D. L. Eisler, A. Alikhani, V. W. Fung, J. Tomblin, W. R. Bowie, and J. L. Isaac-Renton. 2002. Novel Cryptosporidium genotypes in sporadic cryptosporidiosis cases: first report of human infections with a cervine genotype. Emerg. Infect. Dis. 8:263-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pedraza-Diaz, S., C. Amar, A. M. Iversen, P. J. Stanley, and J. McLauchlin. 2001. Unusual Cryptosporidium species recovered from human faeces: first description of Cryptosporidium felis and Cryptosporidium ‘dog type' from patients in England. J. Med. Microbiol. 50:293-296. [DOI] [PubMed] [Google Scholar]
- 17.Pedraza-Diaz, S., C. Amar, and J. McLauchlin. 2000. The identification and characterisation of an unusual genotype of Cryptosporidium from human faeces as Cryptosporidium meleagridis. FEMS Microbiol. Lett. 189:189-194. [DOI] [PubMed] [Google Scholar]
- 18.Peng, M. M., L. Xiao, A. R. Freeman, M. J. Arrowood, A. A. Escalante, A. C. Weltman, C. S. Ong, W. R. Mac Kenzie, A. A. Lal, and C. B. Beard. 1997. Genetic polymorphism among Cryptosporidium parvum isolates: evidence of two distinct human transmission cycles. Emerg. Infect. Dis. 3:567-573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pieniazek, N. J., F. J. Bornay-Llinares, S. B. Slemenda, A. J. da Silva, I. N. Moura, M. J. Arrowood, O. Ditrich, and D. G. Addiss. 1999. New Cryptosporidium genotypes in HIV-infected persons. Emerg. Infect. Dis. 5:444-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sulaiman, I. M., L. Xiao, C. Yang, L. Escalante, A. Moore, C. B. Beard, M. J. Arrowood, and A. A. Lal. 1998. Differentiating human from animal isolates of Cryptosporidium parvum. Emerg. Infect. Dis. 4:681-685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.U.S. Fish and Wildlife Service. 1999. Draft environmental impact statement: resident Canada goose management. U.S. Fish and Wildlife Service, Washington, D.C.
- 22.Xiao, L., C. Bern, M. Arrowood, I. Sulaiman, L. Zhou, V. Kawai, A. Vivar, A. A. Lal, and R. H. Gilman. 2002. Identification of the Cryptosporidium pig genotype in a human patient. J. Infect. Dis. 185:1846-1848. [DOI] [PubMed] [Google Scholar]
- 23.Xiao, L., C. Bern, J. Limor, I. Sulaiman, J. Roberts, W. Checkley, L. Cabrera, R. H. Gilman, and A. A. Lal. 2001. Identification of 5 types of Cryptosporidium parasites in children in Lima, Peru. J. Infect. Dis. 183:492-497. [DOI] [PubMed] [Google Scholar]
- 24.Xiao, L., I. M. Sulaiman, U. M. Ryan, L. Zhou, E. R. Atwill, M. L. Tischler, X. Zhang, R. Fayer, and A. A. Lal. 2002. Host adaptation and host-parasite co-evolution in Cryptosporidium: implications for taxonomy and public health. Int. J. Parasitol. 32:1773-1785. [DOI] [PubMed] [Google Scholar]
- 25.Xiao, L. H., U. M. Morgan, J. Limor, A. Escalante, M. Arrowood, W. Shulaw, R. C. A. Thompson, R. Fayer, and A. A. Lal. 1999. Genetic diversity within Cryptosporidium parvum and related Cryptosporidium species. Appl. Environ. Microbiol. 65:3386-3391. [DOI] [PMC free article] [PubMed] [Google Scholar]