Abstract
Virus enumeration by epifluorescence microscopy (EFM) is routinely done on preserved, refrigerated samples. Concerns about obtaining accurate and reproducible estimates led us to examine procedures for counting viruses by EFM. Our results indicate that aldehyde fixation results in rapid decreases in viral abundance. By 1 h postfixation, the abundance dropped by 16.4% ± 5.2% (n = 6), and by 4 h, the abundance was 20 to 35% lower. The average loss rates for glutaraldehyde- and formaldehyde-fixed samples over the first 2 h were 0.12 and 0.13 h−1, respectively. By 16 days, viral abundance had decreased by 72% (standard deviation, 6%; n = 6). Aldehyde fixation of samples followed by storage at 4°C, for even a few hours, resulted in large underestimates of viral abundance. The viral loss rates were not constant, and in glutaraldehyde- and formaldehyde-fixed samples they decreased from 0.13 and 0.17 h−1 during the first hour to 0.01 h−1 between 24 and 48 h. Although decay rates changed over time, the abundance was predicted by using separate models to describe decay over the first 8 h and decay beyond 8 h. Accurate estimates of abundance were easily made with unfixed samples stained with Yo-Pro-1, SYBR Green I, or SYBR Gold, and slides could be stored at −20°C for at least 2 weeks or, for Yo-Pro-1, at least 1 year. If essential, samples can be fixed and flash frozen in liquid nitrogen upon collection and stored at −86°C. Determinations performed with fixed samples result in large underestimates of abundance unless slides are made immediately or samples are flash frozen. If protocols outlined in this paper are followed, EFM yields accurate estimates of viral abundance.
The realization that viruses are abundant in natural waters and have major effects on the mortality of heterotrophic and autotrophic microbial communities (reviewed in references 9, 20, 24, and 25) has provided the impetus to develop procedures to rapidly and accurately enumerate viral particles in cultures and natural samples. Originally, these efforts focused on using electron microscopy (1, 3, 4, 18), but this was soon superseded by more rapid and accurate methods based on epifluorescence microscopy (EFM) (11, 12, 21, 23) and, most recently, flow cytometry (5, 6, 13).
Enumeration of viruses by EFM and flow cytometry is based on using highly fluorescent nucleic acid dyes. In the initial methods the approach used for enumerating bacteria in natural samples (17) was modified by staining the virus particles with DAPI (4′,6-diamidino-2-phenylindole) and enumerating them on filters by EFM (11, 21, 23). Subsequently, the approach was modified by replacing DAPI with Yo-Pro-1 (12). Although Yo-Pro-1 has significant advantages over DAPI in that the fluorescence is more stable and the fluorescence yield and the binding coefficient for nucleic acids are higher, it has the disadvantages of relatively long staining times and interference by aldehyde-based fixatives. Hence, Yo-Pro-1-stained slides need to be made with freshly collected samples. Nonetheless, the accuracy and precision of viral counts obtained by EFM are considerably greater than the accuracy and precision of viral counts obtained by electron microscopy, and the procedure can be done in the field, making it a desirable approach for estimating viral abundance (14, 23).
In many instances it is necessary to preserve samples in the field and return them to the laboratory for analysis of viral abundance; consequently, efforts were made to modify the original Yo-Pro-1 method so that it could be used with fixed samples. One suggested approach was to use microwave radiation to allow rapid Yo-Pro-1 staining of fixed samples (26), while another modification was to replace Yo-Pro-1 with SYBR Green I (16) or SYBR Gold (7). Unfortunately, the introduction of the alternative dyes was not accompanied by data on the effect of fixation and storage of samples on estimates of viral abundance or by a comparison of the accuracy of the counts to the accuracy of the Yo-Pro-1 method counts. Subsequently, Bettarel et al. (2) compared the original Yo-Pro-1 method (12) with the modified methods involving microwave treatment (26) and replacement of Yo-Pro-1 by SYBR Green I (16). Their conclusion was that the SYBR Green I modification produced less stable fluorescence and yielded lower estimates of viral abundance but that the original Yo-Pro-1 and microwave-modified approaches produced estimates that were not significantly different.
In our laboratory, EFM is routinely used to estimate viral abundance in cultures and natural samples, and over the years we have counted thousands of samples. Although we typically employed the original Yo-Pro-1 method, we also used the modified methods. However, in our hands, the results were inconsistent. In particular, we experimented with the SYBR Green I modification and became suspicious that glutaraldehyde fixation as recommended in the protocol was affecting our results. In particular, we noticed decreases in viral abundance over time when viral isolates were preserved with aldehydes and stored at 4°C.
Aldehyde fixatives are routinely used to preserve natural samples, which are then typically stored at 4°C in the dark before slides are prepared for EFM. The length of time between fixation of samples and preparation of slides can vary from minutes to months or even years. Several studies have shown, however, that the abundance of bacteria decreases when the bacteria are fixed with glutaraldehyde, formaldehyde, or Lugol's iodine and stored at 4°C (10, 15, 22). Workers have also reported decreases in the estimates of viral abundance in aldehyde-preserved water (13, 14, 26, 27) and sediment (8) samples, although the data are quite limited. A recent study of viral isolates and natural marine samples by flow cytometry indicated that storage of aldehyde-fixed samples decreased by as much as 90% in just a few hours (5). This finding led to the suggestion that viruses used for enumeration by flow cytometry should be fixed with 0.5% glutaraldehyde for 15 to 30 min and then flash frozen in liquid nitrogen before they are stored at −80°C.
In this study, our goal was to firmly establish protocols that allow accurate estimation of viral abundance in natural waters by EFM. Specifically, we wished to determine whether aldehyde fixation of samples was responsible for inconsistent estimates of viral abundance in SYBR Green I-stained samples and, if so, what the magnitude of the error was over time. Second, we wished to establish a satisfactory protocol for the preservation of field samples so that accurate estimates of viral abundance can be obtained by EFM.
MATERIALS AND METHODS
Sample collection.
Plastic bottles, rinsed with water from the collection sites, were used to collect water samples from the following locations around Vancouver, British Columbia, Canada: Jericho Beach Pier (49°11′N, 123°10′W), the north arm of the Fraser River (49°12.3′N, 123°03.1′W), and a depth of 50 m in Burrard Inlet (49°18.95′N, 123°8.35′W) at the water intake system of the Vancouver Aquarium. Samples were brought back to the lab and processed immediately for slide preparation, fixation, and storage experiments.
EFM.
For all experiments viral abundance was determined by EFM by using an Olympus AX70 epifluorescence microscope with a wide-blue filter set (excitation at 450 to 480 nm, 515-nm cutoff) and a UPlanApo ×100 objective. The water samples were collected and either processed immediately or fixed with 0.5% glutaraldehyde (electron microscopy [EM] grade) or with filtered (pore size, 0.1 μm) 2.0% formaldehyde. Subsamples were either filtered directly or diluted with filtered (pore size, 0.02 μm) MilliQ water to reduce the number of viruses per field when preparations were observed under the microscope for ease of counting. Slides were made by filtering 800 μl of whole or diluted samples, either fixed or unfixed, through 0.02-μm-pore-size AlO3 filters (Anodisc; Whatman). The filters were immediately stained with SYBR Green I, SYBR Gold, or Yo-Pro-1 (Molecular Probes). SYBR Green I slides were prepared by using a modified method of Noble and Fuhrman (16). The filters were stained sample side up for a minimum of 15 min in the dark with diluted SYBR Green I (a 4 × 10−2 dilution in filtered [pore size, 0.02 μm] MilliQ water) in a plastic petri dish. Each filter was then mounted on a glass slide with 0.1% p-phenylenediamine (made freshly from a frozen 10% aqueous stock; Sigma Chemical Co.) in 50% glycerol-50% phosphate-buffered saline (0.05 M Na2HPO4, 0.85% NaCl; pH 7.5). The SYBR Gold slides were prepared by using the same method that was used for the SYBR Green I slides (7). Yo-Pro-1 slides were prepared by the method of Hennes and Suttle (12) for unfixed samples only. The filters were stained sample side up for 48 h in 100 μl of 50 mM Yo-Pro-1 in a 2 mM NaCN solution in a petri dish. The filters were then rinsed twice, with filtration, with 800 μl of filtered (pore size, 0.02 μm) MilliQ water and mounted onto glass slides with glycerol. The slides were either frozen or counted immediately by EFM. For frozen-storage experiments, the slides were stored at −20°C in a frost-free freezer until they were counted. In all cases, a minimum of 200 virus-like particles in at least 20 random fields were counted (19).
Preliminary experiments.
A series of preliminary experiments were conducted to determine if the effects of aldehyde preservation were dependent on the concentration of glutaraldehyde used for fixation. Duplicate samples were fixed in 0.5, 2, and 4% glutaraldehyde and stored at 4°C. Slides were prepared after 30 min and 1 and 2 days and then every 2 days until 12 days. Slides were counted immediately following preparation.
Glutaraldehyde versus formaldehyde.
To examine differences in preservation between glutaraldehyde and formaldehyde, a seawater sample collected from Jericho Beach was divided into two sets of triplicate subsamples. One set of subsamples was fixed in 0.5% glutaraldehyde (EM grade), and the other set was fixed in filtered 2.0% formaldehyde before the subsamples were incubated at 4°C in the dark for 30 min. Eight slides were then prepared from each subsample by using SYBR Green I as described above. One slide from each set was counted immediately, while the remaining slides were kept in a frost-free freezer at −20°C. On days 1, 2, 4, 7, 9, 13, and 16, one slide was prepared from each of the subsamples, which remained at 4°C in the dark, and immediately counted. One previously prepared frozen slide from each set was counted on days 1, 2, 7, 13 and 16 to compare the effects of storage of fixed samples at 4°C with the effects of storage of frozen slides on estimates of viral abundance. To examine short-term changes in viral abundance in fixed samples, this experiment was repeated, and slides for viral enumeration were prepared at 0, 1, 2, 4, 7, 12, 25, and 48 h.
Comparison of nucleic acid stains and storage methods.
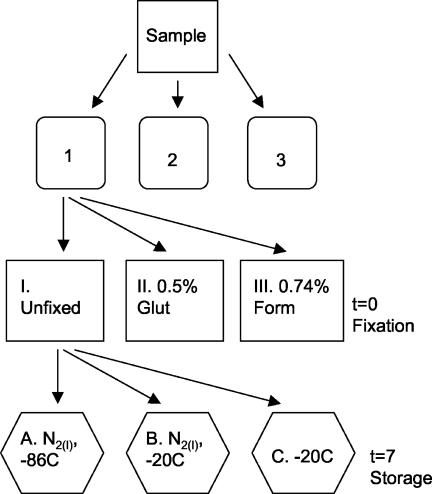
Water samples were collected and divided into three sets of triplicate subsamples (designated sets I, II, and III) (Fig. 1). One set (set I) was left unfixed; the other two sets were fixed in 0.5% EM-grade glutaraldehyde (set II) or 0.74% filtered (pore size, 0.1 μm) formaldehyde (2% formalin) (set III) and stored at 4°C in the dark for 30 min. Immediately after fixation a slide was prepared from each triplicate subsample and stained with SYBR Green I or SYBR Gold. One Yo-Pro-1-stained slide was also made with each subsample of the unfixed triplicate subsamples. All the slides were counted on the same day by EFM. At least 400 virus-like particles were counted in 48 fields for this experiment. While the slides were being made, three 5-ml subsamples (subsamples A, B, and C) were taken from each treatment. Subsample A was flash frozen in liquid nitrogen and stored at −86°C by following the recommended storage procedures for flow cytometry (6), Subsample B was flash frozen and stored at −20°C in a non-frost-free freezer, and subsample C was frozen at −20°C. After 7 days, the subsamples were thawed in a 37°C water bath, and slides stained with SYBR Green I or SYBR Gold were made. At the same time, Yo-Pro-1-stained slides were made from the unfixed set of subsamples. The slides were kept frozen at −20°C in a frost-free freezer and counted within 7 days (Fig. 1). Samples were also stored at 4°C in the dark and sampled after 7 days.
FIG. 1.
Flow chart of comparison of nucleic acid stains and storage experiment. Natural water samples were collected in plastic bottles that were rinsed with water from collection sites. The samples were dispensed into three sets of triplicate containers and either left untreated (set I) or fixed with 0.5% glutaraldehyde (set II) or 0.74% formaldehyde (set III). Each of the triplicate subsamples was split into three treatments and either flash frozen in liquid nitrogen [N2(l)] and stored at −86°C (subsample A) or −20°C (subsample B) or stored directly at −20°C (subsample C). The remaining samples from sets II and III were stored at 4°C. Slides were prepared for enumeration of viral abundance at time zero (t=0) and 7 days (t=7). Glut, glutaraldehyde; Form, formaldehyde.
Analysis of decay.
For both types of fixative, linear, quadratic, and exponential models were fitted to the short- and long-term viral decay data from the Jericho Beach Pier samples by using Sigmaplot (SPSS) or JMP IN (SAS Institute). The models were evaluated based on their r2 values. The predictive ability of the models was tested against the data from the Fraser River and Burrard Inlet samples after 7 days of fixation.
RESULTS AND DISCUSSION
Concerns about the accuracy of our estimates of viral abundance led us to conduct a series of experiments to examine the effects of aldehyde fixation and other preservation methods on the counts of viral particles obtained by EFM. Our goal was to establish protocols that yield accurate estimates of viral abundance in natural water samples. These experiments demonstrated that aldehyde fixation of water samples results in a rapid decrease in the number of viral particles observable by EFM. In order to obtain accurate estimates, slides must be made immediately and kept frozen at −20°C, or fixed samples must be flash frozen in liquid nitrogen and stored at −86°C. Consequently, determinations of viral abundance made with fixed samples should be considered large underestimates unless stained slides were made immediately after fixation or the samples were flash frozen in liquid nitrogen and stored at −86°C. These results and their implications are presented in detail below.
Effect of aldehyde fixation and storage at 4°C.
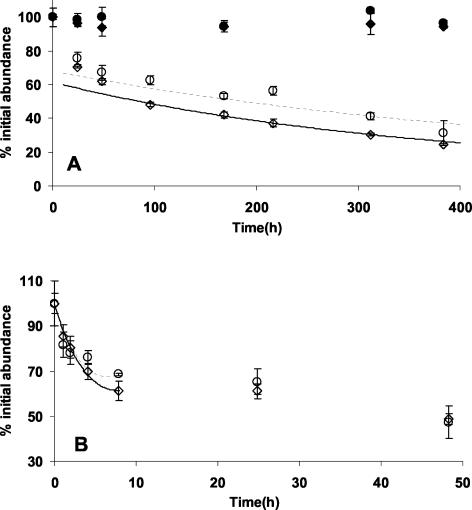
One of the major benefits reported for the use of SYBR Green I was its compatibility with fixatives (16). However, in both 16-day and 48-h experiments viral abundance decreased dramatically and rapidly in glutaraldehyde- or formaldehyde-fixed natural water samples stored at 4°C (Fig. 2). By 1 h postfixation, the abundance had decreased an average of 16.4% ± 5.2% (n = 6), and by 4 h, the viral abundance was 20 to 35% lower than that at zero time (Fig. 2B). Different concentrations of fixative did not result in significant differences in decay rates (P < 0.05, as determined by the slopes test; n = 41) (data not shown). The average rates of decay for glutaraldehyde- and formaldehyde-fixed samples over the first 2 h were 0.12 and 0.13 h−1, respectively. The rate of decay decreased markedly with time and was almost 1 order of magnitude less after 8 h than during the period immediately following fixation. By 16 days the average viral abundance had decreased by 72.2% (standard deviation, 6%; n = 6) (Fig. 2A). The amounts of the decrease were not significantly different between the fixatives for the 48-h and 16-day experiments (P = 0.163 and P = 0.865, respectively, as determined by the paired t test). These results indicate that aldehyde fixation of samples followed by storage at 4°C for even a few hours results in large underestimates of viral abundance by EFM. This may be one of the reasons that the Yo-Pro-1 method was found to yield estimates of viral abundance that were 1.2- to 7.1-fold (average, 3.0-fold; n = 12) higher than estimates based on transmission electron microscopy (12), whereas the estimates obtained by using SYBR Green I averaged only 1.28-fold higher than transmission electron microscopy-based counts (16). It is possible that microwave radiation may reduce the decay (26), although we did not test this in our study; moreover, Xenopoulos and Bird caution that even when microwave radiation is used, the samples must be processed within a few days.
FIG. 2.
Percentages of the initial virus abundance for 0.5% glutaraldehyde-fixed (⋄) and 2.0% formaldehyde-fixed (○) samples collected from Jericho Beach Pier and stored at 4°C. Subsamples were collected over time, and the viruses were enumerated by using SYBR Green I staining. The percentages of the initial virus abundance estimates are shown over time for slides prepared immediately and frozen at −20°C for 0.5% glutaraldehyde-fixed (⧫) and 2.0% formaldehyde-fixed (•) samples. Models of decay for glutaraldehyde- and formaldehyde-fixed samples are indicated by solid and dashed lines, respectively (see text for equations). (A) Long-term experiment conducted over 16 days. (B) Short-term experiment conducted over 48 h. The error bars indicate standard deviations for triplicate samples.
The loss of bacteria from aldehyde-fixed samples has been well documented (10, 22), and losses of up to 50% of bacterial cells within 1 month of fixation have been reported (10). There have also been reports that viral abundance declines in aldehyde-preserved samples. Marie et al. (13), using flow cytometry, observed that the number of viruses infecting the microalga Phaeocystis pouchetii decreased in samples preserved in glutaraldehyde or paraformaldehyde and stored at 4°C. Also, Danovaro et al. (8) found that the abundance of viruses in extracted sediment samples declined by more than 20% after only 24 h of storage and by 30 to 40% after 7 days of storage. Brussaard (5) found that the abundance of virus isolates fixed with glutaraldehyde decreased within a few hours. Different isolates decayed at different rates, but by 14 days following fixation the decreases were as great as 80%.
In our experiments the loss of viruses over time was not well described by a single rate constant. For example, the rate of loss of viruses in glutaraldehyde- and formaldehyde-fixed samples decreased from 0.13 and 0.17 h−1 during the first hour to ca. 0.01 h−1 between 24 and 48 h. This suggests either that a combination of mechanisms contributed to the decrease in viral abundance in fixed samples or that specific viruses decay at different rates.
Correcting for decay in fixed samples.
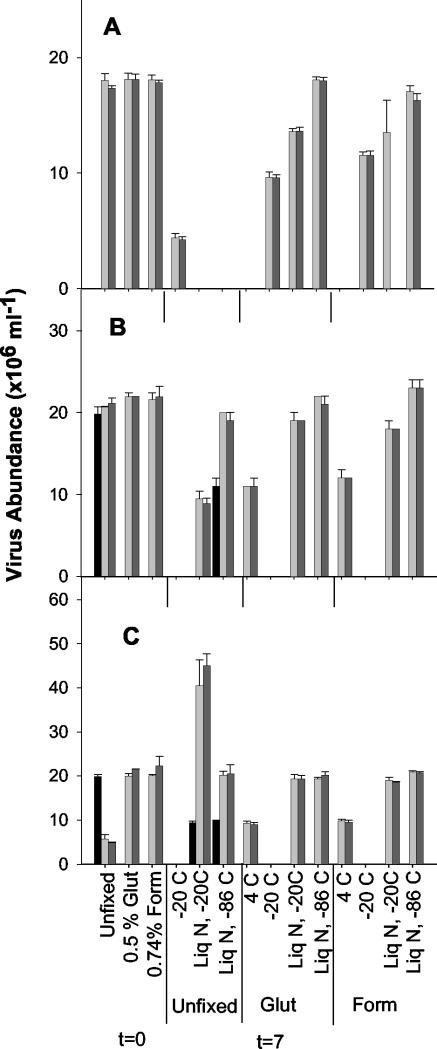
Although the rates of loss of the viruses changed with storage time, the rates were similar for viral communities collected from different environments. For example, 7 days after fixation ca. 40 to 60% of the viruses remained in refrigerated samples from the Jericho Beach Pier (Fig. 2A), the Fraser River (Fig. 3B), and Burrard Inlet (Fig. 3C). Given that the viral communities were likely different in the different habitats, this finding suggests that the rate constants for decay in aldehyde-fixed samples, although variable over time, might be quite robust among samples. This is consistent with previous findings (22) which showed that decreases in bacterial cell numbers in glutaraldehyde-preserved seawater samples are predictable.
FIG. 3.
Virus abundance estimates for slides prepared at time zero (t=0) from unfixed, 0.5% glutaraldehyde-fixed, and 0.74% formaldehyde-fixed samples. Estimates of abundance are also shown for day 7 (t=7) for samples fixed for 30 min or left unfixed and stored at −20°C or flash frozen in liquid nitrogen and stored at −20 or −86°C. Fixed samples were also stored at 4°C. Light grey bars, SYBR Green I-stained slides; dark gray bars, SYBR Gold-stained slides; black bars, Yo-Pro-1-stained slides. Where data are not shown, the treatment was not performed with the sample. The error bars indicate standard deviations for triplicate subsamples. (A) Jericho Beach Pier; (B) Fraser River; (C) Burrard Inlet. Glut, glutaraldehyde; Form, formaldehyde; Liq N, liquid nitrogen.
Although viral decay in the fixed samples was not described by a single rate constant, changes in viral abundance over time were approximated by using models calculated for periods less than and greater than 8 h (Fig. 2). The models were based on data from several experiments conducted with water samples from Jericho Beach Pier. Over the first 8 h, viral decay in glutaraldehyde- and formaldehyde-fixed samples was approximated by the following equations: ln(percent remaining) = 0.0082h2 − 0.1261h + 4.6017 (r2 = 0.90, n = 17) and ln(percent remaining) = 0.0101h2 − 0.1252h + 4.6017 (r2 = 0.83, n = 18), respectively. After the first 8 h, the decay of viruses was estimated by using exponential models. For glutaraldehyde-fixed samples the decay was approximated by ln(percent remaining) = 4.118e−0.0006 h (r2 = 0.93, n = 27), and for formaldehyde-fixed samples the decay was approximated by ln(percent remaining) = 4.216e−0.0004 h (r2 = 0.67, n = 30). We tested these models against independent decay data collected for samples from the Fraser River and Burrard Inlet that were fixed with either 0.5% glutaraldehyde or 0.74% formaldehyde and stored at 4°C. When the model for glutaraldehyde-fixed viral decay was applied to virus abundance data collected at 168 h, the percentages of viruses remaining were underestimated, resulting in overestimation of the initial viral abundance by 23 and 8.0% for the Fraser River and Burrard Inlet samples, respectively. When the model was applied to formaldehyde-fixed samples, the percentage of viruses remaining for the Fraser River sample was underestimated, which resulted in a 9% overestimate of the initial viral abundance, while the percentage of the viruses remaining for the Burrard Inlet sample was overestimated, which resulted in a 10% underestimate of the initial viral abundance. Consequently, these models can be used to approximate initial viral abundance in samples stored for relatively short periods of time. Beyond 1 or 2 weeks of storage, extrapolation to initial abundance would likely yield increasingly inaccurate results given that 25% or less of the initial viral abundance would remain. Caution should be used when these models are applied to viral abundance in stored samples, as over- or underestimates of as much as 20% could affect conclusions regarding the environment studied.
There are several plausible explanations for the decreases in viral abundance in stored fixed samples. Proteases may break down viral proteins, as has been suggested for bacteria in glutaraldehyde-fixed samples (10). Alternatively, aldehydes may cross-link the viral proteins and interfere with stain penetration. Finally, viruses may attach to each other, bacterial cells, or other particulates, and this may reduce the number of free viruses. Regardless of the mechanism, the viruses can no longer be distinguished as individual particles by EFM or flow cytometry (5, 13).
The high rate of loss of viruses in aldehyde-fixed samples is likely the reason that we obtained variable results and other workers have reported lower estimates of viral abundance when the protocol for SYBR Green I staining (16) was used. For example, Bettarel et al. (2) indicated that the estimates of viral abundance made by using the original Yo-Pro-1 method (12) were invariably and significantly higher than those obtained by using SYBR Green I. However, their slides were made from samples fixed in 0.74% formaldehyde and stored for up to 1 week.
Processing of unfixed samples and storage of slides.
We found no significant difference (P > 0.01) between estimates of viral abundance made with fixed and estimates of viral abundance made with unfixed samples, provided that the slides were made immediately. However, it was not possible to use SYBR Green I to stain all unfixed samples (e.g., Burrard Inlet samples) due to occasional sample-dependent high background fluorescence. Counts from time zero for natural samples from the three locations analyzed by using analysis of variance showed no significant difference among stains (P > 0.01), demonstrating that accurate estimates of viral abundance can be made with aldehyde-fixed samples, if the slides are made immediately.
There was no decline in viral abundance during the 16-day experiment on Yo-Pro-1-or SYBR Green I-stained slides stored at −20°C. This finding agrees with studies showing that Yo-Pro-1-stained slides can be stored at −20°C for at least 4 months (12) and with our unpublished data showing that accurate estimates of viral abundance can be made with Yo-Pro-1-stained slides stored for 1 year. Hence, slides do not need to be processed immediately after preparation.
Alternative preservation methods.
As storage of aldehyde-fixed samples at 4°C resulted in rapid decay of viral particles, we explored freezing at −20°C, flash freezing in liquid nitrogen and storage at −20°C, and flash freezing and storage at −86°C as alternative methods to preserve unfixed and fixed samples. Of the three approaches, only flash freezing followed by storage at −86°C produced acceptable results. This is also the protocol that has been recommended for storage of samples for viral enumeration by flow cytometry (6, 13). This method of storage has been found to provide reliable estimates of abundance by flow cytometry for up to 6 months of storage (6). Viral abundance decreased in samples that were flash frozen and stored at −20°C or were stored directly at −20°C (Fig. 3). An exception was unfixed samples from Burrard Inlet that were flash frozen and stored at −20°C, for which the estimates of viral abundance were about double those at time zero (Fig. 3C). The reason for this difference is unknown. Estimates of viral abundance determined by using SYBR Green I or SYBR Gold staining did not decrease for unfixed or 0.5% glutaraldehyde-fixed samples that were flash frozen in liquid nitrogen and stored at −86°C. The viral abundance for samples from Jericho Beach and the Fraser River that were fixed in 0.74% formaldehyde, flash frozen, and stored at −86°C was significantly different from the viral abundance for unfixed and 0.5% glutaraldehyde-fixed samples that were stored in the same way (P < 0.01). This finding is similar to the findings of experiments performed with viral isolates in which the flow cytometric counts decreased for formaldehyde-fixed samples (5). Flash-frozen unfixed samples showed lower abundance when they were stained with Yo-Pro-1 than when they were stained with SYBR Green I or SYBR Gold.
Recommendations.
Whenever possible, slides should be prepared immediately with either unfixed or fixed samples and frozen at −20°C until they are processed, because viral abundance declines rapidly in fixed samples stored at 4°C. Slides made immediately with Yo-Pro-1, SYBR Green I, and SYBR Gold yielded estimates of viral abundance that were indistinguishable from each other. Nonetheless, the suitability of the staining procedures should be checked for the type of samples being analyzed because not all stains were suitable for all samples. If immediate slide preparation is impractical, flash freezing 0.5% glutaraldehyde-fixed or unfixed water samples in liquid nitrogen, followed by storage at −86°C, allows accurate estimates of viral abundance to be made by using SYBR Green I or SYBR Gold staining. Consequently, determinations of viral abundance made with fixed samples should be considered large underestimates unless stained slides were made immediately after fixation or the samples were flash frozen in liquid nitrogen and stored at −86°C. If the protocols outlined above are carefully followed, EFM provides practical and accurate estimates of viral abundance in natural samples.
Acknowledgments
The Vancouver Aquarium water sample was provided by Skip Young of the Vancouver Marine Science Center. The assistance of Howard Chang with statistical calculations is greatly appreciated. Comments of the anonymous reviewers and Steven Wilhelm were greatly appreciated.
This study was supported by a grant from the Natural Science and Engineering Research Council of Canada to C.A.S.
REFERENCES
- 1.Bergh, O., K. Y. Borsheim, G. Bratbak, and M. Heldal. 1989. High abundance of viruses found in aquatic environments. Nature 340:467-468. [DOI] [PubMed] [Google Scholar]
- 2.Bettarel, Y., T. Sime-Ngando, C. Amblard, and H. Laveran. 2000. A comparison of methods for counting viruses in aquatic systems. Appl. Environ. Microbiol. 66:2283-2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borsheim, K. Y., G. Bratbak, and M. Heldal. 1990. Enumeration and biomass estimation of planktonic bacteria and viruses by transmission electron microscopy. Appl. Environ. Microbiol. 56:352-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bratbak, G., and M. Heldal. 1993. Total counts of viruses in aquatic environments, p. 135-142. In P. F. Kemp, B. F. Sherr, E. B. Sherr, and J. J. Cole (ed.), Handbook of methods in aquatic microbial ecology. Lewis, Boca Raton, Fla.
- 5.Brussaard, C. P. D. 2004. Optimization of procedures for counting viruses by flow cytometry. Appl. Environ. Microbiol. 70:1506-1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brussaard, C. P. D., M. Dominique, and G. Bratbak. 2000. Flow cytometric detection of viruses. J. Virol. Methods 85:175-182. [DOI] [PubMed] [Google Scholar]
- 7.Chen, F., J. R. Lu, B. J. Binder, Y. C. Liu, and R. E. Hodson. 2001. Application of digital image analysis and flow cytometry to enumerate marine viruses stained with SYBR Gold. Appl. Environ. Microbiol. 67:539-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Danovaro, R., A. Dell'anno, A. Trucco, M. Seresi, and S. Vanuci. 2001. Determination of virus abundance in marine sediments. Appl. Environ. Microbiol. 67:1384-1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fuhrman, J. A. 1999. Marine viruses and their biogeochemical and ecological effects. Nature 399:541-548. [DOI] [PubMed] [Google Scholar]
- 10.Gundersen, K., G. Bratbak, and M. Heldal. 1996. Factors influencing the loss of bacteria in preserved seawater samples. Mar. Ecol. Prog. Ser. 137:305-310. [Google Scholar]
- 11.Hara, S., K. Terauchi, and I. Koike. 1991. Abundance of viruses in marine waters: assessment by epifluorescence and transmission electron microscopy. Appl. Environ. Microbiol. 57:2731-2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hennes, K. P., and C. A. Suttle. 1995. Direct counts of viruses in natural waters and laboratory cultures by epifluorescence microscopy. Limnol. Oceanogr. 40:1050-1055. [Google Scholar]
- 13.Marie, D., C. P. D. Brussard, R. Thyrhaug, G. Bratbak, and D. Vaulot. 1999. Enumeration of marine viruses in culture and natural samples by flow cytometry. Appl. Environ. Microbiol. 65:45-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Middelboe, M., T. G. Nielsen, and P. K. Bjornsen. 2002. Viral and bacterial production in the North Water: in situ measurements, batch-culture experiments and characterization and distribution of a virus-host system. Deep-Sea Res. 49:5063-5079. [Google Scholar]
- 15.Nishino, S. F. 1986. Direct acridine orange counting of bacteria preserved with acidified Lugol iodine. Appl. Environ. Microbiol. 52:602-604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Noble, R. T., and J. A. Fuhrman. 1998. Use of SYBR Green I for rapid epifluorescence counts of marine viruses and bacteria. Aquat. Microb. Ecol. 14:113-118. [Google Scholar]
- 17.Porter, K., and Y. Feig. 1980. The use of DAPI for identifying and counting aquatic microflora. Limnol. Oceanogr. 25:943-948. [Google Scholar]
- 18.Proctor, L. M., and J. A. Fuhrman. 1990. Viral mortality of marine bacteria and cyanobacteria. Nature 343:60-62. [Google Scholar]
- 19.Suttle, C. A. 1993. Enumeration and isolation of viruses, p. 121-134. In P. F. Kemp (ed.), Handbook of methods in aquatic microbial ecology. CRC Press, Boca Raton, Fla.
- 20.Suttle, C. A. 1994. The significance of viruses to mortality in aquatic microbial communities. Microb. Ecol. 28:237-243. [DOI] [PubMed] [Google Scholar]
- 21.Suttle, C. A., A. M. Chan, and M. T. Cottrell. 1990. Infection of phytoplankton by viruses and reduction of primary productivity. Nature 347:467-469. [Google Scholar]
- 22.Turley, C. M., and D. J. Hughes. 1992. Effects of storage on direct estimates of bacterial numbers of preserved seawater samples. Deep-Sea Res. 39:375-394. [Google Scholar]
- 23.Weinbauer, M. G., and C. A. Suttle. 1997. Comparison of epifluorescence and transmission electron microscopy for counting viruses in natural marine waters. Aquat. Microb. Ecol. 13:225-232. [Google Scholar]
- 24.Wilhelm, S. W., and C. A. Suttle. 1999. Viruses and nutrient cycles in the sea. Bioscience 49:781-788. [Google Scholar]
- 25.Wommack, K. E., and R. R. Colwell. 2000. Virioplankton: viruses in aquatic ecosystems. Microbiol. Mol. Biol. Rev. 64:69-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xenopoulos, M. A., and D. F. Bird. 1997. Virus a la sauce Yo-Pro: microwave-enhanced staining for counting viruses by epifluorescence microscopy. Limnol. Oceanogr. 42:1648-1650. [Google Scholar]
- 27.Yager, P. L., T. L. Connelly, B. Mortazavi, K. E. Wommack, N. Bano, J. E. Bauer, S. Opsahl, and J. T. Hollibaugh. 2001. Dynamic bacterial and viral response to an algal bloom at subzero temperatures. Limnol. Oceanogr. 46:790-801. [Google Scholar]