Abstract
Salmonid rickettsial septicemia, caused by Piscirickettsia salmonis, causes major mortalities in Chilean salmonid aquaculture and is an increasing problem in Atlantic salmon in Ireland and Scotland. Analysis of 16S-to-23S internal transcribed sequences and 16S ribosomal DNA (rDNA) shows that Irish isolates of P. salmonis form two new groups of the organism while Scottish isolates cluster together with Norwegian and Canadian isolates from Atlantic salmon.
Salmonid rickettsial septicemia (SRS), or piscirickettsiosis, is a systemic disease of cultured salmonids initially recognized in coho salmon in Chile (1), where it causes significant losses in aquaculture (4). Subsequently, disease has been recorded in Canada (3), Norway (19), Ireland (21), and Scotland (A. N. Grant, A. G. Brown, D. I. Cox, T. H. Birkbeck, and A. A. Griffen, Letter, Vet. Rec. 423:138, 1996), but the impact in the Northern Hemisphere (0.6 to 18% mortalities) has not yet reached the levels experienced in Chile (30 to 90% mortalities) (1). Coho salmon appear to be more susceptible to SRS than Atlantic salmon, which has altered the balance of species cultured in Chile (18). Rickettsia-like organisms have been observed globally in a wide range of marine organisms (7), but difficulty of culture means their gram-negative classification and usual intracellular location within host cells are often all that is known of them. The etiologic agent of SRS was isolated (8) and later named Piscirickettsiosis salmonis as a new genus and species (9). The 16S ribosomal DNA (rDNA) sequence of the P. salmonis type strain, LF-89, indicated taxonomic placement within the γ subdivision of the Proteobacter group, rather than alongside true rickettsiae in the α subdivision (9). Ensuing 16S rDNA analysis of isolates from Chile, Canada, and Norway by PCR and restriction fragment length polymorphism revealed a homogeneous group with the presence of one outlier, Chilean strain EM-90 (16), which was substantiated from the 16S rDNA sequence (17). More subtle differences between strains are illuminated by analyzing the 16S-to-23S intergenic transcribed spacer (ITS), which possesses a higher level of sequence diversity (17), meaning that ITS sequence data can be used to infer evolutionary and phylogenetic relationships not evident from 16S rDNA. Both 16S rDNA and ITS data can be complicated by the presence of multiple rDNA operons, documented in P. salmonis strains LF-89 and EM-90 (4). However, the P. salmonis-specific ITS primers described by Marshall et al. (14) are reported to amplify only one ITS region, allowing strain-to-strain comparisons (4). Of the isolates studied by Mauel et al. (17)—four Chilean isolates, one Canadian isolate, and one Norwegian isolate—ITS data analysis indicates phylogenetic diversity between isolates from different geographic regions. Here we apply 16S and ITS sequence data analysis to the widest range of SRS isolates yet to be studied, representing the first report of sequences from Irish and Scottish isolates.
Infected fish samples.
Samples were obtained from seven Scottish farms on the mainland and the islands of Mull, Orkney, Skye, and South Uist. The SCO-95A and SCO-98B isolates were received as infected CHSE-214 cell cultures and SCO-98C was received as methanol-fixed kidney material. In 2002, fresh brain and kidney samples dissected from moribund Atlantic salmon were transported on ice to our laboratory for analysis. Samples from Ireland comprised fresh infected tissue (IRE-99A and IRE-99B) and archival material consisting of fish organ samples or tissue culture preparations stored at −80°C in 10% dimethyl sulfoxide.
DNA purification.
DNA was purified from fish tissue and infected CHSE-214 tissue culture preparations. Sections of tissue (approximately 5 mm3) or pelleted CHSE-214 cells harvested from culture flasks were homogenized aseptically in 1 to 2 ml of sterile seawater. A cell pellet was collected from the homogenate by centrifugation at 13,000 rpm for 1 min in a benchtop microcentrifuge, and the pellet was washed twice by vortexing and centrifugation in 1 ml of sterile seawater. DNA was extracted by a procedure described by Gonazález et al. (10), using guanidine isothiocyanate and InstaGene matrix (Bio-Rad) followed by phenol-chloroform extraction and alcohol precipitation.
Oligonucleotide primers.
Two sets of previously described P. salmonis-specific primers were used (Table 1). Primers against 16S rDNA amplified a 470-bp fragment, and 16S to 23S ITS primers yielded a 280-bp product. For infected tissue culture preparations, the universal primers 27F and 1525R were used to amplify >1,500 bp of the gene. Oligonucleotides were synthesized by MWG Biotech (https://ecom.mwgdna.com).
TABLE 1.
Descriptions and sources of the primers used in this study
| Primer designation | Sequence (5′→3′) | Primer location | Reference |
|---|---|---|---|
| PS2S | CTAGGAGATGAGCCCGCGTTG | 16S forward (223F)a | 16 |
| PS2AS | GCTACACCTGCGAAACCACTT | 16S reverse (690R)a | |
| RTS1 | TGATTTTATTGTTTAGTGAGAATGA | ITS forward (223F)b | 14 |
| RTS4 | ATGCACTTATTCACTTGATCATA | ITS reverse (459R)b |
Numbering corresponds to E. coli 16S gene.
Numbering corresponds to the P. salmonis LF-89 ITS sequence.
PCR amplification and analysis.
Reactions were performed in 25-μl volumes consisting of (final concentrations) 0.025 U of HotStarTaq polymerase (QIAGEN), 1× polymerase buffer, 0.2 mM deoxynucleoside triphosphate, and 0.5 μM each of the forward and reverse primers. Template DNA was diluted 1/10 in distilled water, and 2 μl was added to the reaction tubes. PCR cycles consisted of denaturation at 94°C for 2 min, 35 amplification cycles of 94°C for 1 min, 57 or 50°C (16S and ITS primer pairs, respectively) for 1 min, and 72°C for 1 min with a final extension period of 10 min at 72°C. PCR products were analyzed by electrophoresis on a 0.8% agarose-1× Tris-borate-EDTA gel containing ethidium bromide and visualized under UV light.
Cloning and sequencing of ITS and 16S rDNA.
PCR products were cloned into pCR TOPO (Invitrogen, Paisley, United Kingdom), and Escherichia coli TOP10 cells were transformed with ligation mixes as described by the manufacturer. Recombinant clones picked from selective agar plates were cultured overnight in 3 ml of Luria-Bertani broth with 50-μg/ml kanamycin prior to preparation of plasmids (QIAGEN Mini-Prep) following the manufacturer's instructions. Selected transformants were sequenced by using m13 forward and reverse primers (22) by DBS Genomics, University of Durham, using the PRIDM DyeDeoxy terminator cycle sequencing kit (Applied Biosystems) and an Applied Biosystems 373A automatic DNA sequencer. Sequences were compared to those in the DDBJ database by using the program BLAST (http://www.ddbj.nig.ac.jp).
Cladistic analysis.
DNA sequences were aligned with the CLUSTAL W function (23) in the BioEdit sequence editor package (11). Cladograms were constructed from the sequence alignments by using programs available in PHYLIP (Phylogeny Inference Package 3.5c), through the Pasteur Institute web interface (http://bioweb.pasteur.fr). The small degree of sequence divergence between isolates and the short sequence lengths allowed trees to be inferred by maximum-parsimony methods (6). The possibility of homoplasies resulting in an erroneous tree was found to be limiting when nucleotide transversions only were considered (transversion parsimony) and the inferred trees were compared. Other tree-drawing methods applied to our data (maximum-likelihood and DNA distance methods) drew trees with a common topology, giving confidence in the parsimony method shown here. With no other data to imply the lineage of the taxa considered here, trees were left unrooted to allow the degree of kinship between isolates to be examined, but no evolutionary path is inferred. Trees were bootstrapped with 1,000 replicates and a consensus tree, with values attached at nodes, was viewed by using TreeView (20).
Results and analyses.
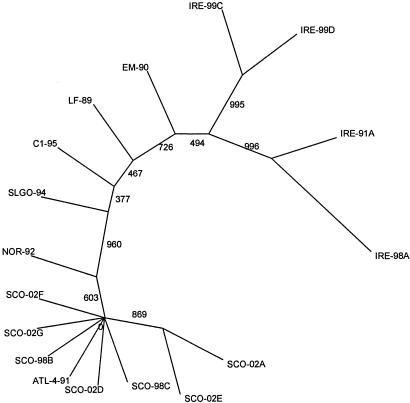
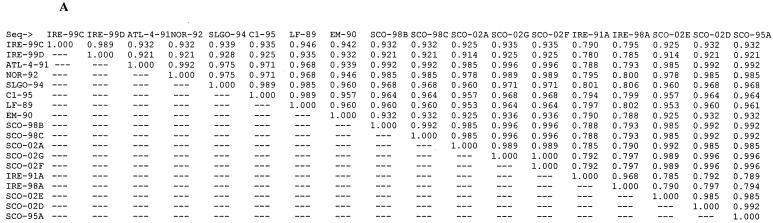
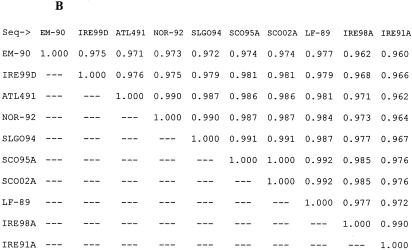
P. salmonis isolates from a number of different hosts and from different geographic locations have previously been studied to ascertain the level of genetic variability between isolates. Our analysis was performed to provide the basis for epidemiological studies of the disease in Europe. Genetic variability between isolates may be one explanation for the major differences between mortality levels in fish, and presumably in virulence of P. salmonis, in Chile and the Northern hemisphere. We report here the first analysis of sequence data from Scottish and Irish isolates causing SRS in farmed Atlantic salmon, where disease incidence has increased since the first reports in 1995 and 1992, respectively. ITS sequences were derived from all samples studied, and a maximum-parsimony cladogram inferred from comparisons of these sequences is shown in Fig. 1. Almost complete sequences for 16S rDNA were obtained for isolates cultured in CHSE-214 cells (SCO-95A, SCO-98B, SCO-02A, IRE-99C, and IRE-99D) and a further sample (SCO-98C) from methanol-fixed tissue. A tree inferred by the same methods from 16S sequence data shared the same overall topology, highlighting the existence of two new groups of isolates, but with less discrimination between closer relatives (data not shown). Similarity matrices showing the level of ITS and 16S sequence identity shared between these isolates and others available in GenBank (Table 2) are shown in Fig. 2A and B, respectively. Previously, only two groups of isolates had been demonstrated, with the Chilean isolate EM-90 alone being easily distinguishable from other P. salmonis isolates (16), displaying ITS sequence identity of 95.2% with ATL-4-91 and 96.5% with the type strain LF-89. A recent study of the relative electrophoretic mobilities of a group of isolates from rainbow trout, coho salmon, and Atlantic salmon in Chile still only separated them into two groups: i.e., the EM-90 and LF-89 clusters (5).
FIG. 1.
ITS cladogram comparing sequences of 287 nucleotide positions. Cladograms were drawn as unrooted trees by maximum-parsimony methods. Bootstrap values of 1,000 replicates are shown.
TABLE 2.
Isolates of P. salmonis compared in this study
| Isolate | Fish host | Yr of isolation | Country of origin | DNA accession no. (EMBL)
|
Reference | |
|---|---|---|---|---|---|---|
| 16S rDNA | ITS | |||||
| LF-89 | Coho salmon | 1989 | Chile | U36941 | U36943 | Mauel et al. (17) |
| EM-90 | Atlantic salmon | 1989 | Chile | U36940 | U36944 | Mauel et al. (17) |
| SLGO-94 | Rainbow trout | 1994 | Chile | U55015 | U62104 | Mauel et al. (17) |
| NOR-92 | Atlantic salmon | 1992 | Norway | U36942 | U36946 | Mauel et al. (17) |
| C1-95 | Coho salmon | 1995 | Chile | U62103 | Mauel et al. (17) | |
| ATL-4-91 | Atlantic salmon | 1991 | Canada | U36915 | U36945 | Mauel et al. (17) |
| SCO-95A | Atlantic salmon | 1995 | Scotland | AY498636 | AY498621 | Grant et al. (1996)a |
| SCO-98B | Atlantic salmon | 1998 | Scotland | AY498630 | This study | |
| SCO-98C | Atlantic salmon | 1998 | Scotland | AY498629 | This study | |
| SCO-02A | Atlantic salmon | 2002 | Scotland | AY498635 | AY498628 | This study |
| SCO-02D | Atlantic salmon | 2002 | Scotland | AY498622 | This study | |
| SCO-02E | Atlantic salmon | 2002 | Scotland | AY498623 | This study | |
| SCO-02F | Atlantic salmon | 2002 | Scotland | AY498626 | This study | |
| SCO-02G | Atlantic salmon | 2002 | Scotland | AY498627 | This study | |
| IRE-91A | Atlantic salmon | 1991 | Ireland | AY498633 | AY498625 | This study |
| IRE-98A | Atlantic salmon | 1998 | Ireland | AY498634 | AY498624 | This study |
| IRE-99C | Atlantic salmon | 1999 | Ireland | AY498632 | This study | |
| IRE-99D | Atlantic salmon | 1999 | Ireland | AY498637 | AY498631 | This study |
Letter, Vet. Rec. 138:423, 1996.
FIG. 2.
Sequence identity matrices drawn from aligned sequences. The matrices show the proportion of identical residues between all of the sequences in the alignment. The reported number represents the ratio of identities to the length of the longer of the two sequences after positions at which both sequences contain a gap are removed. (A) ITS matrix comparing sequences of 287 nucleotide positions. (B) 16S matrix comparing sequences of 1,309 nucleotide positions.
The tree inferred from ITS sequence data (Fig. 1) demonstrates the existence of new groups of isolates collected from infected farmed Atlantic salmon in Scotland and Ireland. Comparison of the ITS and 16S rDNA sequence identity matrices (Fig. 2A and B) confirms the higher level of sequence diversity occurring in the ITS gene sequence. The ITS cladogram discriminates between four different clades or groups within the tree. Working around the tree in an anticlockwise manner, the first group encountered is the furthest outlying group and has as members Irish samples taken from repeat outbreaks of disease sampled from the same site in 1991 and 1998 (IRE-91A and IRE-98A), which share 97% ITS sequence identity. The second group contains two further Irish samples collected from different sites in 1999 (IRE-99C and IRE-99D). High bootstrap values at these nodes indicate the confidence with which these two clusters can be drawn. However, a low bootstrap value (49%) linking these to the next isolate to diverge from the central branch, EM-90 (Atlantic salmon, Chile), implies that a linkage with this isolate, which has been identified as an outlier in previous papers, can be assumed. The separation of the Chilean EM-90 isolate from the next to break off from the central branch is inferred with slightly more certainty (bootstrap value of 73%) and depicts the removal of EM-90 from the type strain, LF-89. However, the data suggest that LF-89, C1-95, and SLGO-94 (all Chilean isolates: the former two from coho salmon and the latter from rainbow trout) share closer kinship with each other. The next deviation from the central branch shows all the Scottish isolates (Atlantic salmon) and Norwegian and Canadian Atlantic salmon isolates (NOR-92 and ATL-4-91) to be confidently removed from all others (bootstrap value of 96%). Within this cluster, a degree of further separation appears, with NOR-92 and SCO-02A and -E being slightly distal, but it remains apparent that isolates in this “Atlantic” cluster are more closely related to each other than they are to any of the Chilean or Irish isolates. The close clustering of three Scottish 2002 isolates with two Scottish 1998 isolates and Canadian ATL-4-91 implies that these are all samples of the same strain that have been collected at different locations over an 11-year period.
While an unrooted tree, in the strictest terms, cannot be used to determine phylogeny, the placement of EM-90 in the tree does draw one to make inferences. As a Chilean isolate infecting Atlantic salmon, it sits between the Irish Atlantic salmon isolates and the Chilean coho salmon isolates, balanced by almost equally uncertain bootstrap values. This placement in the middle ground possibly indicates that there has been transfer of the agent from one of these locations to the other during movement of aquaculture stocks. Near neighbors to the Scottish isolates are samples taken from infected Atlantic salmon in Norway and British Columbia, which may again suggest transfer of P. salmonis with fish stocks. Bravo and Campos (2) have previously suggested shipment of eggs as the origin of SRS in Chile, and P. salmonis has been found associated with the gonads of infected fish and in fertilized ova (13).
Different groups of isolates appear to be localized geographically. For example, the ITS cladogram shows that the Scottish isolates collected from seven different sites over a period of 7 years cluster to form a group of closely related organisms. The Chilean strains isolated from fish other than Atlantic salmon also appear more closely related to each other than other isolates. Their connection could result from the geographic proximity of the sampling locations in Chile or may indicate a specificity of these isolates for non-Atlantic salmonids with which they may have coevolved. Similar focusing of isolates around a geographic locus has been reported by Heath et al. (12) in an analysis of ITS fragments by denaturing gradient gel electrophoresis. By this method, samples collected from five sites around one location in Chile were indistinguishable from the type strain of P. salmonis (LF-89). The focus of disease incidents at particular locations suggests that local sources of P. salmonis may act as holding reservoirs of infection. Alternatively, aquaculture fish stocks may carry P. salmonis in a subclinical or latent form, and other environmental factors may then induce overt disease. It is likely that P. salmonis and the reservoir(s) have coevolved on an evolutionary scale, such that levels of overt infection in a reservoir are low. P. salmonis has perhaps always been able to transfer from this reservoir to infect wild fish sporadically, but farmed fish held close to a reservoir of infection present better conditions for disease to burgeon. Following a report that P. salmonis-like DNA can be detected in bacterioplankton DNA extracted from coastal waters (15), further studies are required to identify possible reservoirs and vectors of P. salmonis that may exist in the environment and the impact of environmental factors in mediating disease in farmed salmonids.
The Chilean isolates share an ability to inflict much higher mortalities upon farmed fish than do the Scottish isolates. However, a direct comparison of the virulence of a range of isolates of P. salmonis in both Atlantic and coho salmon is required to determine whether this is the case or if environmental factors, such as water temperature, are more significant factors in determining mortality.
Nucleotide sequence accession numbers.
Novel sequences were submitted to GenBank (http://www.ncbi.nih.gov/GenBank/) and given accession no. AY498621 to AY498632 (ITS sequences) and AY498633 to AY498637 (16S sequences) (Table 2).
Acknowledgments
We thank D. Cox (Fish Vet Group, Inverness, Scotland), F. Geoghegan (Marine Institute, Dublin, Ireland), A. Laidler (Marine Harvest, Lochailort, Scotland), and K. Murphy (Galway, Ireland) for providing samples and B. Adam for technical assistance.
This work was supported by the Natural Environment Research Council.
REFERENCES
- 1.Branson, E. J., and D. Nieto Diaz-Munoz. 1991. Description of a new disease condition occurring in farmed coho salmon, Oncorhynchus kisutch (Walbaum) in South America. J. Fish Dis. 14:147-156. [Google Scholar]
- 2.Bravo, S., and M. Campos. 1989. Coho salmon syndrome in Chile. Am. Fish. Soc. Fish Health Sect. Newsl. 17:3. [Google Scholar]
- 3.Brocklebank, J. R., T. P. T. Evelyn, D. J. Speare, and R. D. Armstrong. 1993. Rickettsial septicemia in farmed Atlantic and chinook salmon in British Columbia: clinical presentation and experimental transmission. Can. Vet. J. 34:745-748. [PMC free article] [PubMed] [Google Scholar]
- 4.Casanova, A., J. C. Obreque, A. M. G. Sandino, and M. Jashés. 2001. tRNA genes were found in Piscirickettsia salmonis 16S-23S rDNA spacer region (ITS). FEMS. Microbiol. Lett. 197:19-22. [DOI] [PubMed] [Google Scholar]
- 5.Casanova, A., J. R. Obreque, A. Gaggero, E. Landskron, A. M. Sandino, and M. Jashés. 2003. Electrophoretic analysis of ITS from Piscirickettsia salmonis Chilean isolates. FEMS Microbiol. Lett. 225:173-176. [DOI] [PubMed] [Google Scholar]
- 6.Felsenstein, J. 1989. PHYLIP—Phylogeny Inference Package (version 3.2). Cladistics 5:164-166. [Google Scholar]
- 7.Fryer, J. L., and C. Lannan. 1994. Rickettsial and chlamydial infections of freshwater and marine fishes, bivalves and crustaceans. Zool. Stud. 33:95-107. [Google Scholar]
- 8.Fryer, J. L., C. Lannan, L. Garcés, J. Larenas, and P. Smith. 1990. Isolation of a Rickettsiales-like organism from diseased coho salmon (Oncorhynchus kisutch) in Chile. Fish Pathol. 25:107-114. [Google Scholar]
- 9.Fryer, J. L., C. Lannan, S. J. Giovannoni, and N. Wood. 1992. Piscirickettsia salmonis gen. nov., sp. nov., the causative agent of an epizootic disease in salmonid fishes. Int. J. Syst. Evol. Microbiol. 42:120-126. [DOI] [PubMed] [Google Scholar]
- 10.González, I., T. García, A. Fernández, B. Sanz, P. E. Hernández, and R. Martín. 1999. Rapid enumeration of Escherichia coli in oysters by a quantitative PCR-ELISA. J. Appl. Microbiol. 86:231-236. [DOI] [PubMed] [Google Scholar]
- 11.Hall, A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 12.Heath, S., S. Pak, S. Marshall, E. M. Prager, and C. Orrego. 2000. Monitoring Piscirickettsia salmonis by denaturant gel electrophoresis and competitive PCR. Dis. Aquat. Org. 41:19-29. [DOI] [PubMed] [Google Scholar]
- 13.Larenas, J., C. Astorga, J. Contreras, and P. Smith. 1996. Detección de Piscirickettsia salmonis en ovas fertilizadas provenientes de trucha arco iris (Oncorhynchus mykiss) experimentalmente infectadas. [Piscirickettsia salmonis in ova obtained from rainbow trout (Oncorhynchus mykiss) experimentally inoculated.] Arch. Med. Vet. 28:161-166. [Google Scholar]
- 14.Marshall, S., S. Heath, V. Henríquez, and C. Orrego. 1998. Minimally invasive detection of Piscirickettsia salmonis in cultivated salmonids via the PCR. Appl. Environ. Microbiol. 64:3066-3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mauel, M. J., and J. L. Fryer. 2001. Amplification of a Piscirickettsia salmonis-like 16S rDNA product from bacterioplankton DNA collected from the coastal waters of Oregon, USA. J. Fish Dis. 26:251-262. [Google Scholar]
- 16.Mauel, M. J., S. J. Giovannoni, and J. L. Fryer. 1996. Development of polymerase chain reaction assays for detection, identification and differentiation of Piscirickettsia salmonis. Dis. Aquat. Org. 26:189-195. [Google Scholar]
- 17.Mauel, M. J., S. J. Giovannoni, and J. L. Fryer. 1999. Phylogenetic analysis of Piscirickettsia salmonis by 16S, internal transcribed spacer (ITS) and 23S ribosomal DNA sequencing. Dis. Aquat. Org. 35:115-123. [DOI] [PubMed] [Google Scholar]
- 18.Mauel, M. J., and D. L. Miller. 2002. Piscirickettsiosis and piscirickettsiosis-like infections in fish: a review. Vet. Microbiol. 87:279-289. [DOI] [PubMed] [Google Scholar]
- 19.Olsen, A. B., Ø. Evensen, L. Speilberg, H. P. Melby, and T. Håstein. 1993. ‘Ny’ laksesykdom forårsaket av rickettsie. Nor. Fiskeoppdrett. 12:40-41. [Google Scholar]
- 20.Page, R. D. M. 1996. TREEVIEW: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 21.Rodger, H. D., and E. M. Drinan. 1993. Observation of a rickettsia-like organism in Atantic salmon, Salmo salar L., in Ireland. J. Fish Dis. 16:361-370. [Google Scholar]
- 22.Sambrook J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 23.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]