Abstract
The worldwide epidemic of antibiotic resistance is in danger of ending the golden age of antibiotic therapy. Resistance impacts on all areas of medicine, and is making successful empirical therapy much more difficult to achieve. Staphylococcus aureus demonstrates a unique ability to quickly respond to each new antibiotic with the development of a resistance mechanism, starting with penicillin, until the most recent, linezolid and daptomycin. Methicillin resistant S. aureus (MRSA) has become endemic today in hospitals worldwide. Resistance to the newer antimicrobial-agents — linezolid, vancomycin, teicoplanin, and daptomycin are been reported and also the fear of pandrug-resistance. This study was carried out to know the antimicrobial resistant pattern of MRSA to newer antibiotic, to know any isolates are extensively-drug resistant (XDR)/pandrug resistant (PDR), inducible macrolide-lincosamide streptogramin B (iMLSB), and mupirocin resistance. Thirty-six MRSA isolates resistant to the routinely tested antibiotic were further tested for list of antibiotic by a group of international experts. Isolates were tested for iMLSB and mupirocin resistance by the disk diffusion method. Of 385 MRSA, 36 (9.35%) isolates of MRSA were resistant to the routinely tested antibiotic. Among these 36 MRSA isolates, none of our isolates were XDR/PDR or showed resistant to anti-MRSA cephalosporins (ceftaroline), phosphonic acids, glycopeptides, glycylcyclines, and fucidanes. Lower resistance was seen in oxazolidinones (2.78%), streptogramins (5.56%), lipopeptide (5.56%). Thirty-four (94.44%) isolates showed constitutive MLSB (cMLSB) resistance and two (5.56%) iMLSB phenotypes. High- and low-level mupirocin resistance were seen in 13 (36.11%) and six (16.67%), respectively. In our study, none of our isolates were XDR or PDR. No resistance was observed to ceftaroline, telavancin, teicoplanin, and vancomycin; but the presence of linezolid resistance (1, 2.28%) and daptomycin resistance (2, 5.56%) in our rural set-up is a cause of concern.
Keywords: Ceftaroline, Daptomycin, inducible clindamycin resistance, Linezolid, Mupirocin resistance, Newer antibiotic, PDR, Telavancin, XDR
MICROBIOLOGY REPORT
T he emergence of resistance to antibiotics in Gram-positive pathogens has become a major international problem as there are fewer, or even sometimes no, effective antimicrobial agents available for infections caused by these bacteria.[1] The problem of increasing antimicrobial resistance is even more threatening when considering the very limited number of new antimicrobial agents that are in development. As rapidly as new antibiotics are introduced, Staphylococci have developed efficient mechanisms to neutralize them; inevitably this has left fewer effective bactericidal antibiotics to treat these often life-threatening infections.
Multidrug-resistant (MDR) Staphylococci pose a growing problem for human health. The rise of drug-resistant virulent strains of Staphylococcus aureus, particularly methicillin-resistant S. aureus (MRSA) is a serious problem in the treatment and control of Staphylococcal infections.[2,3] Methicillin-resistant Staphylococci (MRS) cause hard-to-treat infections. The most striking situation is that MRSA strains have emerged with concomitant resistance to many commonly used antibiotics of groups, aminoglycosides, macrolides, fluoroquinolones, chloramphenicol, and tetracycline.[4]
A special rule has been applied in defining antimicrobial resistance in S. aureus. Once a S. aureus isolate is characterized as an MRSA, it is instantly classified as an MDR, because resistance to oxacillin or cefoxitin infers nonsusceptibility to all categories of β-lactam antimicrobials listed in this document (i.e., all categories of penicillins, cephalosporins, β-lactamase inhibitors, and carbapenems).[5] Thus, MDR-MRSA is the new or rather a continually evolving paradigmatic pathogen.[6]
MDR-MRSA strains, the silently violent incarnations of S. aureus widespread in community and hospital environments have posed serious clinical imbroglio.[4] MRSA strains are labeled as a “superbug” in the heath domain.[6]
First reported in 1960, the growing problem in the Indian scenario is that MRSA prevalence has increased from 12% in 1992 to 80.89% in 1999.[7] MRSA has become endemic today in hospitals covertly worldwide, and 30% of S. aureus isolates are MDR, 2 decades ago, as conjectured from surveillance in the US.[8]
At all the time, the full range of antibiotics is available but, an utterly sad fact today is that more than 95% MRSA worldwide do not respond to the first-line antibiotics, due to artifices in bacterial genomes confirming the law that simple genomes evolve faster than complex genomes.[9] The following antibacterial agents have been approved during the last 5 years: Quinupristin/dalfopristin, linezolid, daptomycin, and tigecycline. Novel lipoglycopeptide agents liketelavancin, novel cephalosporins (ceftaroline) with enhanced activity against MRSA are in the pipeline.
Several studies have reported the resistance to the newer antimicrobial agents like linezolid, vancomycin, teicoplanin, and daptomycin.[10,11,12] The fear of pandrug-resistance (resistance to all antibiotics and drugs in present use), as cautioned in Gram-negative pathogens,[13] cannot be ruled out in S. aureus and our return to a post-antibioticera. The true extent of antimicrobial resistance in MRSA in our area is unknown. Thus, this study was carried out to with the aim:
To know the antimicrobial resistant pattern of MRSA to newer antibiotic.
To know the number of isolates showing extensively-drug resistant (XDR)/pandrug resistant (PDR).
To find out the percentage of MRSA having constitutive (constitutive macrolide-lincosamide-streptogramin B (cMLSB) phenotype) and inducible (iMLSB) using D-test.
To find out the percentage of MRSA isolates having high- and low-level mupirocin resistance in MRSA by disk diffusion.
A prospective study was carried out in the Department of Microbiology during the period January 2012-August 2014. Approval was obtained from the Institutional Ethics Committee IEC/204, local review board for carrying out the study. A total of 335 nonduplicate MRSA isolated from various clinical specimens like pus, blood, urine, central venous catheters tips, tracheal aspirates, and sputum were randomly selected. MRSA isolates were identified by standard microbiological techniques. This isolates was tested for routine antibiotic by Kirby–Bauer disk diffusion method on Mueller–Hinton agar as per Clinical and Laboratory Standards Institute (CLSI). The antibiotics tested were ciprofloxacin, cefotaxime, gentamycin, oflaxacin, amoxicillin+clavulanic acid, amikacin, ampicillin, tobramycin, cefuroxime, and erythromycin.[4]
Sample size — Thirty-six MRS Aisolates resistant to the routinely tested antibiotic were further tested for list of antibiotic by a group of international experts: A joint initiative by the European Centre for Disease Prevention and Control (ECDC) and the Centers for Disease Control and Prevention (CDC) [Table 1].[5]
Table 1.
List of antibiotic by a group of international experts: Antimicrobial categories and agents[5]

A bacterial isolate was considered resistant, intermediate, or nonsusceptible to an antimicrobial agent by using interpretive criteria provided by European Committee on Antimicrobial Susceptibility Testing (EUCAST), the CLSI, and/or the Food and Drug Administration (FDA).[5]
The isolates were tested for inducible clindamycin resistance (iMLSB) and mupirocin resistance by the disk diffusion method.[4] Statistical analysis was done by using standard normal test (z-test). Antibiotic disk were obtained from HiMedia Laboratories Pvt Ltd, Mumbai, India; Oxoid Limited, UK; and Mast group, UK.
Out of 385 MRSA isolated from various clinical specimen, 36 (9.35%) isolates of MRSA were resistant to the routinely tested antibiotic. [Table 2]. The distribution of the MRSA (resistant to routinely tested antimicrobial agents) was higher among female (19, 52.78%) than males (17, 147.22%). Gender-wise no statistical significance was noted. Ward-wise higher distribution of MRSA (resistant to routinely tested antimicrobial agents) was observed in Obstetrics and Gynecology (Obgy; 12, 33.33%) and Surgery (11, 30.56%) followed by Medicine (7, 19.44%), Intensive Care Unit (ICU;4, 11.11%), and one each from Pediatrics (Ped;2.78%) and Skin (2.78%) [Figure 1].
Table 2.
Distribution of MRSA among the specimen
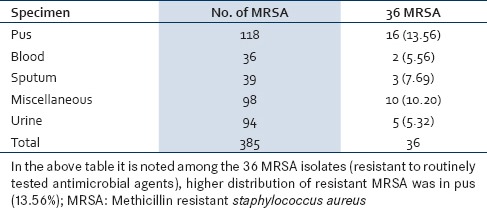
Figure 1.
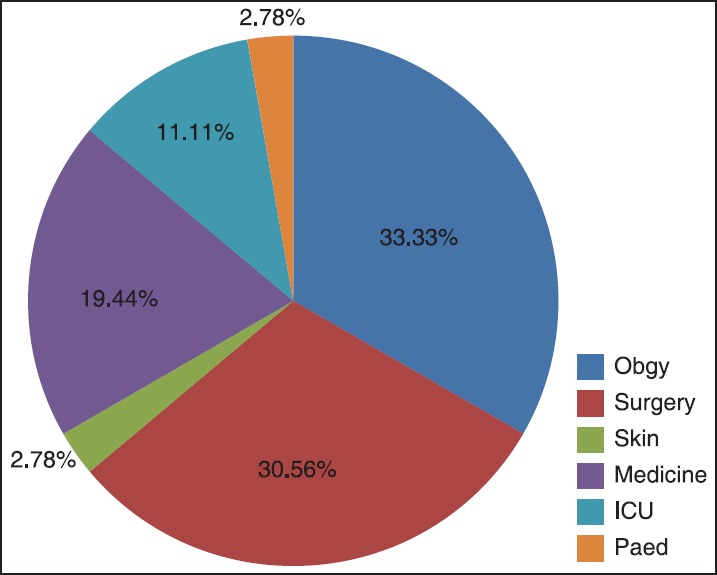
Ward-wise distribution of 36 MRSA. The above chart depicts higher distribution of MRSA isolates (resistant to routinely tested antimicrobial agents) in Obgy (33.33%) and Surgery (30.56%). MRSA: Methicillin resistant Staphylococcus aureus, ICU: intensive care unit, Obgy: Obstetrics and Gynecology, Ped: Pediatrics
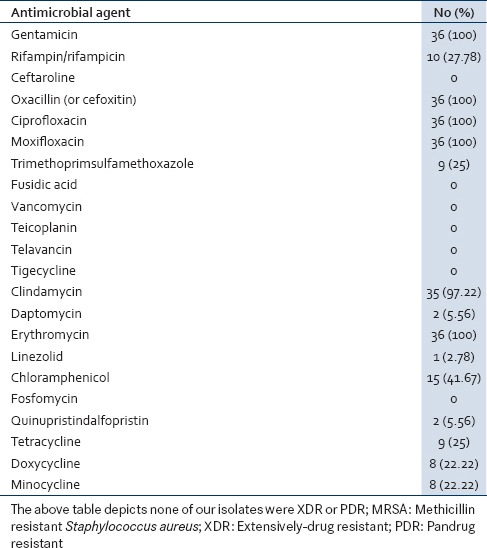
Among these 36 MRSA isolates, none of our isolates were XDR or PDR. Also none of the 36 isolates were resistant to antimicrobial category like anti-MRSA cephalosporins (ceftaroline), phosphonic acids, glycopeptides, glycylcyclines, and fucidanes. But 100% resistance was noted to fluoroquinolones, aminoglycosides, macrolides, and lincosamides showed 97.22% resistance. Lower resistance was seen in oxazolidinones (2.78%), streptogramins (5.56%), and lipopeptides (5.56%) [Table 3].
Table 3.
Antibiotic resistant pattern of 36 isolates of MRSA (resistant to the routinely tested antibiotic)
In our study, erythromycin resistance was noted to the tune of 100%. These isolates were subjected to D-test. We observed 34(94.44%) isolates who showed constitutive MLSB resistance and two (5.56%) iMLSB phenotypes. Of 36 MRSA isolates, low-level resistance in six (16.67%) and high-level mupirocin resistance was observed in 13 (36.11%).
Antibiotic resistance among pathogenic bacteria is a well-documented phenomenon that has severe consequences for the treatment of infections in the hospital setting and increasingly in the community. The Infectious Diseases Society of America recently published a hit list of bacterial pathogens whose antibiotic resistance severely impacts the ability to treat infections in the US hospital setting.[14]
In our study, out of 385 MRSA isolated from various clinical specimens, 36 (9.35%) isolates of MRSA were resistant to the routinely tested antibiotic. In our study, we analyzed the antimicrobial resistance pattern as per the classification of an international expert proposal for interim standard definitions for acquired resistance by Magiorakos et al., and found none of our isolates were XDR or PDR.[5]
Out of 36 MRSA isolates (resistant to routinely tested antimicrobial agents) none of the isolates were resistant to antimicrobial category like anti-MRSA cephalosporins (ceftaroline), phosphonic acids, glycopeptides, glycylcyclines, and fucidanes. Whereas, 100% resistance was noted to fluoroquinolones, aminoglycosides, macrolides, and lincosamides showed 97.22% resistance. Lower resistance was seen in oxazolidinones (2.78%), streptogramins (5.56%), and lipopeptides (5.56%).
S. aureus demonstrates a unique ability to quickly respond to each new antibiotic with the development of a resistance mechanism, starting with penicillin and methicillin, until the most recent, linezolid and daptomycin.[15]
DNA restriction enzyme polymorphism of t-6 ribosomal RNA gene is reported to be distributed throughout S. aureuschromosome, which has the methicillin resistant determinant (mec) as an episome (or even as a plasmid); the mecsequences could enter into the bacterial sequences at three points in the genome of S. aureus.[16] Unfortunately, the plasmid/episome confers rapid resistance to a lot of antibiotics of different classes/generations. Gene transfers through conjugation involving the transposon, “Tn-1546” with the gene mecA, encoding a modified penicillin binding protein confers resistance to the methicillin and other penicillin derivatives was reported. The mecA gene encodes the penicillin-binding protein PBP2a, which cannot be bound by β-lactam antibiotics and in turn prevents the disruption of cell wall formation by these antibiotics. This mec-gene is located on mobile genetic element called the, “staphylococcal cassette chromosome-mec” (SCC-mec).[17]
In fact, when an antibiotic binds to the protein that prevents the synthesis of peptidoglycan in the bacterial cell wall, the resistance is conferred. Some bacteria can produce a “modified penicillin binding protein” that ceases to bind to the antibiotic, which eventually prevents the targeted effects of the antibiotic.[18] Indeed, resistance of S. aureus to β-lactam antibiotics is attributed to the presence of the mecA gene.
Ceftaroline fosamil, the prodrug of the active metabolite, ceftaroline, is a new, broad-spectrum cephalosporin recently approved in the USA for the treatment of acute bacterial skin and skin structure infections (ABSSSIs) and community-acquired bacterial pneumonia (CABP). Ceftaroline has potent in vitro activity against Gram-positive organisms, including MRSA and Streptococcus pneumoniae, as well as common Gram-negative organisms.[19] In our study, we observed none of our isolates showed resistance to ceftaroline. Clark et al., in their study concluded, prolonged selection in the presence of ceftaroline demonstrated no evidence of resistance development for the majority of isolates and lack of cross-resistance with other antibiotic classes among tested species important in complicated skin and soft-structure infections (cSSSI) and community-acquired pneumonia (CAP).[20] While Laudano in his in vivo comparison of ceftaroline fosamil with daptomycin showed the greatest reduction in bacterial titers in vegetations after 4 days of treatment. Only ceftaroline demonstrated 100% sterilization of vegetations in all strains. Resistance to ceftaroline is expected to be limited, as demonstrated in multistep resistance selection studies.[19,21]
Vancomycin, teicoplanin, and daptomycin resistance pattern was also studied. We found none of our isolates were resistant to vancomycin and teicoplanin, but two isolates were found resistant to daptomycin. Susceptibility breakpoint for daptomycin was considered as <1 μg/ml for Staphylococci as recommended by the CLSI.[4]
Dubey in their study reported, out of 390 MRSA strains isolated from outpatient department (OPD), 80 (20.51%) were vancomycin resistant (vancomycin resistant S. aureus (VRSA)) and 173 (44.35%) strains were vancomycin intermediate S. aureus (VISA) and the rest 137 (35.12%) strains were sensitive to vancomycin. Similarly from nosocomial sources, out of 461 MRSA isolates, 110 (23.86%) strains were VRSA and 208 (45.11%) were VISA strains; while 143 (31.01%) strains were totally sensitive to vancomycin; whereas, out of 363 MRSA isolates obtained from ICU and NICU, 61 (16.8%) VRSA strains and 164 (45.17%) VISA strains were found; whereas, the rest 138 (38.01%) isolates were vancomycin sensitive.[6]
Biedenbach et al., in their study reported a 3.2% rate of tolerance to vancomycin and a 31.6% rate of tolerance to teicoplanin for 76 MRSA isolates from a collection of SENTRY Antimicrobial Surveillance Program strains collected from eight medical centers in the Asia-Pacific region.[22]
Reduced susceptibility to vancomycin has been reported to be associated with reduced susceptibility to daptomycin. Diederen et al., reported seven of the 17 VISA isolates to have daptomycin minimum inhibitory concentration (MIC) of 2 μg/ml and one isolate to have MIC 4 μg/ml. However, such association was not seen in our study.[12]
Maria et al., observed daptomycin were more potent in vitro than either vancomycin or teicoplanin against MRSA according to its MIC for 90% of isolates (MIC90) and was more bactericidal according to its minimum bactericidal concentration for 90% isolates (MBC90) and MBC/MIC ratios. A total of 6.1% (29/479) and 18.8% (90/479) of the strains tested exhibited tolerance to vancomycin and teicoplanin, while tolerance to daptomycin was not observed for any of the 479 isolates. Twenty-four (5%) of all strains were tolerant to both vancomycin and teicoplanin.[10]
Celikbilek et al., on the base of MIC90 values, in their study observed daptomycin was four to 16 times more effective than vancomycin, teicoplanin, and linezolid.[23]
In our study, we observed that one (2.78%) of our MRSA isolates was resistant to linezolid. Gu et al., systematically reviewed the published literature and observed <1% of S. aureus and 2% of coagulase-negative Staphylococcus (CoNS) are linezolid resistant. Nonetheless, multifocal outbreaks of linezolid-resistant Staphylococcus (LRS) have been reported and both vertical and horizontal transmission. The most common mechanisms for linezolid resistance were mutation (G2576T) to the 23S rRNA or by acquisition of a plasmid-borne ribosomal methyltransferase gene, cfr.[24] Linezolid resistance in Staphylococcus is defined by both the CLSI and the EUCAST as a linezolid MIC of ≥8 mg/L. The majority of LRS were isolated from patients in North America and Europe. Overall, 46.2% (30/65) of LRSA were reported in North America, 30.8% (20/65) in Europe, 20.0% (13/65) in Asia, and 3.1% (2/65) in South America. Linezolid susceptibility among clinically significant isolates is monitored through two surveillance programs, the global Zyvox Annual Appraisal of Potency and Spectrum (ZAAPS), and the USA Linezolid Experience and Accurate Determination of Resistance (LEADER).[24]
In a study by Kevin et al., stelavancin was consistently more active than vancomycin and teicoplanin against all organisms tested and showed potency equal to or greater than daptomycin and linezolid against all strain types except VanA-type vancomycin-resistant Enterococci (VRE).[11]
In our study, erythromycin resistance was noted to the tune of 100%. This isolates were subjected to D-test, where we found 34 (94.44%) isolates as cMLSB resistance and two (5.56%) iMLSB phenotypes. The resistance to macrolide can be mediated by msrAgene or via erm gene encoding for enzymes that confer inducible or constitutive resistance to macrolide, lincosamide, and type B streptogramin. Nagarajan et al., observed all MRSA ST239 isolates showed high level mupirocin resistance and inducible clindamycin resistance.[25]
Fokas et al., found 3.5% S. aureus isolates had inducible, 60% had constitutive MLS resistance.[26] Interestingly, in a study by Angel et al., there has been no constitutive MLSB resistance.[27]
Mupirocin (pseudomonic acid A) derived from Pseudomonas fluorescens is a topical antibiotic widely used for treatment of MRSA-associated skin and soft-tissue infections. Prolonged, widespread, or uncontrolled and multiple courses of mupirocin are all associated with the development of mupirocin resistance.
Of 36 MRSA isolates, low-level mupirocin resistance was noted in six (16.67%) and high-level mupirocin resistance in 13(36.11%). Nicholson et al., observed the higher prevalence of low- and high-level resistance to mupirocin to the tune of 30 and 24%, respectively.[28]
Low-level mupirocin resistance is usually associated with point mutations in the chromosomally encoded ileSgene; whereas, high-level resistance is generally due to a plasmid-mediated gene, mupA. The mupAgene is typically located on mobile genetic elements, these plasmids typically carry resistance determinants to other antimicrobial agents, including macrolides, gentamicin, tetracycline, and trimethoprim.[29] Suggesting that mupirocin use could select for increased drug resistance in S. aureus.
The emergence and spread of antibiotic resistance remains a global public health concern. A core function of all clinical laboratories is to determine the antibiotic susceptibility pattern of bacterial isolates. This would guide the clinicians in treating the patient infected with MRSA and also in formulation of a definite antibiotic policy; and in effective hospital infection control policy.
CONCLUSION
In our study, none of the isolates showed resistance to ceftaroline, telavancin, teicoplanin, and vancomycin; but the presence of linezolid (1,2.28%) and daptomycin resistance (2, 5.56%) in our rural set-up is cause of concern.
Higher resistance was noted to gentamicin, ciprofloxacin, moxifloxacin, clindamycin, and chloramphenicol. This could be selective antibiotic pressure. But to the relief, none of our isolates were XDR or PDR. To prevent the emergence of pandrug resistance isolates, we recommend:
MRSA strains may spread readily in hospitals from colonized or infected persons. Colonized employees are generally asymptomatic, although they are a potential reservoir of infections acquired by patients. Colonized or infected hospital personnel (healthcare workers) may serve as reservoir and disseminator of MRSA in hospitals. So screening for MRSA among healthcare workers and patients.
The selection of antimicrobial agent should be based on in vitro susceptibility and the hospital-based antibiotic policies must be strictly followed and constant surveillance of drug resistance for all bacterial pathogens is needed.
Curtail the large amount of unnecessary antibiotic use in many areas of life.
National surveillance of antibiotic resistance and antibiotic use. Setting up and/or strengthening infection control committees in hospitals.
Antimicrobial stewardship programs can be implemented to reduce inappropriate use of antimicrobials, thereby controlling the development of resistance.
Biomedical waste management: Genetic recombination mechanisms-conjugation and transformation occur more likely than expected in untreated hospital sewage system, because all sorts of bacteria with grading levels of antibiotic resistance are physically together, thereby inducing cell-to-cell contact or DNA intake from some lysed pathogenic strain.[30]
ACKNOWLEDGEMENTS
I am grateful to the Management of MIMER Medical College, Talegaon Dabhade, Pune for their support and encouragement. Thanks to Mrs. Hemwati Kaple for technical support.
Footnotes
Source of Support: Nil.
Conflict of Interest: None declared.
REFERENCES
- 1.Patel H, Shah A, Mistry M, Chanda S. In vitro antimicrobial susceptibility study in clinical isolates of Streptococci and Enterococci. Afr J Micro Res. 2011;5:1374–8. [Google Scholar]
- 2.Livermore DM. Antibiotic resistance in Staphylococci. Int J Antimicrob Agents. 2000;16:S3–10. doi: 10.1016/s0924-8579(00)00299-5. [DOI] [PubMed] [Google Scholar]
- 3.Zapun A, Contreras-Martel C, Vernet T. Penicillin-binding 2. Proteins and beta-lactam resistance. FEMS Microbiol Rev. 2008;32:361–85. doi: 10.1111/j.1574-6976.2007.00095.x. [DOI] [PubMed] [Google Scholar]
- 4.Wayne, PA, USA: Clinical and Laboratory Standards Institute (CLSI). M100-S21: Performance standards for antimicrobial susceptibility testing. 2012; 32. 21st Informational Supplement. [Google Scholar]
- 5.Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18:268–81. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 6.Debasmita D, Shakti R, Mahesh CS, Lolly P, Debata NK, Rabindra NP. Surveillance of infection status of drug resistant Staphylococcus aureus in an Indian teaching hospital. Asian Pac J Trop Dis. 2013;3:133–42. [Google Scholar]
- 7.Verma S, Joshi S, Chitnis V, Hemwani N, Chitnis D. Growing problem of methicillin resistant Staphylococci: Indian Scenario. Indian J Med Sci. 2000;54:535–40. [PubMed] [Google Scholar]
- 8.Elixhauser A, Steiner C. Rockville, MD: Agency for Health care Research and Quality; 2007. Infections with Methicillin-resistant Staphylococcus Aureus (MRSA) in U.S. Hospitals, 1993–2005 HCUP Statistical Brief #35 July. http://www.hcup-us.ahrq.gov/reports/statbriefs/sb35.pdf . [PubMed] [Google Scholar]
- 9.David MZ, Daum RS. Community-associated methicillin resistant Staphylococcus aureus: Epidemiology and clinical consequences of an emerging epidemic. Clin Microbiol Rev. 2010;23:616–87. doi: 10.1128/CMR.00081-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maria MT, Bradley DK, Judith NS, Steven DB. Inhibitory and bactericidal activities of daptomycin, vancomycin, and teicoplanin against methicillin-resistant Staphylococcus aureus isolates collected from 1985 to 2007. Antimicrob Agents Chemother. 2009;53:1735–8. doi: 10.1128/AAC.01022-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krause KM, Renelli M, Difuntorum S, Wu TX, Debabov DV, Benton BM. In Vitro activity of telavancin against resistant gram-positive bacteria. Antimicrob Agents Chemother. 2008;52:2647–52. doi: 10.1128/AAC.01398-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diederen BM, Duijn IV, Willemse P, Klutymans JW. In vitro activity of daptomycin against methicillin resistant Staphylococcus aureus including heterogeneously glycopeptide resistant strains. Antimicrob Agents Chemother. 2006;50:3189–91. doi: 10.1128/AAC.00526-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Falagas ME, Bliziotis IA. Pandrug-resistant Gram-negative bacteria: The dawn of the post antibiotic era? Int J Antimicrob Agent. 2007;29:630–6. doi: 10.1016/j.ijantimicag.2006.12.012. [DOI] [PubMed] [Google Scholar]
- 14.Boucher HW, Talbot GH, Bradley JS, Edwards JE, Gilbert D, Rice LB, et al. Bad bugs: No drugs: No ESKAPE! An update from the Infectious Diseases Society of America. Clin Infect Dis. 2009;48:1–12. doi: 10.1086/595011. [DOI] [PubMed] [Google Scholar]
- 15.Pantosti A, Sanchini A, Monaco M. Mechanisms of antibiotic resistance in Staphylococcus aureus. Future Microbiol. 2007;2:323–34. doi: 10.2217/17460913.2.3.323. [DOI] [PubMed] [Google Scholar]
- 16.Garrison MW, Nuemiller JJ. In vitro activity of tigecycline against quinolone-resistant Streptococcus pneumoniae, methicillin resistant Staphylococcus aureus and vancomycin-resistant Enterococci. Int J Antimicrob Agent. 2007;29:191–6. doi: 10.1016/j.ijantimicag.2006.08.048. [DOI] [PubMed] [Google Scholar]
- 17.Deurenberg RH, Vink C, Kalenic S, Friedrich AW, Bruggeman CA, Stobberingh EE. The molecular evolution of methicillin-resistant Staphylococcus aureus. Clin Microbiol Infect. 2007;13:222–35. doi: 10.1111/j.1469-0691.2006.01573.x. [DOI] [PubMed] [Google Scholar]
- 18.Slonczewski JL, Foster JW. Microbiology, an evolving science. 1st Edition. New York: WW Norton; 2009. pp. 1–1039. [Google Scholar]
- 19.Laudano JB. Ceftaroline fosamil: A new broad-spectrum cephalosporin. J Antimicrob Chemother. 2011;66(Suppl 3):iii11–8. doi: 10.1093/jac/dkr095. [DOI] [PubMed] [Google Scholar]
- 20.Clark C, McGhee P, Appelbaum PC, Kosowska-Shick K. Multistep resistance development studies of ceftaroline in gram-positive and -negative bacteria. Antimicrob Agents Chemother. 2011;55:2344–51. doi: 10.1128/AAC.01602-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clark C, McGhee P, Appelbaum PC, Kosowska-Shick K. Washington, DC, USA: American Society for Microbiology; Multistep resistance development studies of ceftaroline (CPT) with Streptococcus pneumoniae, Streptococcus pyogenes, staphylococci, and enterococci. In: Abstracts of the Fiftieth Interscience Conference on Antimicrobial Agents and Chemotherapy, Boston, MA, 2010. Poster #E-813. [Google Scholar]
- 22.Biedenbach DJ, Bell JM, Sader HS, Fritsche TR, Jones NR, Turnidge JD. Antimicrobial susceptibility of gram-positive bacterial isolates from the Asia-Pacific region and an in vitro evaluation of the bactericidal activity of daptomycin vancomycin and teicoplanin: A SENTRY program report 2003-2004) Int J Antimicrob Agents. 2007;30:143–9. doi: 10.1016/j.ijantimicag.2007.03.015. [DOI] [PubMed] [Google Scholar]
- 23.Celikbilek N, Ozdem B, Gurelik FC, Guvenman S, Guner HR, Acikgoz ZC. Invitro susceptibility pattern of methicillin-resistant Staphylococcus aureus to Vancomycin, teicoplanin, linezolide and daptomycin. Mikrobiyol Bul. 2011;45:512–8. [PubMed] [Google Scholar]
- 24.Gu B, Kelesidis T, Tsiodras S, Hindler J, Humphries RM. The emerging problem of linezolid- resistant Staphylococcus. J Antimicrob Chemother. 2012;68:4–11. doi: 10.1093/jac/dks354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nagarajan A, Saravanan M, Padma K. Emergence of Methicillin-Resistant Staphylococcus aureus ST239 with High Level Mupirocn and Inducible Clindamycin resistance in a Tertiary care centre in Chennai, South India. J Clin Microbiol. 2012;50:3412–3. doi: 10.1128/JCM.01663-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fokas S, Fokas S, Tsironi M, Kalkani M, Dionysopouloy M. Prevalence of inducible clindamycin resistance in macrolide-resistant Staphylococcus spp. Clin Microbiol Infect. 2005;11:337–40. doi: 10.1111/j.1469-0691.2005.01101.x. [DOI] [PubMed] [Google Scholar]
- 27.Angel MR, Balaji V, Prakash J, Brahmandathan KN, Mathews MS. Prevalence of inducible clindamycin resistance in gram positive organisms in a tertiary care centre. Indian J Med Microbiol. 2008;26:262–4. doi: 10.4103/0255-0857.42041. [DOI] [PubMed] [Google Scholar]
- 28.Nicholson AM, Thoms C, Wint H, Didier M, Willis R, McMorris N, et al. The detection of mupirocin resistance and the distribution of methicillin resistant Staphylococcus aureus at the University Hospital of the West Indies, Jamaica. West Indian Med J. 2010;59:509–13. [PubMed] [Google Scholar]
- 29.McDougal LK, Fosheim G, Patel JB. Washington, DC: Program and abstracts of the 48th Annual ICAAC/IDSA 46th Annual Meeting; 2008. Team ABCs. Emergence of resistance among USA 300 MRSA isolates causing invasive disease in the US [abstract C1-166] p. 103. [Google Scholar]
- 30.Perron GG, Lee AE, Wang Y, Huang WE, Barraclough TG. Bacterial recombination promotes the evolution of multi-drug-resistance in functionally diverse populations. Proc Biol Sci. 2012;279:1477–84. doi: 10.1098/rspb.2011.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]


