Abstract
Background
The aim of this study was to investigate the distribution of epidermal growth factor receptor (EGFR)vIII mutation in Chinese non-small cell lung cancer (NSCLC) patients and to explore the likely relationship between EGFRvIII mutation and response to EGFR-tyrosine kinase inhibitors (TKIs) in squamous cell carcinoma (SCC).
Methods
Samples were derived from two patient cohorts: (i) 114 early-stage NSCLC who received surgical resection; and (ii) 31 advanced-stage SCC who received EGFR-TKI EGFRvIII. EGFR and V-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog (KRAS) mutations were detected by reverse transcription polymerase chain reaction (RT-PCR), denaturing high-performance liquid chromatography, and PCR-restriction fragment length polymorphism, respectively. The associations of EGFRvIII, EGFR, and KRAS mutations with clinical outcome of EGFR-TKI treatment were evaluated using the Kaplan-Meier method, descriptive analysis, and multi-variable Cox regression analysis.
Results
In the first cohort, EGFRvIII mutation was detected in eight of 114 (7.0%) patients, including 11.1% (6/54) SCC and 3.6% (2/55) adenocarcinomas (ADC) (P = 0.269). In the second cohort, five (16.1%) and 10 out of 31 advanced SCC presented EGFRvIII and EGFR mutations, respectively. No appreciable discrepancy of progression-free survival or disease control rate was detected between the patients with and without EGFRvIII mutation (P > 0.05). However, longer median overall survival (OS) was observed in patients harboring EGFRvIII compared to those without EGFRvIII, although the difference did not reach statistical significance.
Conclusion
The frequency of EGFRvIII mutation in SCC was higher than in ADC. SCC patients harboring EGFRvIII mutations had a tendency for prolonged OS.
Keywords: EGFR-TKI, EGFRvIII mutation, NSCLC
Introduction
Lung cancer is the leading cause of cancer-related death, both in China and worldwide. Over the last decade, the treatment decision for non-small cell lung cancer (NSCLC) patients has changed from empiric to genotypic personalized therapy. Lung cancers with sensitizing epidermal growth factor receptor (EGFR) mutations and echinoderm microtubule-associated protein-like 4-anaplastic lymphoma kinase translocations (EML4-ALK) can have remarkable responses to EGFR and ALK inhibitors, respectively, leading to significant clinical outcome improvements.1–6 However, these advances in the therapeutic field have mainly occurred for non-squamous NSCLC. Squamous cell carcinoma (SCC) patients seldom benefit from the major advances in the development of targeted therapeutics. For example, the frequency of EGFR mutation in Chinese SCC patients is 10–15%,7–9 a higher frequency than is found in Caucasians.10 However, the response rate of EGFR mutation in SCC to EGFR-tyrosine kinase inhibitors (TKIs) is not as good as the response rate in lung ADC with EGFR mutations,11 which suggests that EGFR mutation might not be a predominant biomarker for SCC. Therefore, it is urgent for oncologists to search for the meaningful biomarkers that are specific to SCC.
Numerous studies have confirmed that activation of the EGFR signaling pathway is related to cellular proliferation and resistance to apoptosis in cancer cells.12,13 In addition to intracellular mutations of the EGFR gene, deletions in the extracellular domain also seem to activate the receptor. The most common of these truncated receptors is the type III EGFR deletion mutant (variously named EGFRvIII, de2-7 EGFR, or ΔEGFR), which lacks a portion of the extracellular ligand binding domain (exon 2–7) and can auto-phosphorylate independent of EGFR-binding ligands, subsequently activating downstream signaling cascades to promote tumor cell proliferation and inhibit apoptosis.14,15 The EGFRvIII mutation has been reported to frequently occur in several malignant tumors, including glioblastoma, head and neck tumors, and ovarian, breast, and lung cancers.16,17 Ji et al.18 reported a difference between the EGFRvIII mutation in SCC and ADC (5.36% vs. 0%), and subsequent in vivo and in vitro experiments have shown the inhibition of tumor cells harboring EGFRvIII after treatment with erlotinib. This suggests that EGFRvIII might be one of the special biomarkers for SCC and could partially explain why patients with EGFR wild-type SCC respond to EGFR-TKI. However, Ohtsuka et al.19,20 failed to observe a discrepancy in EGFRvIII mutation between SCC and ADC. Currently, the results from previous studies concerning EGFRvIII mutation in lung cancer patients remain controversial and data from Chinese populations is scarce.
To our knowledge, the present report represents the largest screening study for the detection of EGFRvIII mutation in Chinese NSCLC patients. The aim of this retrospective study was to determine the clinical-pathologic characteristics and distribution profiles of EGFRvIII mutation in Chinese NSCLC patients, particularly in SCC, and to explore the relationship between EGFRvIII mutation and the response to EGFR-TKI.
Methods
Patient selection and treatment plan
Two cohorts of patients with NSCLC were enrolled in this study. The first cohort consisted of 114 consecutive Chinese patients with early stage NSCLC who underwent surgery at the Peking University Cancer Hospital between February 2002 and July 2009, and included 55 patients with ADC, 54 patients with SCC, and five patients with adenosquamous carcinoma. The second cohort was comprised of 31 patients diagnosed with advanced SCC of lung who were treated with EGFR-TKI in our center and provided sufficient tissue samples for molecular detection. The study was reviewed and approved by the institutional ethics committee at the Peking University Cancer Hospital. All patients signed informed consent to participate in this study and permit the further use of their tumor tissues.
Patients with advanced SCC of the lung were required to receive EGFR-TKI therapy until the disease progressed, as determined by imaging and aggravated symptoms. Categories of progressive disease (PD), stable disease (SD), partial remission (PR), and complete remission (CR) from the Response Evaluation Criteria in Solid Tumors were used to evaluate a patient's response to EGFR-TKI after receiving therapy for 30 days.
Total ribonucleic acid extraction and epidermal growth factor receptor (EGFR)vIII mutation detection using reverse transcription polymerase chain reaction (RT-PCR)
For the first cohort, total ribonucleic acid (RNA) was extracted from 50–100 mg of each frozen tumor specimen using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) in a tissue grinder. Total RNA was then treated with amplification grade DNase I for 15 minutes at 37°C. For the second cohort, RNA was isolated from formalin-fixed, paraffin embedded specimens using RNeasy extraction protocol (Qiagen, Valencia, CA, USA).
For each patient, one microgram of total RNA was reverse transcribed using Superscript II (Invitrogen, USA) with oligo (dT) priming according to the manufacture's protocol. In the first step of PCR, 2 μL of single strand cDNA (10% of the reverse transcription reaction volume) was used as a template in a 20 μL volume PCR reaction containing 0.8 mM MgSO4, 0.4 μL of each primer, 0.4 mM dNTPs, 1 × High Fidelity PCR buffer, and 0.1 unit of Platinum Taq High Fidelity (Invitrogen, USA). Forward and reverse primer sequences used to specifically amplify EGFRvIII were 5′ CTT CGG GGA GCA GCG ATG CGA C 3′ (spanning the 5′ untranslated region and the beginning of exon 1) and 5′ ACC AAT ACC TAT TCC GTT ACA C 3′ (within exon 9), respectively. PCR cycling conditions began with an initial denaturation step at 95°C for five minutes, followed by 10 cycles of denaturation at 95°C for 30 seconds, annealing at 58°C for 30 seconds, and extension at 72°C for 30 seconds, accompanied with 0.5°C down of annealing temperature in every cycle; then 35 cycles of denaturation at 95°C for 30 seconds, annealing at 53°C for 30 seconds, extension at 72°C for 30 seconds, and extension at 72°C for 10 minutes. In the second step, 2 μL of the first PCR product was used as a template in a 20 μL volume PCR reaction; other ingredients and PCR conditions were the same as those used in the first step. These primers generated a 243 bp PCR product for the EGFRvIII transcript. PCR reactions were analyzed after running running 2 μL of product on a 2% agarose gel and staining with ethidium bromide. In a preliminary experiment, a 200–300 bp positive strand was detected and proven to be an EGFRvIII-positive sample by direct sequencing. This sample was used as a positive control and distilled water was used as a template for the negative control. The products of EGFRvIII positive samples were verified by DNA clone sequencing (Fig 1).
Figure 1.
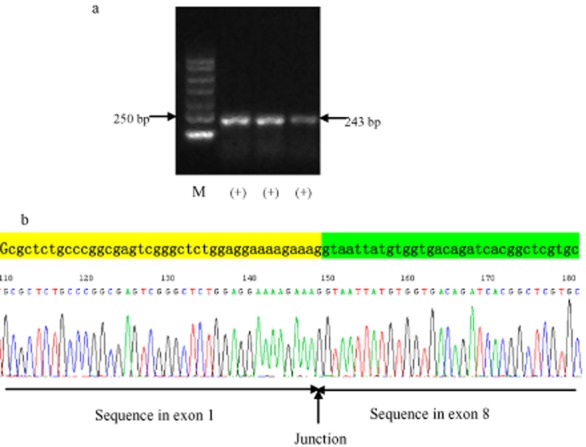
Representative examples of epidermal growth factor receptor (EGFR)vIII by reverse transcription polymerase chain reaction.(a) EGFRvIII electropherogram; (b) EGFRvIII sequencing. M, marker.
EGFR mutation detection using denaturing high-performance liquid chromatography (DHPLC)
Sequences of 19 and 21 exons of EGFR tyrosine kinase domains were detected by denaturing high-performance liquid chromatography (DHPLC). Detailed methods and procedures have been elaborated in our previous research.6 Both the EGFRvIII positive samples in the first cohort and the advanced SCC samples in the second cohort were tested for EGFR mutation status.
KRAS mutation detection using PCR-restriction fragment length polymorphism
KRAS mutation was detected by PCR-restriction fragment length polymorphism. The detailed method and procedure has been reported in our previous research.21 Only the advanced SCC samples in the second cohort were tested for K-RAS mutation status.
Statistical analysis
The relationship between clinicopathologic factors and EGFRvIII was analyzed using Pearson's χ2 or Fisher's exact tests, as appropriate. All statistical tests were two-sided, with significance defined as P < 0.05. Kaplan-Meier curves were used to estimate progression-free survival (PFS) and overall survival (OS). Multi-variable Cox regression analysis was used to identify independent factors of PFS and OS. General data analysis was conducted using SPSS Version 17.0 (IBM, Chicago, IL, USA).
Results
EGFRvIII mutation in post-surgery non-small cell lung cancer patients
A total of 114 post-surgical specimens of adequate quality for detection of EGFRvIII and EGFR mutations were collected, and their clinicopathologic characteristics were recorded (Table 1). The study participants included 65 men and 49 women, and the median patient age was 61 years (range: 37–80 years). The most common histologic subtypes were ADC (55, 48.25%) and SCC (54, 47.37%). According to the 2009 American Joint Committee on Cancer staging for NSCLC, there were 56 patients in stage I, 28 in stage II, and 30 patients in stage III. Among the 114 patients, 61 patients were never smokers and 53 patients were smokers or former smokers.
Table 1.
Clinicopatholical characteristics of 114 NSCLC patients
| Number | Percent (%) | |
|---|---|---|
| Age (year) | ||
| Median (range) | 61 (37–80) | |
| Gender | ||
| Male | 65 | 57.0 |
| Female | 49 | 43.0 |
| Pathology | ||
| Adenocarcinoma | 55 | 48.3 |
| SCC | 54 | 47.4 |
| Adenosquamous carcinoma | 5 | 4.3 |
| Stage | ||
| Stage I | 56 | 49.1 |
| Stage II | 28 | 24.6 |
| Stage III | 30 | 26.3 |
| Smoking status | ||
| Never smoker | 61 | 53.5 |
| Smoker or former smoker | 53 | 46.5 |
NSCLC, non-small cell lung cancer; SCC, squamous cell carcinoma.
In the cohort of 114 post-surgery patients, eight patients (7.02%) were detected as having an EGFRvIII mutation (Table 2) (Fig 1), including six SCC and two ADC patients. The EGFRvIII-positive rates were 11.11% (6/54) in SCC, 3.64% (2/55) in adenocarcinoma, and 0 (0/5) in adenosquamous carcinoma. According to our data, EGFRvIII mutation was more likely to occur in men, smokers or former smokers, and SCC patients, than in women, never smokers, and ADC patients; however, the difference did not reach statistical significance (P = 0.113, P = 0.142, P = 0.269, respectively) (Table 3). To determine the correlation between EGFRvIII and EGFR mutations, we further analyzed the EGFR mutation status in eight EGFRvIII-positive samples by DHPLC. Unfortunately, no EGFR mutation was detected in these post-surgery samples, for either EGFR exon 19 or 21 mutation.
Table 2.
Clinicopathological characteristics of EGFRvIII-positive patients
| Gender | Pathology | PS | Smoking status | P-stage | EGFRvIII expression | EGFR mutation | DFS (m) | OS (m) | Follow-up (m) |
|---|---|---|---|---|---|---|---|---|---|
| M | SCC | 0 | Smoker | I | (+) | (−) | No relapse | Alive | 80 |
| M | SCC | 0 | Never smoker | I | (+) | (−) | No relapse | Alive | 99 |
| M | SCC | 0 | Smoker | I | (+) | (−) | No relapse | Alive | 39 |
| M | SCC | 0 | Smoker | I | (+) | (−) | 6 | 12 | 12 |
| M | SCC | 1 | Smoker | II | (+) | (−) | No relapse | Alive | 46 |
| M | ADC | 0 | Smoker | II | (+) | (−) | 30 | 30 | 30 |
| M | SCC | 0 | Smoker | III | (+) | (−) | No relapse | Alive | 45 |
| F | ADC | 0 | Never smoker | III | (+) | (−) | 15 | 26 | 26 |
ADC, adenocarcinoma; DFS, disease-free survival; EGFR, epidermal growth factor receptor; F, female; M, male; OS, overall survival; PS, performance status; SCC, squamous cell carcinoma.
Table 3.
Correlation between EGFRvIII expression and pathology
| Pathology | Total | |||
|---|---|---|---|---|
| SCC | ADC | AdCa | ||
| EGFRvIII (−) | 48 | 53 | 5 | 106 |
| EGFRvIII (+) | 6 | 2 | 0 | 8 |
| Total | 54 | 55 | 5 | 114 |
Fisher's exact test: P = 0.269. ADC, adenocarcinoma; AdCa, adenosquamous carcinoma; EGFR, epidermal growth factor receptor; SCC, squamous cell carcinoma.
Correlation between EGFRvIII mutation and EGFR-tyrosine kinase inhibitor treatment outcome in advanced squamous cell carcinoma patients
From January 2004 to December 2010, a total of 520 patients were diagnosed with advanced SCC in the Department of Thoracic Oncology at Peking University Cancer Hospital. Thirty-one of these patients who received EGFR-TKI therapy could provide adequate tissue specimens for determining EGFRvIII mRNA level, EGFR mutation, and KRAS mutation status. This subgroup comprised 24 men and seven women, and the median age was 66 years (range, 32–78 years). Regarding smoking status, 20 patients were either smokers or former smokers, and 11 patients had never smoked. There were seven patients diagnosed as stage IIIb and 24 stage IV according to clinical stage when the patient started treatment with EGFR-TKI. Among these, six patients with EGFR mutation received EGFR-TKI as a first-line therapy, 15 patients received EGFR-TKI as second-line therapy, and 10 patients received EGFR-TKI as third-line or further. Detailed information concerning patient EGFR-TKI treatment and gene detection results are shown in Table 4.
Table 4.
Clinicopathology and gene status characteristics of 31 SCC patients
| Number | Percent (%) | |
|---|---|---|
| Age (year) | ||
| Median (range) | 66 (32–78) | |
| Gender | ||
| Male | 24 | 77.4 |
| Female | 7 | 22.6 |
| EGFR-TKI treatment line | ||
| 1-line | 6 | 19.4 |
| 2-line | 15 | 48.4 |
| ≥3-line | 10 | 32.2 |
| Stage | ||
| Stage IIIb | 7 | 22.6 |
| Stage IV | 24 | 77.4 |
| Smoking status | ||
| Never smoker | 11 | 35.5 |
| Smoker or former smoker | 20 | 64.5 |
| EGFR status | ||
| Mutation type | 10 | 32.3 |
| Wild type | 21 | 67.7 |
| EGFRvIII type | ||
| Positive | 5 | 16.1 |
| Negative | 26 | 83.9 |
EGFR, epidermal growth factor receptor; SCC, squamous cell carcinoma; TKI, tyrosine kinase inhibitor.
EGFR sensitive and EGFRvIII mutations were detected in 10 and five cases of 31 SCC patients, respectively. The median PFS, OS, and disease control rate (DCR) were 2.03 months, 10.2 months, and 54.8% (17/31), respectively. Overall response rate, DCR, and PFS for mutated SCC were 10%, 70% and 2.0 months, respectively. The patients carrying EGFRvIII mutations possessed similar PFS and DCR to those without EGFRvIII mutation (P > 0.05). However, longer median OS was observed in patients’ harboring EGFRvIII mutations compared to those without EGFRvIII (15.0 vs. 7.3 months, adjusted hazard ratio = 0.18, P = 0.114), although the difference did not reach statistical significance. Of the five EGFRvIII-positive mutation patients, there were three patients concurrently harboring an EGFR mutation, and two patients were EGFR wild type. PFS in the two patients with EGFRvIII-positive/EGFR wild type were 1.2 and 23 months. However, in the three EGFRvIII-positive/EGFR mutation-positive patients, PFS was 1.3, 2.0, and 19.3 months. Unfortunately EGFRvIII mutation did not predict the response to EGFR-TKI in SCC patients. KRAS mutation was detected in four cases of 31 SCC patients. The patients with a KRAS mutation experienced shorter PFS and OS than those without the KRAS mutation, but the difference did not reach statistical significance (mPFS: 1.3 vs. 2.0 months, P = 0.198; mOS: 2.1 vs. 11.5 months, P = 0.082). Only one patient harbored EGFRvIII, EGFR, and KRAS mutations simultaneously; their response to EGFR-TKI was PD, and PFS was only one month. Detailed information concerning the treatment of EGFRvIII-positive patients and results of gene detection are shown in Table 5.
Table 5.
Clinical characteristics of five EGFRvIII positive patients treated with EGFR-TKI
| Gender | TKI line | Smoking status | C-stage | EGFR mutation | KRAS mutation | Response | PFS (m) | OS (m) | Status |
|---|---|---|---|---|---|---|---|---|---|
| M | 2 | Smoker | IV | (−) | (−) | PD | 1.2 | 8.0 | Died |
| M | 3 | Smoker | IV | (−) | (−) | SD | 23.0 | NE | Alive |
| M | 4 | Smoker | IV | (+) | (+) | PD | 1.3 | 13.5 | Died |
| F | 3 | Never smoker | IV | (+) | (−) | SD | 2.0 | 15.0 | Died |
| F | 2 | Never smoker | IV | (+) | (−) | SD | 19.3 | 19.3 | Died |
EGFR, epidermal growth factor receptor; NE, not evaluable; PD, progressive disease; PR, partial disease; SD, stable disease; TKI, tyrosine kinase inhibitor.
Discussion
An increasing number of studies have identified serial biomarkers in lung ADC, such as EGFR mutations1–4 and EML4-ALK,5,22 and corresponding targeted agents, which have been extensively used in the determination of treatment options. However, these biomarkers are not predictive for the therapeutic response of SCC, and a powerful biomarker for use in treating this disease remains elusive. In the present study, we used RT-PCR to detect EGFRvIII and are the first to report the frequency of EGFRvIII in Chinese lung cancer patients. We also found that patients with EGFRvIII were more likely to have SCC, were male or former/current smokers.
In our study, the frequency of EGFRvIII in post-surgery and advanced SCC patients ranged from 11.1–16.1%, respectively, which was higher than in ADC. However, our results on EGFRvIII frequency in lung cancer were inconsistent with previous studies. Garcia et al. and Okamoto et al. reported relatively low EGFRvIII frequencies, with 2.8% and 3.2%, respectively.16,23 Htsuka et al.19,20 found 14% of EGFRvIII mutations in SCC, but failed to observe EGFRvIII mutation in other histologic types, including lung ADC and adenosquamous cell carcinomas. The possible reason for the inconsistency might be that distinct methods were used to detect EGFRvIII mutation. The relatively low frequency of EGFRvIII in lung cancer might also contribute to the inconsistency, as these are based on small samples and, therefore, add statistical bias. The frequency of EGFRvIII needs to be further investigated in large samples. In the present study, we found that the EGFRvIII mutation mainly existed in men or smokers, similar to previous reports. Based on both our findings and those of previous research, screening male lung SCC patients who were former/current smokers might enrich a subset with the EGFRvIII mutation.
Several studies have reported that EGFR-TKI might inhibit the proliferation of tumor cells (such as malignant glioma and lung cancer cells) harboring EGFRvIII in vivo and in vitro, which implies that EGFRvIII might be a predictor to EGFR-TKI.18,24 Unfortunately, a total of eight patients with EGFRvIII expression in the first cohort in the present study were not treated with EGFR-TKI after their disease relapsed. Therefore, we screened 31 advanced patients with SCC who had received EGFR-TKI therapy for EGFRvIII analysis, and found that the median PFS and DCR in EGFRvIII positive and negative patients were similar. However, patients with the EGFRvIII mutation tended to have a prolonged OS compared with EGFRvIII negative cases.
The sensitizing EGFR mutation is a powerful predictor of EGFR-TKI. However, we performed multi-factor regression analysis and found that the EGFR mutation had no interactive influence on EGFRvIII. We also analyzed clinical features which might affect PFS and OS in SCC patients; no difference was obtained between positive and negative EGFRvIII mutant patients, EGFR mutation and EGFR wild-type, different gender, age or stage. According to our data, we concluded that the EGFRvIII mutation might be a prognostic tool, but is not a predictive factor in lung SCC. Further study is required to confirm this conclusion.
The EGFR mutation results of advanced SCC patients in this retrospective study were higher than reported in previous research.7–10 In clinical practice, only mutated SCC patients have the opportunity to receive EGFR-TKI as a first-line therapy. In second-line or further settings, EGFR mutation status is often an important factor to determine therapeutic strategy for advanced SCC patients. Therefore, this cohort of patients were selected based on EGFR-TKI treatment and could not represent real EGFR mutation status of SCC. However, mPFS for mutated SCC was only 2.0 months, which was the same as EGFR wild-type SCC. The efficacy of EGFR-TKI treatment was significantly inferior in EGFR mutated SCC compared to mutated adenocarcinoma patients, consistent with results reported by Shukuya et al. and Fang W et al.11,25
KRAS is an important molecule in the downstream signaling network of EGFR. KRAS mutation results in the inhibition of GTPase activity, leading to the constitutive activation of the RAS protein, which may render tumor cells independent of EGFR signaling. The clinical significance of KRAS mutation remains controversial. Several studies have testified that KRAS mutation is associated with primary drug resistance to EGFR-TKI,26,27 but a recent pooled analysis of 1543 patients from four studies indicated that neither KRAS wild-type nor codon 12 mutations had predictive value. The predictive value of KRAS codon 13 mutations requires validation.28 The present study showed that the patients who harbored a KRAS mutation had a shorter mPFS and mOS than those with KRAS wild type, but the difference did not reach statistical significance. Furthermore, the KRAS mutation had no interaction with the EGFRvIII mutation.
Our results imply that neither EGFR nor EGFRvIII mutations can predict the response to EGFR-TKI through multi-variable Cox regression analysis. More and more studies focus on SCC because of its poor response to targeted therapy. Recently, a comprehensive genomic analysis of lung SCC demonstrated the complex genomic alterations on the core cellular pathway of SCC. Phosphoinositide 3-kinase/receptor tyrosine kinase/RAS signaling possessed 69% of alteration, which might affect the efficacy of EGFR-TKI.29 It is necessary to explore further in vivo and in vitro studies in order to understand the mechanism of SCC, which could reveal more effective targeted therapy for SCC.
There were limitations to our study. None of the patients in the first cohort (n = 114) received EGFR-TKI therapy at disease recurrence. There were only five patients with positive EGFRvIII in the second cohort treated with EGFR-TKI therapy (n = 31), resulting from the small sample size. Therefore, the value of using EGFRvIII alone or in combination with EGFR mutation status to predict the response to EGFR-TKI therapy could not be determined.
Conclusion
In conclusion, EGFRvIII mutation is a rare event in Chinese NSCLC patients; however occurs more frequently in a selective population of men, smokers, and SCC patients. EGFRvIII mutation might be a prognostic, but not a predictive, factor for survival. Further larger scale studies are warranted.
Acknowledgments
We wish to thank the Tissue Bank at the Peking University Cancer Hospital for providing frozen surgery samples; Dr. Ning Wang, radiologist, from the Radiology Department of the Peking University Cancer Hospital & Institute, for his contribution to response assessment; and Mr. Guoshuang, Feng, statistician, from the Chinese Center for Disease Control and Prevention, for his contribution to statistics analyses. This work was supported by the National Natural Sciences Foundation Distinguished Young Scholars [81025012]; the National Natural Sciences Foundation Key Program [81330062]; the Education Ministry Innovative Research Team Program [IRT13003]; the Peking University-Tsinghua University Joint Center for Life Sciences Clinical Investigator; and Chinese Geriatric Oncology [CGOS-02-2014-1-2-00100].
Disclosure
No authors report any conflict of interest.
References
- Mok T, Wu Y, Thongprasert S, et al. Gefitinib or carboplatin- paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361:947–957. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- Maemondo M, Inoue A, Kobayashi K, et al. Gefitinib or chemotherapy for non-small cell lung cancer with mutated EGFR. N Engl J Med. 2010;362:2380–2388. doi: 10.1056/NEJMoa0909530. [DOI] [PubMed] [Google Scholar]
- Mitsudomi T, Morita S, Yatabe Y, et al. Gefitinib versus cisplatin plus docetaxel in patients with non-small cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): an open-label, randomised phase 3 trial. Lancet Oncol. 2010;11:121–128. doi: 10.1016/S1470-2045(09)70364-X. [DOI] [PubMed] [Google Scholar]
- Zhou C, Wu YL, Chen G, et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): a multicentre, open-label, randomized, phase 3 study. Lancet Oncol. 2011;12:735–742. doi: 10.1016/S1470-2045(11)70184-X. [DOI] [PubMed] [Google Scholar]
- Shaw AT, Yeap BY, Mino-Kenudson M, et al. Clinical features and outcome of patients with non-small-cell lung cancer who harbor EML4-ALK. J Clin Oncol. 2009;27:4247–4253. doi: 10.1200/JCO.2009.22.6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai H, Mao L, Wang HS, et al. Epidermal growth factor receptor mutations in plasma DNA samples predict tumor response in Chinese patients with stages IIIB to IV non-small-cell lung cancer. J Clin Oncol. 2009;27:2653–2659. doi: 10.1200/JCO.2008.17.3930. [DOI] [PubMed] [Google Scholar]
- Duan JC, An TT, Wu MN, et al. Correlation between the efficacy of epidermal growth factor receptor tyrosine kinase inhibitors and EGFR mutations in advanced squamous cell lung cancer. Chin J Tuberc Respir Dis. 2012;35:323–328. (In Chinese.) [PubMed] [Google Scholar]
- Park SH, Ha SY, Lee JI, et al. Epidermal growth factor receptor mutations and the clinical outcome in male smokers with squamous cell carcinoma of lung. J Korean Med Sci. 2009;24:448–452. doi: 10.3346/jkms.2009.24.3.448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu YL, Zhong WZ, Li LY, et al. Epidermal growth factor receptor mutations and their correlation with gefitinib therapy in patients with non-small cell lung cancer: a meta-analysis based on updated individual patient data from six medical centers in mainland China. J Thorac Oncol. 2007;2:430–439. doi: 10.1097/01.JTO.0000268677.87496.4c. [DOI] [PubMed] [Google Scholar]
- Forbes SA, Bhamra G, Bamford S, et al. The catalogue of somatic mutations in cancer (COSMIC) Curr Protoc Hum Genet. 2008 doi: 10.1002/0471142905.hg1011s57. ; Chapter 10: Unit 10.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukuya T, Takahashi T, Kaira R, et al. Efficacy of gefitinib for non-adenocarcinoma non-small-cell lung cancer patients harboring epidermal growth factor receptor mutations: a pooled analysis of published reports. Cancer Sci. 2011;102:1032–1037. doi: 10.1111/j.1349-7006.2011.01887.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontanini G, Vignati S, Bigini D, et al. Epidermal growth factor receptor (EGFR) expression in non-small cell lung carcinomas correlates with metastatic involvement of hilar and mediastinal lymph nodes in the squamous subtype. Eur J Cancer. 1995;31A:178–183. doi: 10.1016/0959-8049(93)00421-m. [DOI] [PubMed] [Google Scholar]
- Chaffanet M, Chauvin C, Lainé M, et al. EGF receptor amplification and expression in human brain tumors. Eur J Cancer. 1992;28:11–17. doi: 10.1016/0959-8049(92)90374-b. [DOI] [PubMed] [Google Scholar]
- Pedersen MW, Meltorn M, Damstrup L, Poulsen HS. The type III epidermal growth factor receptor mutation. Biological significance and potential target for anti-cancer therapy. Ann Oncol. 2001;12:745–760. doi: 10.1023/a:1011177318162. [DOI] [PubMed] [Google Scholar]
- Haley J, Whittle N, Bennet P, Kinchington D, Ullrich A, Waterfield M. The human EGF receptor gene: structure of the 110kb locus and identification of sequences regulating its transcription. Oncogene Res. 1987;1:375–396. [PubMed] [Google Scholar]
- Garcia de Palazzo IE, Adams GP, Sundareshan P, et al. Expression of mutated epidermal growth factor receptor by non-small cell lung carcinomas. Cancer Res. 1993;53:3217–3220. [PubMed] [Google Scholar]
- Moscatello DK, Holgado-Madruga M, Godwin AK, et al. Frequent expression of a mutant epidermal growth factor receptor in multiple human tumors. Cancer Res. 1995;55:5536–5539. [PubMed] [Google Scholar]
- Ji H, Zhao X, Yuza Y, et al. Epidermal growth factor receptor variant III mutations in lung tumorigenesis and sensitivity to tyrosine kinase inhibitors. Proc Natl Acad Sci USA. 2006;103:7817–7822. doi: 10.1073/pnas.0510284103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsuka K, Ohnishi H, Furuyashiki G, et al. Clinico- pathological and biological significance of tyrosine kinase domain gene mutations and overexpression of epidermal growth factor receptor for lung adenocarcinoma. J Thorac Oncol. 2006;1:787–795. [PubMed] [Google Scholar]
- Ohtsuka K, Ohnishi H, Fujiwara M, et al. Abnormalities of epidermal growth factor receptor in lung squamous-cell carcinomas, adenosquamous carcinomas, and large-cell carcinomas: tyrosine kinase domain mutations are not rare in tumors with an adenocarcinoma component. Cancer. 2007;109:741–750. doi: 10.1002/cncr.22476. [DOI] [PubMed] [Google Scholar]
- Wang S, An T, Wang J, et al. Potential clinical significance of a plasma-based KRAS mutation analysis in patients with advanced non-small cell lung cancer. Clin Cancer Res. 2010;16:1324–1330. doi: 10.1158/1078-0432.CCR-09-2672. [DOI] [PubMed] [Google Scholar]
- Kwak EL, Bang YJ, Camidge DR, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med. 2010;363:1693–1703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto I, Kenyon LC, Emlet DR, et al. Expression of constitutively activated EGFRvIII in non-small cell lung cancer. Cancer Sci. 2003;94:50–56. doi: 10.1111/j.1349-7006.2003.tb01351.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellinghoff IK, Wang MY, Vivanco I, et al. Molecular determinants of the response of glioblastomas to EGFR kinase inhibitors. N Engl J Med. 2005;353:2012–2024. doi: 10.1056/NEJMoa051918. [DOI] [PubMed] [Google Scholar]
- Fang W, Zhang J, Liang W, et al. Efficacy of epidermal growth factor receptor-tyrosine kinase inhibitors for Chinese patients with squamous cell carcinoma of lung harboring EGFR mutation. J Thorac Dis. 2013;5:585–592. doi: 10.3978/j.issn.2072-1439.2013.09.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pao W, Wang TY, Riely GJ, et al. KRAS mutations and primary resistance of lung adenocarcinomas to gefitinib or erlotinib. PLoS Med. 2005;2:e17. doi: 10.1371/journal.pmed.0020017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han SW, Kim TY, Hwang PG, et al. Predictive and prognostic impact of epidermal growth factor receptor mutation in non-small-cell lung cancer patients treated with gefitinib. J Clin Oncol. 2005;23:2493–2501. doi: 10.1200/JCO.2005.01.388. [DOI] [PubMed] [Google Scholar]
- Shepherd FA, Domerg C, Hainaut P, et al. Pooled analysis of the prognostic and predictive effects of KRAS mutation status and KRAS mutation subtype in early-stage resected non-small-cell lung cancer in four trials of adjuvant chemotherapy. J Clin Oncol. 2013;31:2173–2181. doi: 10.1200/JCO.2012.48.1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature491: Nature. 2012;489:519–525. doi: 10.1038/nature11404. (Published erratum appears in 2012; 288) [DOI] [PMC free article] [PubMed] [Google Scholar]


